Abstract
Any one model system, be it culture or animal, only recapitulates one aspect of the viral life cycle in the human host. By providing recent examples of animal models for Epstein Barr Virus and Kaposi Sarcoma-associated Herpesvirus, we would argue that multiple animal models are needed to gain a comprehensive understanding of the pathogenesis associated with human oncogenic herpesviruses. Transgenic mice, homologous animal herpesviruses, and tumorgraft and humanized mouse models all complement each other in the study of viral pathogenesis. The use of animal model systems facilitates the exploration of novel antiviral and anti-cancer treatment modalities for diseases associated with oncogenic herpesviruses.
Keywords: KSHV, EBV, Kaposi Sarcoma, primary effusion lymphoma, LANA, Burkitt lymphoma, nasopharyngeal carcinoma, multicentric Castleman's disease, herpesvirus
Introduction
Herpesviruses are ubiquitous in the human population and establish lifelong persistence in the body. Their evolutionary strategy is to be disseminated through prolonged and intimate contact among their hosts. For this transmission strategy to have evolved, the predominant phenotype of the infected carrier has to subtle – otherwise no other potential host would come close; the most dramatic phenotype must manifest itself only after a long period of asymptomatic shedding, typically after the next generation of hosts has been infected. Indeed, herpesviruses are normally and predominantly “silently” transmitted from mother to child. Mother-to-child transmission in infancy is the predominant mode for acquiring Epstein-Barr Virus (EBV) and Kaposi sarcoma-associated herpesvirus (KSHV) in endemic healthy populations[1,2].
Cancers associated with these two human viruses manifest themselves only in a minor fraction of infected individuals and only in the context of co-factors that affect the latent reservoirs, e.g. malaria-associated B cell activation in the case of EBV. EBV is associated with multiple cancers including Burkitt lymphoma, nasopharyngeal carcinoma, a subset of gastric cancers, a subset of Hodgkin disease, non-Hodgkin lymphoma localized to the central nervous system, and post-transplant lymphoproliferative disease (PTLD) in the context of iatrogenic immunosuppression. Furthermore, EBV is associated with other non-neoplastic diseases including infectious mononucleosis, oral hairy leukoplakia, lupus [3] and X- linked immunodeficiency [4]. These different EBV disease states represent a variety of infected cell types, gene expression states, and levels of host immune activation. Hence, it would not be possible to recapitulate all these mechanistically different outcomes of infection in just one animal model.
KSHV, also known as human herpesvirus 8, causes Kaposi Sarcoma (KS), primary effusion lymphoma, a variant of multicentric Castleman's disease and an acute replication syndrome associated with inflammation [5]. It is instructive to enumerate the many different scenarios in which KS has been observed, each representing a different host/infection stage.
-
(a)
Classic KS is predominantly a disease of older men and speculatively associated with diminished immune control due to aging. It can also be thought of as the result of an ever-expanding reservoir of latent B cells in response to environmental stimuli. As this latent reservoir increases, so does the likelihood that deleterious mutations occur in the infected B cells.
-
(b)
Iatrogenic KS is the result of chemical immunosuppression. Interestingly, switching from the T cell-selective immunosuppressant cyclosporine/FK506 to the T and B cell immunosuppressant rapamycin/sirolimus is associated with KS regression.
-
(c)
Endemic KS, prior to the emergence of human immunodeficiency virus (HIV), is a disease of children in a specific geographic locale in Sub-Saharan Africa, which also sees a geographic clustering of malaria and Burkitt lymphoma.
-
(d)
AIDS-KS is associated with diminished immune function due to HIV infection and enhanced KSHV transmission in high-risk populations. To date, almost 20 years after the introduction of anti-retroviral therapy (ART),KS remains the most common cancer in people living with HIV/AIDS both in Sub-Saharan Africa and in the US/Europe.
-
(e)
KS now also develops in latent HIV-infected patients on long-term ART, i.e. in the absence of active HIV replication and despite a reasonable number of CD4 cells (>200). These patients tend to be older men and may represent the intersection of incomplete immune repertoire restoration after HIV exposure, long-term immune activation due to microbial translocation, and diminished immune function due to aging.
-
(f)
In Sub-Saharan Africa, KS is also observed in children that acquired both HIV and KSHV at infancy from their mother[1].
-
(g)
Organ and bone marrow transplantation of HIV-positive patients is now routine, and failure rates in ART-adherent recipients are no worse than in HIV-negative patients with comparable co-morbidities. This makes sense intuitively since transplant-associated immunosuppressants suppress replicating CD4 T cells, the preferred vehicle for lytic HIV replication. Some of these transplant recipients or the organ donors also carry KSHV and as a consequence KSmay develop.
The above disease types each represent a different genesis from the primary infection event to fulminant disease, each deserving and necessitating a different animal model to capture and recreate the salient features of tumor development.
Kaposi sarcoma-associated herpesvirus (KSHV)
There is no animal model for KSHV. In fact there is no one, perfect model for any human virus. By their nature and design, all models whether a tissue culture model, 3D organ culture, or animal model system, represent one aspect of diseasebut never the complete human infection cycle. A good animal model is one that faithfully represents a part of the viral life cycle, or a stage of carcinogenesis, for which no other experimental systems exist. A good animal model system is also one that is inexpensive and easy to manipulate. It fills a gap in our understanding of pathogenesis and allows for the testing of anti-viral or anti-cancer agents.
KSHV does not replicate in any species except homo sapiens. Even most primates cannot be productively infected by KSHV. Even in infected humans, KSHV viral loads are diminutive in comparison to EBV and some other herpesviruses. This is perhaps due to the multitude of cell innate restrictions for this virus[6,7]. Models of primary KSHV infection are limited to humanized mouse models[8]. These serve to reveal cellular tropism (CD19 B cells, macrophages), tissue preference (spleen), viral latency, interactions with other viruses, host immune responses, and sensitivity to replication inhibitors, such as ganciclovir. Viral replication in these models is extremely limited and the input dose is rarely amplified. No serial transmission has been demonstrated in these animals to date.
KSHV can persist in non-human primates and in rare cases causes KS-like lesions[9]. Again, viral replication is limited and shedding is not observed. For all intents and purposes, non-human primates can be considered dead-end hosts for KSHV.
Non-human Rhadinoviruses
In the absence of an infection model for the human virus, homologous viruses and transgenic models have been explored. Each of these mimics different aspects of the disease or the phenotype of a subset of viral gene products. Each of these has been successful and must be considered significant in its own right.
KSHV is part of the rhadinovirus sub-group of gammaherpesviruses, and rhadinoviruses are divided into two lineages (reviewed in [10]). One lineage is represented by KSHV and a primate virus named retroperitoneal fibromatosisherpesvirus (RFHV), and the other lineage is represented by herpesvirussaimiri (HVS) and rhesus monkey rhadinovirus (RRV). RRV has served as a robust animal model system for KSHV. Two independent strains of RRV have been sequenced and their genomes are very similar to each other and KSHV. RRV replicates to high titers in cell culture and the virus is readily detectable in rhesus macaques. A breakthrough development was the creation of RRV recombination systems, which allowed for viral genetics and the exploration of individual RRV genes in the context of animal infections, as well as for the development of RRV as a vaccine vector[11-13]. In the context of simian immunodeficiency virus (SIV), RRV can induce lymphomathough KS-like skin lesions have not been observed. HerpesvirusSaimiri (HVS) was the first rhadinovirus isolated from primates and the HVS shares some molecular mechanism and host cell targets with KSHV [10,14], although this virus infects and transforms T cells in culture. While KSHV has the predilection to establish latency in almost all environments, the primate rhadinoviruses such as RRV and HVS readily enter the lytic replication phase and produce plaques on primary fibroblasts. Unlike KSHV, most primate rhadinoviruses exhibit population seropositivity rates above 80%, i.e. similar to the alpha- and beta herpesviruses.
The murine homolog of KSHV is murineherpesvirus 68 (MHV68). Prior to discovery of KSHV, MHV68 was used as a mouse model for EBV. MHV-68 replicates to high titers in culture and in wild-type mice (lung, spleen) it establishes latency in CD19 B cells and the myeloid compartment, and can be reactivated from latency. It does not form lymphoma or skin lesions upon natural infection of wild-type mice; however, MHV68 can immortalize and transform fetal liver-derived murine B cells[15]. The advantage of mouse models is the ability to engineer mutations in the murine genome. Thus, this model has been utilized with great success to study tissue tropism, latency, and the immune response to MHV68 infection in the context of mice lacking certain host genes[16-18]. Most recently, the outcome of co-infections of MHV68 with other parasites has also been reported[19,20].
Genetically engineered mouse models (GEMM) for KSHV
GEMM have contributed substantially to our understanding of non-viral human cancers as well as polyomavirus and papillomavirus disease (papillomavirus animal models are reviewed in[21]). In fact, SV40 T antigen expressed from the SV40 promoter and the human insulin promoter was the second genetic mouse cancer model after the ras transgenic “oncomouse”. GEMM allow for the study of individual viral genes in a normal tissue and developmental context. In the case of KSHV, which establishes a very tight latency pattern in B cells, all latency genes are known: the KSHV cyclin homolog, vCyc, vFLIP, kaposin, LANA and all viral micro RNAs are regulated by a common latency promoter, which is B cell specific in mice. Moreover, vIL6, K1, K15 each are regulated by their own promoters and the viral interferon regulatory factors (vIRF-1, −2, −3, −4) are grouped in an “operon” fashion. GEMM exist for vIL6[22], K1, vCyc, vFLIP[23,24], LANA and the viral micro RNAs[25, 26]. Each of these recapitulates a particular aspect of viral biology in the intact and developing animal (Figure 1B). By crossing KSHV GEMM to defined mutations in the host genome, conserved genetic interactions have been demonstrated in vivo, e.g. complementation of mir-155 by the KSHV mir-K12-11 ortholog[26]. With the exception of vGPCR and vIL6[27], most single KSHV gene transgenic mice form tumors after long latency and at low penetrance. Lymphoma development in KSHV GEMM can be increased by modulating host tumor pathways, such as p53 or Myc[28].
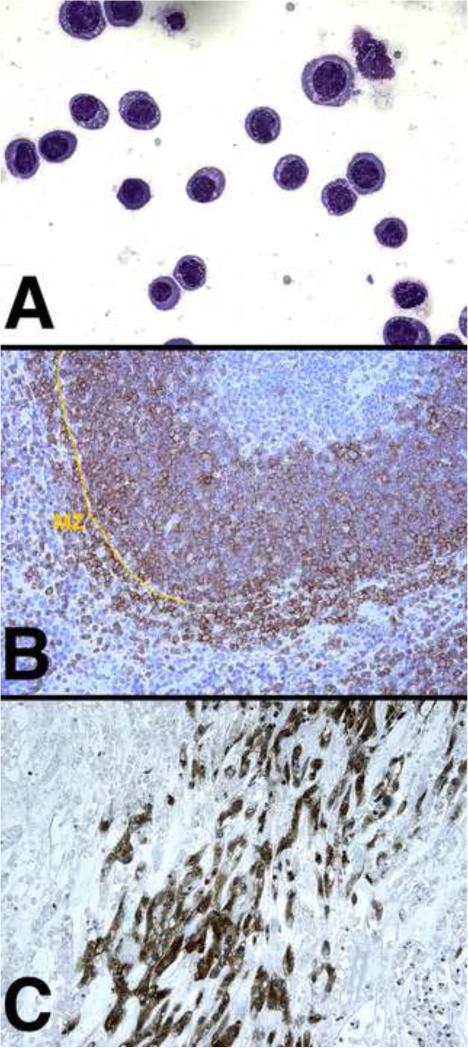
Figure 1.
(A) Exudate after intraperitoneal growth of a PEL cell line (BCBL-1) in immunodeficient (C.B.17-SCID) mice Wright-giemsastain at 400× magnification. (B) Spleen section of KSHV-latency locus transgenic mice stained for the B cell marker B220 (red) and counterstained with hematoxylin ; MZ marginal zone (200× magnification).
(C)KS-like lesion induced by the KSHV positive, human L1T2 (ATCC cat# VR1802) tumor cells under the skin of immunodeficient mice. VEGF-receptor2 (VEGF2) stain at 100× magnification.
Lastly, xenograft models have been explored to understand KSHV tumorigenesis and to evaluate anti-cancer agents (Figure 1A). These have the advantage of containing the entire genetic make-up of the KSHV-associated tumor, including tumor-specific host polymorphisms and mutations that contribute to the human disease phenotype. Both KSHV-infected primary effusion lymphoma (PEL) and KS tumor models have been developed (Figure 1A and C). These recapitulate clinical drug efficacy data, e.g. response tomTOR inhibitors[29-31]. Next generation, triple gene immunodeficient mice, e.g. NOD/scid/γ2ko (NGS), enabled the propagation of patient PEL as ascites, i.e. without intermediate culture [32] and with perhaps fewer culture-acquired changes. A potential drug candidate is typically profiled across multiple cell lines in culture. The same precaution should be taken for xenograft and primary graft animal models. None of the KSHV-tumor xenograft models generate infectious virions. Hence, theyare most often used to study agents that target tumors with a latent viral gene expression profile or agents that block paracrine or autocrine factors induced by KSHV.
Epstein Barr Virus (EBV)
EBV is a gammaherpesvirus of the lymphocryptovirus lineage. EBV infects CD19-positive B cells and establishes latency (type I or 0) in CD38-positive memory B cells. During type 0 latency only the viral non-coding RNAs (EBERs and micro RNAs) are expressed; in type I latency the EBNA1 protein is expressed as well. Whereas the principal cell lineages of KSHV lytic replication are endothelial cells and B cells, EBV can replicatelyticallyin epithelial cells and B cells. EBV causes types of B cell lymphoma and carcinomas (nasopharyngeal and a subset of gastric), which are neoplasms of epithelial cells. The biggest biological difference between EBV and KSHV is that EBV can transform mature CD19-positive B cells in culture, whereas KSHV cannot. These EBV-drivenlymphoblastoid lines (LCL) are immortal and tumorigenic in NGS mice.
EBV does not infect mice or other rodents (although there does exist literature of EBV causing disease in New Zealand white rabbits). Models of primary EBV infection are thus limited to humanized mouse models, several of which have been described and used to characterize viral mutants[33,34]. In these models, particularly in T cell deficient humanized mouse models, EBV induces fatal lymphoma with a latency type III gene expression pattern, similar to PTLD. Lytic amplification and transmission through mouse saliva has not been reported, though this has not been exhaustively studied.
Non-human lymphocryptoviruses
Lymphocryptoviruses including EBV are found in all non-human primates. Rhesus lymphocrytovirus (rhLCV), which shows homology to human EBV, is used as a model system to study EBV[35,36]. ArhLCMV bacterial artificial chromosome (BAC) has become available for rhLCMV[37]. The widespread introduction of BAC technologies for human and animal herpesviruses in the past years has revolutionized genetic approaches to these viruses in culture as well as in animal models. The effect of deletions of certain rhLCV genes on viral replication and persistence hasbeen very informative[38]. MHV68 is also used as a mouse homolog of EBV (in addition to KSHV), and has been used in immunological studies. MHV68 mimics certain signature aspects of EBV biology, e.g. the presence of small RNAs[39-41], which can be explored in the context of the authentic lytic infection cycle.
GEMM of EBV
Similar to KSHV, single transgene GEMM of EBV have revolutionized our understanding of EBV lymphomas as well as EBV-associated carcinomas. Ectopic expression of the viral transforming genes LMP1 and LMP2 induce drastic changes in B cell development if directed to the B cell lineage [3,42] and significant epithelial cell hyperplasia when driven by a keratin promoter. These studies have highlighted the fact that LMP1 is a homolog of CD40. Co-expression of LMP1 and LMP2 together, and with cellular oncogenes such as Myc (which is translocated in the majority of Burkittlymphoma) allowed for new insights into B cell reprogramming by these proteins[43-46].
Xenograft models of EBV-associated tumors have been essential to our understanding of EBV tumorigenesis. The most representative model of nasopharyngeal carcinoma (NPC) relies on serial in vivo passage of tumor explants, since the EBV episome is rapidly lost from NPC tumor cells upon growth in culture. The same phenotype of episome loss upon culture is also observed in KS-explant cell lines, but never in human tumors or the animal xenograft. This phenomenon underscores the idea that growth in immunodeficient mice provides a microenvironment and unique stromal-tumor cell interactions similar to human tumors, and that much of the oncogenic functions of these two large DNA tumor viruses evolved to engage neighboring, uninfected and untransformed cells. At this point these functions cannot be explored in cell culture models.
Conclusions
There is no one superior, animal model for any human virus. By their nature and by their design all animal models represent one aspect of the disease progress and not the complete lifecycle of a human tumor virus. Each of these animal models reveals a different and important aspect of viral infection more clearly than in the human patient. Each of these models offers the opportunity for experimental manipulation and hypothesis testing in contrast to human clinical studies, which are only correlative by nature. The ever increasing ease of germline manipulations in mice combined with the integration of genomic approaches in clinical and experimental studies make for an exciting future of investigating human disease through animal models.
Highlights.
Due to the fact that any particular model system only recapitulates one aspect of viral pathogenesis, we postulate that multiple animal models are needed to fully understand the biology of oncogenic herpesviruses.
Transgenic mice, homologous animal viruses, tumorgraft and humanized mouse models of infection complement each other for the comprehensive study of viral oncogenesis.
Acknowledgements
We would like to apologize in advance for excluding many excellent references due to the limit on both the number and recent publication of references required by the journal. In keeping with the tradition of Current Opinion essays, we restricted our citations to the most recent work. We apologize for not including many of the seminal works in the field. We thank Anthony Eason for critical reading. Workin the authors’ laboratories was supported in part by public health service grants CA163217, CA180097, DE023946, and CA019014.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Olp LN, Minhas V, Gondwe C, Kankasa C, Wojcicki J, Mitchell C, West JT, Wood C. Effects of Antiretroviral Therapy on Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Transmission Among HIV-Infected Zambian Children. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piriou E, Asito AS, Sumba PO, Fiore N, Middeldorp JM, Moormann AM, Ploutz-Snyder R, Rochford R. Early age at time of primary Epstein-Barr virus infection results in poorly controlled viral infection in infants from Western Kenya: clues to the etiology of endemic Burkitt lymphoma. J Infect Dis. 2012;205:906–913. doi: 10.1093/infdis/jir872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minamitani T, Yasui T, Ma Y, Zhou H, Okuzaki D, Tsai CY, Sakakibara S, Gewurz BE, Kieff E, Kikutani H. Evasion of affinity-based selection in germinal centers by Epstein-Barr virus LMP2A. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1514484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaigne-Delalande B, Li FY, O'Connor GM, Lukacs MJ, Jiang P, Zheng L, Shatzer A, Biancalana M, Pittaluga S, Matthews HF, et al. Mg2+ regulates cytotoxic functions of NK and CD8 T cells in chronic EBV infection through NKG2D. Science. 2013;341:186–191. doi: 10.1126/science.1240094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uldrick TS, Wang V, O'Mahony D, Aleman K, Wyvill KM, Marshall V, Steinberg SM, Pittaluga S, Maric I, Whitby D, et al. An interleukin-6-related systemic inflammatory syndrome in patients co-infected with Kaposi sarcoma-associated herpesvirus and HIV but without Multicentric Castleman disease. Clin Infect Dis. 2010;51:350–358. doi: 10.1086/654798. [Description of a new clinical phenotype associated with uncontrolled KSHV lytic replication.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A. 2015;112:E4306–4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang LX, Kang G, Kumar P, Lu W, Li Y, Zhou Y, Li Q, Wood C. Humanized-BLT mouse model of Kaposi's sarcoma-associated herpesvirus infection. Proc Natl Acad Sci U S A. 2014;111:3146–3151. doi: 10.1073/pnas.1318175111. [The latest in a series of studies over the years to utilize humanized mouse models for the study of KSHV.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang H, Wachtman LM, Pearson CB, Lee JS, Lee HR, Lee SH, Vieira J, Mansfield KG, Jung JU. Non-human primate model of Kaposi's sarcoma-associated herpesvirus infection. PLoS Pathog. 2009;5:e1000606. doi: 10.1371/journal.ppat.1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damania BaEC. Fields Virology. Wolters Kluwer Publishers Inc.; 2013. Kaposi sarcoma-associated herpesvirus. pp. 2080–2128. Chapter 65. [Google Scholar]
- 11.Estep RD, Rawlings SD, Li H, Manoharan M, Blaine ET, O'Connor MA, Messaoudi I, Axthelm MK, Wong SW. The rhesus rhadinovirus CD200 homologue affects immune responses and viral loads during in vivo infection. J Virol. 2014;88:10635–10654. doi: 10.1128/JVI.01276-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin G, Robinson BA, Rogers KS, Wong SW. A Rhesus Rhadinovirus Viral Interferon (IFN) Regulatory Factor Is Virion Associated and Inhibits the Early IFN Antiviral Response. J Virol. 2015;89:7707–7721. doi: 10.1128/JVI.01175-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilello JP, Manrique JM, Shin YC, Lauer W, Li W, Lifson JD, Mansfield KG, Johnson RP, Desrosiers RC. Vaccine protection against simian immunodeficiency virus in monkeys using recombinant gamma-2 herpesvirus. J Virol. 2011;85:12708–12720. doi: 10.1128/JVI.00865-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo YE, Riley KJ, Iwasaki A, Steitz JA. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol Cell. 2014;54:67–79. doi: 10.1016/j.molcel.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang X, Paden CR, Morales FM, Powers RP, Jacob J, Speck SH. Murine gamma-herpesvirus immortalization of fetal liver-derived B cells requires both the viral cyclin D homolog and latency-associated nuclear antigen. PLoS Pathog. 2011;7:e1002220. doi: 10.1371/journal.ppat.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cieniewicz B, Dong Q, Li G, Forrest JC, Mounce BC, Tarakanova VL, van der Velden A, Krug LT. Murine Gammaherpesvirus 68 Pathogenesis Is Independent of Caspase-1 and Caspase-11 in Mice and Impairs Interleukin-1beta Production upon Extrinsic Stimulation in Culture. J Virol. 2015;89:6562–6574. doi: 10.1128/JVI.00658-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sewatanon J, Liu H, Ling PD. Promyelocytic leukemia protein modulates establishment and maintenance of latent gammaherpesvirus infection in peritoneal cells. J Virol. 2013;87:12151–12157. doi: 10.1128/JVI.01696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, et al. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol. 2015;194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matar CG, Anthony NR, O'Flaherty BM, Jacobs NT, Priyamvada L, Engwerda CR, Speck SH, Lamb TJ. Gammaherpesvirus Co-infection with Malaria Suppresses Anti-parasitic Humoral Immunity. PLoS Pathog. 2015;11:e1004858. doi: 10.1371/journal.ppat.1004858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reese TA, Wakeman BS, Choi HS, Hufford MM, Huang SC, Zhang X, Buck MD, Jezewski A, Kambal A, Liu CY, et al. Coinfection. Helminth infection reactivates latent gamma-herpesvirus via cytokine competition at a viral promoter. Science. 2014;345:573–577. doi: 10.1126/science.1254517. [19 and 20. are examples of how viral and non-viral pathogens can interact es studied in animal models of co-infection. These experimental models provide plausible mechanisms to explain the long-established epidemiological association between EBV/KSHV disease and parasites in endemic regions of Sub-Saharan Africa.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park JW, Shin MK, Pitot HC, Lambert PF. High incidence of HPV-associated head and neck cancers in FA deficient mice is associated with E7's induction of DNA damage through its inactivation of pocket proteins. PLoS One. 2013;8:e75056. doi: 10.1371/journal.pone.0075056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suthaus J, Stuhlmann-Laeisz C, Tompkins VS, Rosean TR, Klapper W, Tosato G, Janz S, Scheller J, Rose-John S. HHV-8-encoded viral IL-6 collaborates with mouse IL-6 in the development of multicentric Castleman disease in mice. Blood. 2012;119:5173–5181. doi: 10.1182/blood-2011-09-377705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballon G, Akar G, Cesarman E. Systemic expression of Kaposi sarcoma herpesvirus (KSHV) Vflip in endothelial cells leads to a profound proinflammatory phenotype and myeloid lineage remodeling in vivo. PLoS Pathog. 2015;11:e1004581. doi: 10.1371/journal.ppat.1004581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad A, Groshong JS, Matta H, Schamus S, Punj V, Robinson LJ, Gill PS, Chaudhary PM. Kaposi sarcoma-associated herpesvirus-encoded viral FLICE inhibitory protein (vFLIP) K13 cooperates with Myc to promote lymphoma in mice. Cancer Biol Ther. 2010;10:1033–1040. doi: 10.4161/cbt.10.10.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sin SH, Dittmer DP. Viral latency locus augments B-cell response in vivo to induce chronic marginal zone enlargement, plasma cell hyperplasia, and lymphoma. Blood. 2013;121:2952–2963. doi: 10.1182/blood-2012-03-415620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sin SH, Kim YB, Dittmer DP. Latency locus complements MicroRNA 155 deficiency in vivo. J Virol. 2013;87:11908–11911. doi: 10.1128/JVI.01620-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma T, Jham BC, Hu J, Friedman ER, Basile JR, Molinolo A, Sodhi A, Montaner S. Viral G protein-coupled receptor up-regulates Angiopoietin-like 4 promoting angiogenesis and vascular permeability in Kaposi's sarcoma. Proc Natl Acad Sci U S A. 2010;107:14363–14368. doi: 10.1073/pnas.1001065107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sin SH, Kim Y, Eason A, Dittmer DP. KSHV Latency Locus Cooperates with Myc to Drive Lymphoma in Mice. PLoS Pathog. 2015;11:e1005135. doi: 10.1371/journal.ppat.1005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy D, Sin SH, Lucas A, Venkataramanan R, Wang L, Eason A, Chavakula V, Hilton IB, Tamburro KM, Damania B, et al. mTOR inhibitors block Kaposi sarcoma growth by inhibiting essential autocrine growth factors and tumor angiogenesis. Cancer Res. 2013;73:2235–2246. doi: 10.1158/0008-5472.CAN-12-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatt AP, Bhende PM, Sin SH, Roy D, Dittmer DP, Damania B. Dual inhibition of PI3K and mTOR inhibits autocrine and paracrine proliferative loops in PI3K/Akt/mTOR-addicted lymphomas. Blood. 2010;115:4455–4463. doi: 10.1182/blood-2009-10-251082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders P, Bhende PM, Foote M, Dittmer DP, Park SI, Damania B. Dual inhibition of phosphatidylinositol 3-kinase/mammalian target of rapamycin and mitogen activated protein kinase pathways in non-Hodgkin lymphoma. Leuk Lymphoma. 2015;56:263–266. doi: 10.3109/10428194.2014.917639. [29-31 document how the dependency of KSHV on the mTOR signaling pathway can be exploited to treat lymphoma and Kaposi Sarcoma. The efficacy of mTOR inhibitors against forms of KS has been documented in multiple clinical settings.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhatt S, Ashlock BM, Natkunam Y, Sujoy V, Chapman JR, Ramos JC, Mesri EA, Lossos IS. CD30 targeting with brentuximab vedotin: a novel therapeutic approach to primary effusion lymphoma. Blood. 2013;122:1233–1242. doi: 10.1182/blood-2013-01-481713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahl A, Linnstaedt SD, Esoda C, Krisko JF, Martinez-Torres F, Delecluse HJ, Cullen BR, Garcia JV. A cluster of virus-encoded microRNAs accelerates acute systemic Epstein-Barr virus infection but does not significantly enhance virus-induced oncogenesis in vivo. J Virol. 2013;87:5437–5446. doi: 10.1128/JVI.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma SD, Xu X, Plowshay J, Ranheim EA, Burlingham WJ, Jensen JL, Asimakopoulos F, Tang W, Gulley ML, Cesarman E, et al. LMP1-deficient Epstein-Barr virus mutant requires T cells for lymphomagenesis. J Clin Invest. 2015;125:304–315. doi: 10.1172/JCI76357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F. Nonhuman primate models for Epstein-Barr virus infection. Curr Opin Virol. 2013;3:233–237. doi: 10.1016/j.coviro.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skalsky RL, Kang D, Linnstaedt SD, Cullen BR. Evolutionary conservation of primate lymphocryptovirus microRNA targets. J Virol. 2014;88:1617–1635. doi: 10.1128/JVI.02071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohashi M, Orlova N, Quink C, Wang F. Cloning of the Epstein-Barr virus-related rhesus lymphocryptovirus as a bacterial artificial chromosome: a loss-of-function mutation of the rhBARF1 immune evasion gene. J Virol. 2011;85:1330–1339. doi: 10.1128/JVI.01411-10. [The latest technological advancement in rhesus lymphocrytovirus research.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohashi M, Fogg MH, Orlova N, Quink C, Wang F. An Epstein-Barr virus encoded inhibitor of Colony Stimulating Factor-1 signaling is an important determinant for acute and persistent EBV infection. PLoS Pathog. 2012;8:e1003095. doi: 10.1371/journal.ppat.1003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feldman ER, Kara M, Coleman CB, Grau KR, Oko LM, Krueger BJ, Renne R, van Dyk LF, Tibbetts SA. Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. MBio. 2014;5:e00981–00914. doi: 10.1128/mBio.00981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diebel KW, Oko LM, Medina EM, Niemeyer BF, Warren CJ, Claypool DJ, Tibbetts SA, Cool CD, Clambey ET, van Dyk LF. Gammaherpesvirus small noncoding RNAs are bifunctional elements that regulate infection and contribute to virulence in vivo. MBio. 2015;6:e01670–01614. doi: 10.1128/mBio.01670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, Kracker S, Yasuda T, Casola S, Vanneman M, Homig-Holzel C, Wang Z, Derudder E, Li S, Chakraborty T, et al. Immune surveillance and therapy of lymphomas driven by Epstein-Barr virus protein LMP1 in a mouse model. Cell. 2012;148:739–751. doi: 10.1016/j.cell.2011.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shair KH, Raab-Traub N. Transcriptome changes induced by Epstein-Barr virus LMP1 and LMP2A in transgenic lymphocytes and lymphoma. MBio. 2012;3 doi: 10.1128/mBio.00288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fish K, Chen J, Longnecker R. Epstein-Barr virus latent membrane protein 2A enhances MYC-driven cell cycle progression in a mouse model of B lymphoma. Blood. 2014;123:530–540. doi: 10.1182/blood-2013-07-517649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arcipowski KM, Stunz LL, Bishop GA. TRAF6 is a critical regulator of LMP1 functions in vivo. Int Immunol. 2014;26:149–158. doi: 10.1093/intimm/dxt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vrazo AC, Chauchard M, Raab-Traub N, Longnecker R. Epstein-Barr virus LMP2A reduces hyperactivation induced by LMP1 to restore normal B cell phenotype in transgenic mice. PLoS Pathog. 2012;8:e1002662. doi: 10.1371/journal.ppat.1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]