Summary
Our understanding of the genetic basis of systemic lupus erythematosus has progressed rapidly in recent years. While many genetic polymorphisms have been associated with disease susceptibility, the next major step involves integrating these genetic polymorphisms into the molecular mechanisms and cellular immunology of the human disease. In this review, we summarize some recent work in this area, including the genetics of the type I IFN response in SLE, including polygenic and monogenic factors, as well as epigenetic influences. Contributions of both HLA and non-HLA polymorphisms to the complex genetics of SLE are reviewed. We also review recent reports of specific gene deficits leading to monogenic SLE-like syndromes. The molecular functions of common SLE-risk variants are reviewed in depth, including regulatory variations in promoter and enhancer elements and coding-change polymorphisms, and studies which are beginning to define the molecular and cellular functions of these polymorphisms in the immune system. We discuss epigenetic influences on lupus, with an emphasis on micro-RNA expression and binding, as well as epigenetic modifications that regulate the expression levels of various genes involved in SLE pathogenesis and the ways epigenetic marks modify SLE susceptibility genes. The work summarized in this review provides a fascinating window into the biology and molecular mechanisms of human SLE. Understanding the functional mechanisms of causal genetic variants underlying the human disease greatly facilitates our ability to translate genetic associations toward personalized care, and may identify new therapeutic targets relevant to human SLE disease mechanisms.
Keywords: systemic lupus erythematosus, genetics, interferon, autoimmune diseases
1. Introduction
Systemic lupus erythematosus (SLE) is a severe, chronic autoimmune disorder characterized by involvement of multiple organ systems, loss of tolerance to self-antigens and dysregulated interferon responses. It is a highly heterogeneous condition, and different patients exhibit different combinations of symptoms and laboratory features. Humoral autoimmunity is a distinctive feature of SLE and many patients have circulating autoantibodies directed against double stranded DNA (anti-ds-DNA) and/or small nuclear RNA-binding proteins (such as anti-Ro, anti-La, anti-Sm, and anti-RNP). The pathogenesis of SLE is multifactorial, and the irreversible breakdown in immunologic self-tolerance which characterizes the disease can be attributed to the interplay among multiple genetic risk factors and environmental influences. Incidence of SLE is highest in women particularly during the childbearing years (female: male ratio 9:1) however, individuals of all ages, genders, and ancestral backgrounds are susceptible [1, 2]. SLE occurrence is four times higher in African-Americans as compared to European-Americans [3] and various studies have exhibited both genetic and immunologic differences among SLE patients from these ancestral backgrounds [4–7]. Familial aggregation and monozygotic twin studies strongly support the idea of genetic predisposition to SLE. Familial aggregation studies have demonstrated that siblings of SLE patients have greater relative risk for the disease, with sibling risk ratio (λs) as high as 29 compared with the general population [8] . Likewise, there is approximately ten-fold higher risk for SLE in monozygotic twins than in dizygotic twins [9, 10], while first degree relatives of patients with SLE have a 20-fold increased risk of developing SLE as compared with the healthy population [3, 11]. Within the families with multiple affected members, the SLE occurrence does not usually follow a classical Mendelian inheritance pattern. In the majority of cases, genetic susceptibility of SLE follows the common disease-common variant assumption, with polygenic inheritance of multiple alleles with a modest effect size (odds ratios for disease between 1.15 and 2.5) that combine to result in overall genetic risk. While the etiology of most SLE cases appears to be complex genetically, a few cases SLE and SLE-like disorders can be attributed to highly penetrant rare mutations which will be discussed in detail subsequently.
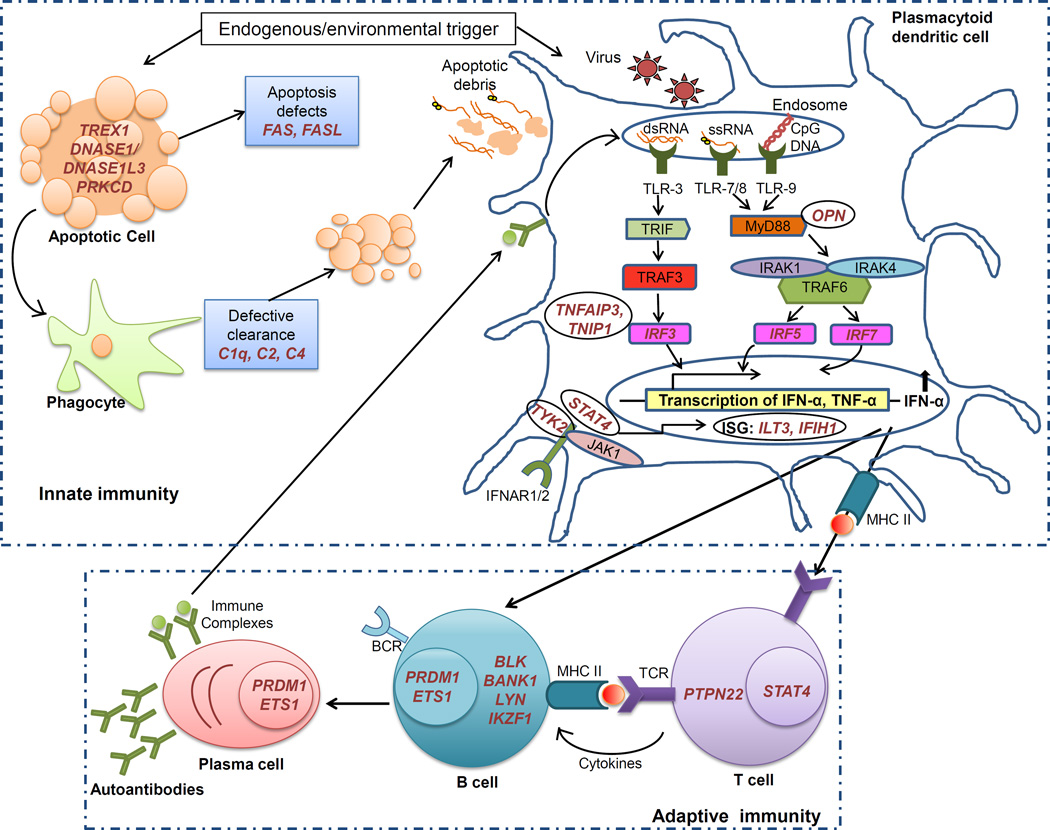
Numerous genome wide association studies (GWAS) have been performed in patients with SLE across various ethnic populations, and currently more than 40 common risk loci have been definitively linked to SLE susceptibility in case-control genetic studies [12, 13]. As expected, the strongest association signal among the common genetic variants obtained from HLA region, while many other non-HLA SLE susceptibility loci are located within or near genes with functional relevance in the immune system. In addition, number novel genetic loci have been associated with SLE susceptibility that may or may not function within immune system pathways and have no known previous relationship to the pathogenesis of SLE. Examining the list of genes associated with SLE, there is a remarkable over-representation of genes involved in type I interferon (IFN) signaling, production, and response [13, 14]. Interferon alpha (IFN-α) is a type I interferon classically involved in viral defense which has the potential to break self-tolerance by activating antigen-presenting cells after the uptake of self-material [15], and is central to the pathogenesis of SLE [16]. IFN-α can be synthesized by many cells in response to viral infection, but plasmacytoid dendritic cells (pDCs) play a specialized role in the production of IFN-α [17]. Circulating IFN-α levels are high in SLE patients [18] and this high IFN phenotype is heritable within SLE families with a complex or polygenic pattern of inheritance [11]. These data suggest that high serum IFN-α is a heritable risk factor for SLE [11]. Also, some individuals treated with recombinant IFN-α have developed de novo SLE, which typically improves when the IFN is discontinued [19, 20]. Many of the genetic polymorphisms associated with SLE susceptibility have been shown to contribute to high IFN levels in human SLE patients [21–23]. These data support the idea that gain-of-function polymorphisms in the IFN pathway are a common pathogenic mechanism in SLE. Additionally, number novel genetic loci have been identified that have an effect on IFN-α levels in SLE patients, supporting the genetic nature of the IFN dysregulation observed in SLE [24–28]. Collectively these data strongly support the causative role of this heritable molecular subphenotype in SLE etiology and pathogenesis. In this review, we will discuss polygenic and monogenic influences on type I IFN. In addition to type I IFN-related genes, genes related to other immune system functions such as B- and T- cell signaling, clearance of dead cellular debris, and cytokine signaling make up a large portion of the genetic loci associated with SLE. In this review, we will discuss polygenic and monogenic influences on type I IFN, as well as the functional significance of some of the other SLE-associated polymorphisms located in immune system genes. These immunogenetic data provide novel insights into the molecular pathogenesis of human SLE. Figure 1 illustrates these major pathways and cell types which are involved in human SLE, and the genes discussed in the review are shown in the cell type in which they are presumed to function.
Figure 1. Proposed cellular location for gene products described in the review.
Major pathways and cell types involved in SLE, with SLE-risk genes shown in the cell type in which they are presumed to function. Panel A shows genes that influence different cell types involved in innate immune response, while panel B shows genes that impact various cell types involved in adaptive immune response.
2. Human leukocyte antigen (HLA) complex in SLE
The classical HLA complex (also referred to as major histocompatibility locus [MHC]) is the most gene-dense region of the genome, encoding more than 200 genes in a 3.6 Mb region on 6p21.3. Many of these genes function in the immune system. The HLA region is subdivided into the telomeric class I region and the centomeric class II and class III regions. The class I and II regions encode highly polymorphic classical HLA genes (HLA-A, -B, -C, -DR, -DQ, -DP) that process and present peptides for T cell recognition and involved in transplant compatibility, and the class III region encodes a variety of important immune system genes (such as C2, C4A, C4B, TNF-α, lymphotoxin–α, heat shock proteins, and CFB). Association between HLA variants and SLE is extensively explored and till today all SLE GWAS in different ethnic populations demonstrated the HLA region as the prominent strongest predictor of genetic risk [13]. Among these HLA regions, genes in class II are dominantly represented as SLE susceptibility loci. Previous studies of the HLA region in SLE have shown that HLA-DRB1 (DRB1*1501 and DRB1*0301) is a robustly associated with SLE [29, 30]. Recent GWAS studies in European and Asian population have further confirmed these associations [13, 31, 32]. Given the importance of the HLA class II molecules in T cell dependent antibody responses, the close association between class II alleles (specifically HLA-DR and HLA-DQ alleles) with autoantibody subsets in SLE patients seems to make sense [29]. A study comparing anti–dsDNA negative SLE cases to healthy controls demonstrated a significant association for a SNP, rs2301271, 9 kb downstream from HLA-DQA2 [33], and a strong association was observed between anti-dsDNA and HLA-DR3 at rs2187668, further supporting the importance of the HLA region in determining autoantibody responses. Similarly, a recent largest SLE sub-phenotype genetic association study demonstrated HLA-DRB1*03:01 not only just influence SLE susceptibility but is also associated with anti-Ro and anti-La autoantibody production [34]. Furthermore, the role of SLE-associated HLA Class II alleles in initiating SLE-relevant autoantibody responses has been shown in humanized mice expressing the HLA-DR3 transgene but not other DR or DQ alleles [35]. An HLA class III gene, super viralicidic activity 2-like (SKIV2L) encoding RNA helicase SKI2W enzyme, was identified as a SLE susceptibility risk gene independent of class II loci [36]. Also, the rs3131379 SNP in MSH5 (HLA class III locus) demonstrated association in a GWAS study [37]. It seems likely given the complexity of the region and the high density of immune system genes, that the multiple association signals reported to date could represent multiple independent risk factors. A recent study using high-density SNP screening of the MHC region supports this idea, reporting multiple independent loci associated with SLE, including HLA-DRB1*0301, DRB1*1401, DRB1*1501 and the DQB2 alleles, CREBL1, MICB and OR2H2 [22]. Together these studies highlight the essential influence of HLA genes in SLE pathogenesis.
3. Polygenic Influences on type I IFN
3.1. IFN Regulatory Factors
Interferon regulatory factors (IRFs) are classically involved in inducing IFN-α and IFN-induced genes downstream of Toll-like receptor (TLR) activation. Additionally they play prominent role in cytokine secretion, cellular apoptosis, immune cell development, tumor suppression, and cell activation and differentiation [38, 39]. Interestingly, genetic variations in three of the nine IRFs (IRF5, IRF7, and IRF8) have been linked to SLE susceptibility, supporting a key role for this family of proteins in SLE pathogenesis. Studies in several different ethnic groups have confirmed IRF5 as a risk locus for SLE, and its association with increased circulating IFN-α levels [22, 40]. Four main functional variants in IRF5 have been reported, a promoter polymorphism which alters binding affinity at the promoter, one at rs2004640 which creates an alternate splice site (exon 1B) in the untranslated first exon, another is a 30-bp in-frame insertion/deletion in exon 6, and the third is a 3’UTR polymorphism which creates an alternate polyadenylation site resulting in shorter and more stable mRNA. It has been demonstrated that these four variants combine to form a SLE risk haplotype in individuals of European ancestry [41] and these various IRF5 haplotypes are also associated with the formation of anti-Ro, anti-La, and anti-dsDNA autoantibodies [42]. As IRF5 activates IFN production, these variants may pose a risk due to their ability to produce excess IFN. Studies on IRF5 in human SLE cohorts have shown that the risk variant predisposes to greater serum IFN-α, supporting the idea that the risk haplotype is a gain-of-function variant [22]. The same risk variant has been associated with autoantibody formation in SLE patients and in healthy individuals, and most of the risk of SLE related to IRF5 genotype is found within the autoantibody positive, high IFN group of patients [42, 43]. IRF7 is another IRF family member which also interacts with the MyD88 adaptor protein downstream of TLR signaling, and is phosphorylated and activated following TLR engagement [38]. Genetic variants in the IRF7 locus have been associated with SLE in various studies [37, 44, 45]. Several SNPs in the IRF7 region were shown to correlate with IFN levels and autoantibody profiles in SLE patients of various ancestral backgrounds [46]. A recent study on human dendritic cells demonstrated a cis-expression quantitative loci (eQTL) SNP located within the SLE-associated haplotype is not only associated with IRF7 expression but also confers the trans-eQTL effect on type I IFN response regulation in activated but not unstimulated dendritic cells [47]. This supports the previous human studies that indicate IRF7 variants are associated with greater IFN-α in circulation. IRF8 has no direct interaction with MyD88, but it does appear to play a role in the TLR pathway as dendritic cells lacking IRF8 do not produce inflammatory cytokines in response to TLR9 ligand [48]. IRF8 deficiency in humans results in an immunodeficiency characterized by the loss of monocytes and dendritic cells, indicating its role in monocyte and dendritic cell development [49]. Genetic variants in the IRF8 gene region are associated with susceptibility to both SLE and multiple sclerosis (MS) [50, 51]. Type I IFN plays a contrasting role in MS and SLE, as while IFN-α is increased and thought to be causal in SLE, type I IFN levels are lower in patients with MS than in healthy controls [38], and recombinant human IFN-β is used as a treatment for MS. A study investigating whether IRF8 alleles were associated with type I IFN levels or serologic profiles in SLE and MS revealed that MS-associated allele downstream of IRF8 (rs17445836G) was associated with decreased activity of the type I IFN pathway in these two different autoimmune diseases and was associated with anti-dsDNA antibodies and increased IRF8 in B cells in the patients with SLE [52]. Taken together, these data support a role for IRF8 variants in modulating type I IFN and humoral autoimmunity in multiple autoimmune conditions.
3.2. STAT4
STAT4 encodes the signal transducer and activator of transcription 4 protein (STAT4) which plays an important role in downstream responses to type I IFN and other cytokines. Functionally, activation and phosphorylation of STAT4 is induced by IL-12, IL-23, and IFN-α which then promotes Th1 as well as Th17 responses [53]. Both candidate gene association studies and multiple GWAS studies using populations from European or Asian ancestry have demonstrated a robust association between SLE and STAT4 [13]. In SLE patients, the risk variant for STAT4 (rs7574865) was associated with increased sensitivity to IFN-α signaling, as evident from its simultaneous association with both lower serum IFN-α activity and greater IFN-α-induced gene expression in PBMCs [21]. Interestingly, the same risk variant was also associated with a more severe SLE phenotype characterized by a higher frequency of nephritis and the presence of anti-dsDNA antibodies [54].
3.3. IFIH1
IFIH1 is an innate immune receptor located in the cytosol which senses dsRNA and promotes IRF3 and 7 phosphorylation, activating transcription of antiviral genes and type I IFN production. Based on the studies in SLE and other various autoimmune conditions it has been proposed that a common coding change variant in IFIH1 (rs1990760, A946T) leads to increased expression and gain-of-function in IFIH1 and subsequent predisposition to human autoimmune disease [13]. This risk variant was also modulated IFN-α-induced gene expression in peripheral blood cells in anti-dsDNA positive SLE patients, and was associated with the production of anti-dsDNA antibodies [55]. Recently, a large-scale multi-ancestral admixture mapping genetic screen revealed three independently associated variants in the IFIH1 gene (one intronic and two missense variants). These variants were associated with increased apoptosis and elevated expression of inflammation-related genes, as well as autoantibody production [56].
3.4. OPN
Osteopontin (OPN, encoded by the SPP1 gene) is overexpressed in humans with SLE and has been involved in the development of murine lupus [57, 58]. A number of studies have linked genetic variants in OPN with SLE susceptibility, clinical manifestations of SLE; and high IFN levels in SLE affected males and young-onset female lupus patients [13, 59]. OPN interacts with the MyD88 adaptor protein downstream of TLR ligation, and is an important molecule for IFN-α production in plasmacytoid dendritic cells (pDCs), thus supporting its role in SLE pathogenesis [60].
4. Monogenic Causes of SLE and SLE-like high-IFN syndromes
4.1. Monogenic deficiencies in SLE
4.1.1. Complement deficiency
Relatively rare primary complement defects such as complete deficiency of the early components of classical complement pathway genes (C1q, C1r/s, C2, C4A and C4B) are strongly associated with increased susceptibility to SLE [61–64]. The occurrence of a lupus-like syndrome or SLE has been demonstrated in 90% of homozygous C1q deficient cases, in more than 50% of cases with C1s/C1r deficiencies, in 10 to 30% of C2 deficient cases (more often a deletion in intron 6 that leads to a premature stop codon in exon 7 resulting in failure to synthesize the protein), and in 75% of C4-deficient cases [65]. Complement is essential for opsonization and clearance of autoantibody-containing immune complexes and clearing apoptotic cells. Interestingly, complement also plays a role in T and B cell activation and complement deficiency may disrupt the balance of lymphoid cell activation. Deficiencies of the classical complement component pathway are likely to affect SLE pathogenesis by decreasing clearance of apoptotic cell debris and immune complexes (IC), resulting in increased self-antigen availability, and dysregulated IC-related TLR signaling or IC induced cytokines such as IFN-α [65]. The genes for complement components C2 and C4 are in linkage disequilibrium with MHC polymorphisms, and these genes are hypothesized to contribute independently to the risk of SLE [66].
4.1.2. Apoptosis defects
Several lines of evidence indicate that abnormalities in the apoptosis (programmed cell death) contribute to the development of SLE, as the elimination of autoreactive T or B cells is impaired in this disease [67]. In addition, it appears that increased lymphocyte apoptosis and delayed clearance of phagocyte-mediated apoptotic cells as evident in SLE patients could contribute to B-cell hyperactivity and subsequent autoantibody production [68]. The accelerated apoptosis of circulating cells observed in SLE patients could serve as a major source of autoantigens in the form of apoptotic blebs and debris [68]. The role of FAS-mediated apoptosis in immunity and elimination of autoreactive lymphocytes are clear [69]. Defects in FAS and FASL provided one of the first murine models of SLE, the MRL/lpr mouse [70]. The monogenic human condition autoimmune lymphoproliferatve syndrome (ALPS) results from FAS or FASL defects, and this condition shares some features of SLE such as lymphadenopathy, positive anti-nuclear antibodies (ANA), autoimmune cytopenias, and sometimes organ system inflammation [71]. However, the role of FAS and FASL gene polymorphisms in the etiology of SLE has been inconclusive [72]. Studies in SLE patients indicated that FAS may be involved in the defective apoptosis of T cells and resistance to the FAS-mediated apoptosis in these cells [69, 73, 74]. Other studies have observed that polymorphisms in FAS and FASL genes could alter their basal expression [75] and thus have been suggested to play important roles in pathogenesis of SLE [72, 74, 76] .
4.1.3. DNASE1/DNASE1L3
Deoxyribonuclease I (DNase I, encoded by DNASE1) is specific endonuclease essential during apoptosis. Decreased DNase I activity may result in increased risk of antinucleosome antibody production, a cardinal feature of SLE. Various studies have reported a link between low DNase I activity and the development of murine or human SLE [77, 78]. Two unrelated cases of juvenile SLE exhibiting very high levels of antinucleosomal antibodies were reported with a mutation in exon 2 of DNAse type 1 [79], however this mutation has not been confirmed in other patient populations. Recently, study of seven consanguineous families with multiple SLE affected children by linkage analysis and exome sequencing identified loss-of-function mutations in DNASE1L3 as a monogenic cause of an SLE-like syndrome [80]. A strict Mendelian autosomal recessive pattern of association was observed between this mutation and SLE. Clinically, the affected individuals presented with hallmarks of SLE such as ANA, anti-dsDNA autoantibodies, anti-neutrophil cytoplasmic antibodies (ANCA) and hypocomplementemia [80]. It is possible that these DNASE mutations facilitate an SLE-like phenotype via inappropriate accumulation of DNA which can then become a neo-antigen and form inflammatory immune complexes with anti-dsDNA antibodies.
4.1.4. PRKCD
Recently, in another study on consanguineous family with three siblings affected with SLE was reported in which the affected individuals all had homozygous inactivating mutations in PRKCD gene [81]. PRKCD encodes protein kinase C δ (PKCδ), which has been implicated in the control of cell proliferation, apoptosis, and B-cell signaling [82, 83]. The identified missense mutation of PRKCD (G510S) resulted in reduced expression and activity of the encoded protein PKCδ, leading to resistance to B cell receptor- and calcium-dependent apoptosis and increased B cell proliferation. B cells from the patients carrying PRKCD mutations demonstrated hyperproliferative responses to stimulation through the BCR, CD40 and TLR9 signaling pathways [81]. Phenotypically, the affected siblings had anti-dsDNA antibodies, nephritis and hypocomplementemia without hypergammaglobulinemia [81].
4.2 Monogenic influences on IFN: SLE-like interferonopathies
4.2.1 TREX1
TREX1, also called, DNAse type III, is the main 3’-5’DNA exonuclease enzyme that proofreads DNA polymerase, also functioning as a DNA degrading enzyme in granzyme-A-mediated apoptosis and as a cytosolic DNA sensor [84]. TREX1 deficiency impairs DNA damage repair, leading to the intracellular accumulation of endogenous DNA. This defective DNA clearance stimulates systemic autoimmunity by inducing TLR-independent IFN-α production. TREX1 mutations in humans lead to Aicardi-Goutieres syndrome (AGS), which shares several features with SLE. This gene is also commonly mutated in SLE [85], as missense variants in TREX1 were observed in 0.5 to 2.7% SLE patients but were nearly absent in healthy controls indicating TREX1 mutations are the most frequent form of monogenic lupus [86, 87]. The TREX1-SLE association supports the importance of defective clearance of DNA in activation of innate immunity and the development of SLE.
4.2.2 STING
TLR-independent cytosolic DNA-sensing pathways that signal via stimulator of interferon genes (STING; also known as “TMEM173,” “MPYS,” “MITA,” and “ERIS”) mediate immunity to pathogens and also promote autoimmune pathology [88]. STING activates host defense by induction of IFN-β through a well-characterized pathway involving TBK1 and interferon regulatory factor 3 (IRF3) [88] and the production of the NF-κB-driven cytokines TNF-α and IL-6 [89]. STING deficiency ameliorates the autoimmune phenotype of TREX1-deficient mice, as the loss of STING reduces the overproduction of IFN and chronic inflammation [90]. Supporting this idea, gain-of-function mutations in TMEM173, gene encoding STING, were recently reported in subjects with a monogenic condition characterized by high IFN levels and vascular and pulmonary inflammation [91, 92]. Loss-of-function polymorphisms for STING are also known, however they have not yet been linked to any human diseases [93].
4.2.3 SAMHD1
SAMHD1 also known as Aicardi- Goutières syndrome type 5 (AGS5), is a putative nuclease encoded by the SAMHD1/ AGS5 gene that is upregulated in response to viral infections and may have a regulatory role in immune system and cerebral vascular homeostasis [94–97]. Similar to TREX1, mutations in SAMHD1 including biallelic null alleles as well as missense mutations have been associated with Aicardi-Goutieres syndrome. At least 16 different mutations in the SAMHD1 gene have been reported in patients with Aicardi-Goutieres syndrome [98–101]. These mutations result in loss-of-function of the SAMHD1 protein. However, how this protein dysfunction leads to immune system abnormalities, inflammatory damage to the brain and skin, and other characteristics of this syndrome is still unknown. Circulating IFN-α is increased in these patients, even though the molecular mechanism by which SAMHD1 deficiency causes IFN overproduction is not known.
4.2.4 TRAP
The acid phosphatase 5 (ACP5) gene encodes tartrate-resistant acid phosphatase (TRAP). Deficiency of the ACP5 gene has been associated with an immuno-osseous dysplasia called spondyloenchondrodysplasia (SPENCD). SPENCD has been regarded primarily as a skeletal dysplasia, but patients with this disease also demonstrates a high frequency of autoimmune phenotypes, including SLE, hemolytic anemia, Sjogren’s syndrome, inflammatory myositis, hypothyroidism and thrombocytopenia [102, 103]. It has been identified that loss-of-function mutations in the ACP5 gene are causal of the disease resulting in elevated serum IFN activity and an IFN signature in SPENCD patients, and [102, 103]. TRAP is expressed in bone and in immune cells, including osteoclasts and dendritic cells. Since OPN is a recognized substrate for osteoclast-derived TRAP and is dephosphorylated by TRAP [104], it is possible that in the absence of TRAP in pDC, OPN would remain phosphorylated and persistently activate IFN-α via TLR9/MyD88 [103]. This hypothesis is supported by the observation that SPENCD patients have higher urinary levels of phosphorylated OPN as compared with controls, suggesting that TRAP is responsible for dephosphorylating OPN, and that this function is defective in patients with SPENCD [103]. Interestingly, as noted above genetic polymorphisms in the SPP1 gene have been associated with SLE susceptibility, and patients carrying SPP1 risk alleles have elevated IFN-α activity and increased OPN protein levels [59, 105]. These data would suggest that in humans TRAP deficiency may result in over-active OPN and type I IFN, and it is possible that this may be particularly important in those patients with SPP1 risk alleles.
5 Regulatory variants associated with SLE susceptibility
5.1 TNFAIP3 and TNIP1
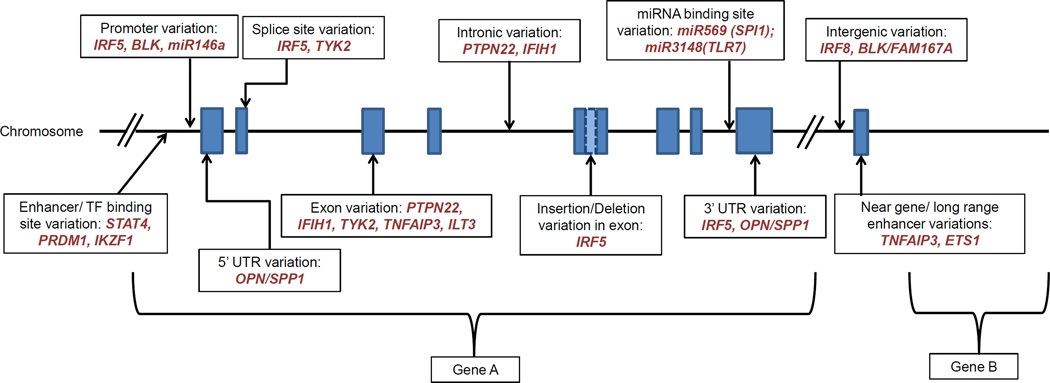
The tumor necrosis factor alpha inducible protein 3 (TNFAIP3) gene, encodes the ubiquitin-modifying enzyme A20, a critical regulator of NF-κB activity downstream of tumor necrosis factor alpha (TNFα), TLRs, and interleukin 1 receptor [106]. TNFAIP3 limits inflammatory signals by diminishing NF-κB signaling. Variants near TNFAIP3 have been associated with susceptibility to multiple polygenic autoimmune disorders including SLE, rheumatoid arthritis, and others [107]. Presumably these autoimmune disease associated variants would be loss-of-function, reducing the ability of TNFAIP3 to limit inflammation. A number of independent genetic associations between SLE and various SNPs spanning the TNFAIP3 region have been reported [108–110]. These include a common coding-change variant in exon 3 (rs2230926), as well as a coding-change variant in the deubiquitination domain that reduces the enzymatic function of TNFAIP3 [111]. Another study further characterized the TNFAIP3 risk haplotype by fine mapping and genomic re-sequencing in multiple world populations, and found a TT>A polymorphic dinucleotide (deletion T followed by a T to A transversion) far downstream of the TNFAIP3 gene as top polymorphism accounting for association between TNFAIP3 and SLE in European ancestry [112]. This polymorphism lies within a long-range enhancer element that binds NF-κB and SATB1, enabling physical interaction of this enhancer with the TNFAIP3 promoter through long-range DNA looping. Impaired binding of NF-κB to the TT>A risk alleles or knockdown of SATB1 expression by shRNA inhibits the looping interaction resulting in reduced A20 expression [113], and the reduced A20 expression would lead to impaired control over inflammatory signaling pathways. TNFAIP3-interacting protein 1 (TNIP1) is another gene that is involved in limiting inflammatory NF-κB signaling, and variants in this gene have also been associated with SLE susceptibility [114, 115]. The risk haplotypes of TNIP1 result in decreased expression of TNIP1 mRNA [115], consistent with the idea that the SLE-risk variant reduces the effect of the immunosuppressive TNIP1 gene product. When an inactive form of the protein encoded by TNIP1 (ABIN1) is knocked into mice, an immune complex nephritis that resembles lupus nephritis develops [116]. The results for both TNFAIP3 and TNIP1 indicate the importance of NF-κB pathway regulation in the pathogenesis of SLE. Figure 2 shows the proposed molecular mechanisms of these genes as well as others discussed in this review in a schematic diagram.
Figure 2.
Schematic showing a generalized gene diagram, with proposed location of functional variants in known regulatory regions, exons, splice sites, introns and intergenic sites discussed in this review. The actual SLE associated variants are located in various regions throughout the genome.
5.2 BLK
BLK encodes a B lymphocyte specific tyrosine kinase, a member of the Src family of kinases that functions in intracellular signaling and regulates cell proliferation and differentiation. A GWAS carried out in the European population revealed that two different BLK SNPs were associated with SLE. One of these polymorphisms was located within the intergenic region of FAM167A and BLK and was associated with reduced expression of BLK but increased expression of FAM167A in SLE patients [37, 117] . Association of BLK polymorphisms with SLE was later confirmed in Asian ancestry as well [31]. Recently, using trans-population fine mapping and sequencing, the SLE-associated interval in the BLK promoter has have been refined to two variations [118]. The first is a common SNP (rs922483) in the proximal BLK promoter that causes decreased promoter activity and modulation in alternative promoter usage, and the second is a tri-allelic variant (rs1382568) in the upstream alternative BLK promoter that results in altered promoter activity in B progenitor cell lines [118]. This study identifies regulatory variations near the BLK gene that modulate the transcription of BLK in B cells, which would presumably alter immune responses and thus contribute to SLE pathogenesis.
5.3 ETS1 and PRDM1
ETS1 is a member of the ETS family of transcription factors that acts as a negative regulator of B cells and T-helper-17 cell differentiation by inhibiting the function of PR domain zinc finger protein 1 (encoded by PRDM1, also known as BLIMP1) [119, 120]. PRDM1 functions as repressor of IFN-β gene expression, and is an important factor in B cell development. GWAS studies in European and Asian populations have demonstrated that PRDM1 is a risk locus for SLE [13]. A recent study has also shown that ETS1 is an SLE susceptibility locus in Asian populations, and this was then also later replicated in European populations [121]. The risk variant identified in these studies was located in the 3’ UTR region of the gene (rs1128334), and has been associated with decreased ETS1 expression levels in PBMCs from healthy individuals [31]. Decreased expression of this suppressive gene would be expected to result in increased immune system activation. Murine models support this idea, as ETS1-deficient mice developed a lupus-like disease distinguished by high titers of autoantibodies and local complement activation [122]. A multi-ancestral fine-mapping study spanning ETS1 loci examined the functional mechanism of ETS1 variants on the basis of their likelihood of affecting transcription factor binding, miRNA binding, or chromatin state [123]. They observed that the ETS1 risk allele rs6590330 caused enhanced binding of pSTAT1 and decreased ETS1 expression [123].
5.4 IKZF1
DNA-binding protein Ikaros (encoded by IKZF1) is a lymphoid-restricted zinc finger transcription factor that controls lymphocyte differentiation and proliferation, as well as self-tolerance through regulation of B-cell receptor signaling [124]. IKZF1 was established as a novel susceptibility gene for SLE GWAS in Chinese Han population and has been replicated in a Europena population [13, 121]. The risk allele of IKZF1 SNP rs4917014 was associated with correlated with lower expression levels of IKZF1 mRNA in PBMCs from SLE patients [125]. Additionally, expression quantitative trait locus (eQTL) studies of IKZF1 showed that this same risk allele also affected the expression of multiple genes in trans such as C1QB and five type I interferon response genes [126]. This study supported both cis and trans transcriptional influences from this IKZF1 SNP, supporting a regulatory role for this polymorphism in SLE.
6 Coding-change polymorphisms
6.1 PTPN22
Tyrosine-protein phosphatase nonreceptor type 22 (PTPN22) is a lymphoid specific phosphatase that controls antigen receptor signal transduction in both T and B lymphocytes [127]. Two functional variants of the PTPN22 gene have been identified— one is a nonsynonymous variant rs2476601 (arg620trp) that increases the risk of SLE, as well as multiple other autoimmune diseases [128]. A loss of function PTPN22 variant (Arg263Gln) reduces the phosphatase activity of PTPN22, and has been associated with protection against SLE in European population [129]. PTPN22 is expressed in most leukocytes, and studies of the coding change polymorphism have documented numerous alterations in the cellular immune system related to these polymorphisms, and it’s likely that the role of this gene in SLE is complex. Roles for the PTPN22 risk alleles in effector T cell and regulatory T cell development have been supported [130]. A study in SLE patients has suggested a link between PTPN22 and innate cytokine production, as the SLE patients who carry the common coding-change risk allele for PTPN22 had higher serum IFN-α activity and lower serum tumor necrosis factor (TNF) levels [131]. Recent work with PTPN22 in model systems also supports a role for PTPN22 in TLR responses and type I IFN production [132]. The common PTPN22 risk allele has also been associated with anti-dsDNA autoantibody production in SLE [33], and abnormalities in the B cell compartment such as expansion of transitional and anergic B cells [133], and decreased removal of autoreactive B cells [134].
6.2 ILT3
Immunoglobulin-like transcript 3 receptor (ILT3) is an immunosuppressive surface receptor that is induced by type I IFNs, and thus could represent a negative feedback pathway regulating type I IFNs in vivo. ILT3 is expressed on dendritic cells and monocyte/macrophage lineage cells [135]. ILT3 expression has been examined in multiple sclerosis patients who were being treated with type I IFN in the form of recombinant IFN-β, and increased ILT3 expression was observed in patients who were responding to treatment [136]. ILT3 polymorphisms have been examined in SLE in a candidate gene study in which coding-change polymorphisms that were likely to alter protein folding were prioritized [26]. The rs11540761 coding-change SNP in the extracellular region of ILT3 was correlated with decreased cell surface expression of ILT3 on circulating MDCs and to a lesser extent PDCs in these patients. The rs1048801 SNP changes an amino acid on the cytoplasmic portion of the ILT3 receptor, and this polymorphism was not associated with a change in expression of ILT3 on dendritic cells but was associated with increased serum levels of TNF-α. Both these loss-of-function polymorphisms were significantly and independently linked with increased levels of serum type I IFN activity in SLE patients, supporting the idea that loss of this suppressive receptor results in decreased control of type I IFN responses in SLE.
7 Epigenetic influences on SLE pathogenesis
Though the recent advances in genetic association studies have identified many SLE predisposing genes, we are not able to fully account for SLE susceptibility with these genetic variations, and epigenetics may account for some of this unexplained heritability. Furthermore, the incomplete disease concordance rates in monozygotic twin studies in SLE would suggest epigenetic influences to SLE development. Studies examining methylation in monozygotic twins discordant for SLE have demonstrated significant differences in these discordant pairs [137]. Differences in epigenetic modifications such as DNA methylation, histone modifications (acetylation, ubiquitination, phosphorylation and citrullination of histone tails) and noncoding RNA, can impact the expression and function of genes involved in SLE pathogenesis.
7.1 Effect of DNA hypomethylation and histone modification changes on type I IFN and SLE pathogenesis
Within DNA, methylation usually occurs at CpG islands sites by methyltransferases that lead to inducive or suppressive effects on gene expression. In SLE patients, DNA from T cells is hypomethylated at multiple sites, which can lead to upregulated expression pro-inflammatory molecules [138]. Various factors like ultraviolet light, SLE-inducing drugs, aging, and altered expression of certain microRNAs can promote DNA hypomethylation [138]. Studies have shown that various methylation-sensitive genes that functionally contribute to SLE development are overexpressed in SLE CD4+ T cells, for example CD11A, perforin, CD70, and CD40L [139]. A recent genome-wide DNA methylation analysis comparing SLE patients and healthy controls demonstrated significant hypomethylation of type I IFN-regulated genes in SLE CD4+ T cells, CD19+ B cells and CD14+ monocytes [140, 141]. Given that type I IFN is increased in the circulation of many SLE patients, it is not clear how much of the epigenetic change in circulating T cells is induced by type I IFN signaling vs. some intrinsic hypomethylation in SLE patient cells. Supporting the idea that some of the changes might be intrinsic, a study in human T cells from healthy individuals with or without the SLE-risk MECP2-IRAK1 haplotype demonstrated lupus-associated variant in the MECP2/IRAK1 locus leads to an increased levels of a specific MECP2 transcript isoform in stimulated T cells and further showed a similar hypomethylation of IFN-regulated genes, providing evidence for a genetic–epigenetic interaction associated with SLE-risk variants [142].
Different combinations of histone modifications can affect DNA replication, transcription and chromatin structure [143], and alterations in histone modifications leading to abnormal gene expressions likely contribute to SLE pathogenesis [12]. For example, a study demonstrated significantly increased H4 acetylation (H4ac) in monocytes from SLE patients, as well as increased expression of genes involved in the type I IFN pathway [144]. Additionally, studies examining epigenetic information in Epstein–Barr virus-transformed lymphoblastoid cell lines exhibited an enrichment of active histone marks in the promoters of SLE and RA susceptibility genes [145, 146]. These studies support the idea that epigenetic methylation marks modulate the genetic variations implicated in SLE and autoimmune disease.
7.2 Influences of microRNAs on type I IFN and SLE pathogenesis
MicroRNAs (miRNAs) are endogenous small non-coding RNA molecules which function in RNA silencing and post-transcriptional regulation of gene expression by binding to specific target messenger RNA (mRNA) transcripts and promoting their degradation/destabilization and/or translational inhibition [147]. Recent studies support a critical role for miRNAs in the function of both innate and adaptive immunity, and miRNA alterations have been associated with autoimmune diseases such as SLE, RA, and Sjogren’s syndrome [148, 149]. It has been observed that several miRNAs play essential role in negative regulation of innate immune responses. For example miR-146a can inhibit type I IFN production by targeting multiple key molecules in the innate signaling pathway such as TLR7, RIG-I pathway [150, 151]. This miRNA also suppresses NFκB activation and related cytokine production by acting on signaling adaptor proteins the TNF receptor-associated family (TRAF)-6 and IL-1 receptor-associated kinase (IRAK)-1 [152]. Another miRNA miR-155/miR-155* was shown to influence regulation of type I interferon production in pDCs [153].
MiRNA expression profiling studies in PBMCs, plasma and different tissues from SLE patients has discovered distinct miRNA signatures as compared with healthy individuals demonstrating association of miRNAs dysregulation with disease activity, organ system involvement, and autoantibody profiles, indicating a possible role for miRNAs as biomarkers in SLE patients [148, 154]. Of the particular interest are studies which have investigated the functional impact of SLE-associated genetic polymorphisms upon miRNA expression or binding. An SLE-risk allele has been reported in the miR146a miRNA. This miRNA is a negative regulator of the IFN pathway, and a promoter variant (rs57095329) was linked to decreased expression of miR-146a [155]. This promoter polymorphism results in reduced protein-binding affinity for the transcription factor ETS1 (also note genetic variants in ETS1 described above), which causes lower levels of miR-146a expression [155]. Alteration of miRNA binding sites in target mRNA represents another mechanism by which inherited genetic variation can impact the ability of miRNAs to regulate gene expression. Some SLE-associated SNPs have been identified that introduce or abolish miRNA binding sites. An SLE associated SNP in the SPI1 gene is in complete linkage with a functional SNP in the 3′ UTR region of this gene (rs1057233) that alters the target sequence of microRNA miR-569 [156]. This genetic variation disrupts the miR-569 binding site, resulting in an increase in SPI1 mRNA level which could contribute to SLE development [156]. A TLR7 SLE-risk allele provides another example of genetic variation altering miRNA binding. The SLE risk allele of TLR7 (rs3853839) is common across multiple ancestral backgrounds, and demonstrates an allelic effect on TLR7 expression [157]. This risk allele results in altered binding of miRNA-3148 and slower degradation of the TLR7 transcript [157]. These studies illustrate how genetic variations that impact miRNA binding sites impact transcript abundance, and subsequently alter immune cells contributing to SLE pathogenesis.
8 Conclusions
In summary, this review illustrates the wide diversity of molecular genetic mechanisms involved in SLE pathogenesis. This is illustrated in both the large number of immune cell types involved in both innate and adaptive immunity (Figure 1), as well as the large diversity in the genetic mechanisms by which these polymorphisms impact the molecular biology of SLE (Figure 2). The work summarized in this review represents a critical area of SLE research which allows for a fascinating window into human SLE pathogenesis. By understanding the functional mechanisms of causal genetic variants underlying the human disease, we will be able to understand the molecular pathogenesis of SLE in humans. This will facilitate our ability to translate genetic associations toward personalized care, and may help us use existing medications in a way that is more directed at pathogenic factors which are relevant for a particular individual, and new therapeutic targets may also emerge as the molecular mechanisms of human SLE are more fully understood.
Highlights.
Genetic risk factors for lupus differ somewhat between world populations
Multiple genes contribute to type I interferon dysregulation in lupus
Lupus-risk genes function via a wide range of molecular and immunologic mechanisms
Micro-RNAs and methylation changes also contribute to the lupus disease process
Acknowledgments
Funding Sources: Y Ghodke-Puranik – none, TB Niewold – Research grants from the NIH (AR060861, AR057781, AR065964, AI071651), Rheumatology Research Foundation, CureJM Foundation, the Mayo Clinic Foundation, and the Lupus Foundation of Minnesota.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures and Conflict of Interest: The authors report no financial conflict of interest.
References
- 1.Lopez P, Mozo L, Gutierrez C, Suarez A. Epidemiology of systemic lupus erythematosus in a northern Spanish population: gender and age influence on immunological features. Lupus. 2003;12:860–865. doi: 10.1191/0961203303lu469xx. [DOI] [PubMed] [Google Scholar]
- 2.Weckerle CE, Niewold TB. The unexplained female predominance of systemic lupus erythematosus: clues from genetic and cytokine studies. Clin Rev Allergy Immunol. 2011;40:42–49. doi: 10.1007/s12016-009-8192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harley JB, Kelly JA, Kaufman KM. Unraveling the genetics of systemic lupus erythematosus. Springer Semin Immunopathol. 2006;28:119–130. doi: 10.1007/s00281-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 4.Ko K, Koldobskaya Y, Rosenzweig E, Niewold TB. Activation of the Interferon Pathway is Dependent Upon Autoantibodies in African-American SLE Patients, but Not in European-American SLE Patients. Frontiers in immunology. 2013;4:309. doi: 10.3389/fimmu.2013.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez E, Comeau ME, Freedman BI, Kelly JA, Kaufman KM, Langefeld CD, et al. Identification of novel genetic susceptibility loci in African American lupus patients in a candidate gene association study. Arthritis and rheumatism. 2011;63:3493–3501. doi: 10.1002/art.30563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko K, Franek BS, Marion M, Kaufman KM, Langefeld CD, Harley JB, et al. Genetic ancestry, serum interferon-alpha activity, and autoantibodies in systemic lupus erythematosus. The Journal of rheumatology. 2012;39:1238–1240. doi: 10.3899/jrheum.111467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma S, Jin Z, Rosenzweig E, Rao S, Ko K, Niewold TB. Widely divergent transcriptional patterns between SLE patients of different ancestral backgrounds in sorted immune cell populations. Journal of autoimmunity. 2015;60:51–58. doi: 10.1016/j.jaut.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alarcon-Segovia D, Alarcon-Riquelme ME, Cardiel MH, Caeiro F, Massardo L, Villa AR, et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52:1138–1147. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- 9.Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35:311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 10.Block SR, Winfield JB, Lockshin MD, D’Angelo WA, Christian CL. Studies of twins with systemic lupus erythematosus. A review of the literature and presentation of 12 additional sets. Am J Med. 1975;59:533–552. doi: 10.1016/0002-9343(75)90261-2. [DOI] [PubMed] [Google Scholar]
- 11.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes and immunity. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Y, Tsao BP. Advances in lupus genetics and epigenetics. Curr Opin Rheumatol. 2014;26:482–492. doi: 10.1097/BOR.0000000000000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghodke-Puranik Y, Niewold TB. Genetics of the type I interferon pathway in systemic lupus erythematosus. International journal of clinical rheumatology. 2013:8. doi: 10.2217/ijr.13.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crow MK. Advances in understanding the role of type I interferons in systemic lupus erythematosus. Curr Opin Rheumatol. 2014;26:467–474. doi: 10.1097/BOR.0000000000000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 16.Niewold TB. Interferon alpha as a primary pathogenic factor in human lupus. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31:887–892. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzgerald-Bocarsly P, Dai J, Singh S. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev. 2008;19:3–19. doi: 10.1016/j.cytogfr.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. The New England journal of medicine. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 19.Niewold TB, Swedler WI. Systemic lupus erythematosus arising during interferon-alpha therapy for cryoglobulinemic vasculitis associated with hepatitis C. Clin Rheumatol. 2005;24:178–181. doi: 10.1007/s10067-004-1024-2. [DOI] [PubMed] [Google Scholar]
- 20.Ronnblom LE, Alm GV, Oberg KE. Possible induction of systemic lupus erythematosus by interferon-alpha treatment in a patient with a malignant carcinoid tumour. J Intern Med. 1990;227:207–210. doi: 10.1111/j.1365-2796.1990.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 21.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009;182:34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis and rheumatism. 2008;58:2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Transl Res. 2010;155:109–117. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kariuki SN, Franek BS, Kumar AA, Arrington J, Mikolaitis RA, Utset TO, et al. Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis research & therapy. 2010;12:R151. doi: 10.1186/ar3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kariuki SN, Ghodke-Puranik Y, Dorschner JM, Chrabot BS, Kelly JA, Tsao BP, et al. Genetic analysis of the pathogenic molecular sub-phenotype interferon-alpha identifies multiple novel loci involved in systemic lupus erythematosus. Genes Immun. 2014 doi: 10.1038/gene.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen MA, Patterson KC, Kumar AA, Kumabe M, Franek BS, Niewold TB. Functional genetic polymorphisms in ILT3 are associated with decreased surface expression on dendritic cells and increased serum cytokines in lupus patients. Annals of the rheumatic diseases. 2013;72:596–601. doi: 10.1136/annrheumdis-2012-202024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pothlichet J, Niewold TB, Vitour D, Solhonne B, Crow MK, Si-Tahar M. A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol Med. 2011;3:142–152. doi: 10.1002/emmm.201000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harley IT, Niewold TB, Stormont RM, Kaufman KM, Glenn SB, Franek BS, et al. The role of genetic variation near interferon-kappa in systemic lupus erythematosus. J Biomed Biotechnol. 2010:2010. doi: 10.1155/2010/706825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connolly JJ, Hakonarson H. Role of cytokines in systemic lupus erythematosus: recent progress from GWAS and sequencing. J Biomed Biotechnol. 2012;2012:798924. doi: 10.1155/2012/798924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nature genetics. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung SA, Taylor KE, Graham RR, Nititham J, Lee AT, Ortmann WA, et al. Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet. 2011;7:e1001323. doi: 10.1371/journal.pgen.1001323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris DL, Fernando MM, Taylor KE, Chung SA, Nititham J, Alarcon-Riquelme ME, et al. MHC associations with clinical and autoantibody manifestations in European SLE. Genes and immunity. 2014;15:210–217. doi: 10.1038/gene.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang C, Deshmukh US, Gaskin F, Bagavant H, Hanson J, David CS, et al. Differential responses to Smith D autoantigen by mice with HLA-DR and HLA-DQ transgenes: dominant responses by HLA-DR3 transgenic mice with diversification of autoantibodies to small nuclear ribonucleoprotein, double-stranded DNA, and nuclear antigens. J Immunol. 2010;184:1085–1091. doi: 10.4049/jimmunol.0902670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernando MM, Stevens CR, Sabeti PC, Walsh EC, McWhinnie AJ, Shah A, et al. Identification of two independent risk factors for lupus within the MHC in United Kingdom families. PLoS Genet. 2007;3:e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nature genetics. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen MA, Niewold TB. Interferon regulatory factors: critical mediators of human lupus. Transl Res. 2015;165:283–295. doi: 10.1016/j.trsl.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao GN, Jiang DS, Li H. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta. 2015;1852:365–378. doi: 10.1016/j.bbadis.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 40.Cham CM, Ko K, Niewold TB. Interferon regulatory factor 5 in the pathogenesis of systemic lupus erythematosus. Clin Dev Immunol. 2012;2012:780436. doi: 10.1155/2012/780436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niewold TB, Kelly JA, Kariuki SN, Franek BS, Kumar AA, Kaufman KM, et al. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Annals of the rheumatic diseases. 2012;71:463–468. doi: 10.1136/annrheumdis-2011-200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherian TS, Kariuki SN, Franek BS, Buyon JP, Clancy RM, Niewold TB. Brief Report: IRF5 systemic lupus erythematosus risk haplotype is associated with asymptomatic serologic autoimmunity and progression to clinical autoimmunity in mothers of children with neonatal lupus. Arthritis and rheumatism. 2012;64:3383–3387. doi: 10.1002/art.34571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suarez-Gestal M, Calaza M, Endreffy E, Pullmann R, Ordi-Ros J, Sebastiani GD, et al. Replication of recently identified systemic lupus erythematosus genetic associations: a case-control study. Arthritis Res Ther. 2009;11:R69. doi: 10.1186/ar2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Q, Zhao J, Qian X, Wong JL, Kaufman KM, Yu CY, et al. Association of a functional IRF7 variant with systemic lupus erythematosus. Arthritis and rheumatism. 2011;63:749–754. doi: 10.1002/art.30193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salloum R, Franek BS, Kariuki SN, Rhee L, Mikolaitis RA, Jolly M, et al. Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis and rheumatism. 2010;62:553–561. doi: 10.1002/art.27182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsujimura H, Tamura T, Kong HJ, Nishiyama A, Ishii KJ, Klinman DM, et al. Toll-like receptor 9 signaling activates NF-kappaB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J Immunol. 2004;172:6820–6827. doi: 10.4049/jimmunol.172.11.6820. [DOI] [PubMed] [Google Scholar]
- 49.Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lessard CJ, Adrianto I, Ice JA, Wiley GB, Kelly JA, Glenn SB, et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. American journal of human genetics. 2012;90:648–660. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chrabot BS, Kariuki SN, Zervou MI, Feng X, Arrington J, Jolly M, et al. Genetic variation near IRF8 is associated with serologic and cytokine profiles in systemic lupus erythematosus and multiple sclerosis. Genes Immun. 2013 doi: 10.1038/gene.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korman BD, Kastner DL, Gregersen PK, Remmers EF. STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep. 2008;8:398–403. doi: 10.1007/s11882-008-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng J, Yin J, Huang R, Petersen F, Yu X. Meta-analysis reveals an association of STAT4 polymorphisms with systemic autoimmune disorders and anti-dsDNA antibody. Hum Immunol. 2013 doi: 10.1016/j.humimm.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 55.Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. Journal of immunology. 2011;187:1298–1303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molineros JE, Maiti AK, Sun C, Looger LL, Han S, Kim-Howard X, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS genetics. 2013;9:e1003222. doi: 10.1371/journal.pgen.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katagiri Y, Mori K, Hara T, Tanaka K, Murakami M, Uede T. Functional analysis of the osteopontin molecule. Ann N Y Acad Sci. 1995;760:371–374. doi: 10.1111/j.1749-6632.1995.tb44660.x. [DOI] [PubMed] [Google Scholar]
- 58.Forton AC, Petri MA, Goldman D, Sullivan KE. An osteopontin (SPP1) polymorphism is associated with systemic lupus erythematosus. Hum Mutat. 2002;19:459. doi: 10.1002/humu.9025. [DOI] [PubMed] [Google Scholar]
- 59.Kariuki SN, Moore JG, Kirou KA, Crow MK, Utset TO, Niewold TB. Age- and gender-specific modulation of serum osteopontin and interferon-alpha by osteopontin genotype in systemic lupus erythematosus. Genes Immun. 2009;10:487–494. doi: 10.1038/gene.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao W, Liu YJ. Opn: key regulator of pDC interferon production. Nat Immunol. 2006;7:441–443. doi: 10.1038/ni0506-441. [DOI] [PubMed] [Google Scholar]
- 61.Pettigrew HD, Teuber SS, Gershwin ME. Clinical significance of complement deficiencies. Ann N Y Acad Sci. 2009;1173:108–123. doi: 10.1111/j.1749-6632.2009.04633.x. [DOI] [PubMed] [Google Scholar]
- 62.Slingsby JH, Norsworthy P, Pearce G, Vaishnaw AK, Issler H, Morley BJ, et al. Homozygous hereditary C1q deficiency and systemic lupus erythematosus. A new family and the molecular basis of C1q deficiency in three families. Arthritis Rheum. 1996;39:663–670. doi: 10.1002/art.1780390419. [DOI] [PubMed] [Google Scholar]
- 63.Provost TT, Arnett FC, Reichlin M. Homozygous C2 deficiency, lupus erythematosus, and anti-Ro (SSA) antibodies. Arthritis Rheum. 1983;26:1279–1282. doi: 10.1002/art.1780261017. [DOI] [PubMed] [Google Scholar]
- 64.Fielder AH, Walport MJ, Batchelor JR, Rynes RI, Black CM, Dodi IA, et al. Family study of the major histocompatibility complex in patients with systemic lupus erythematosus: importance of null alleles of C4A and C4B in determining disease susceptibility. Br Med J (Clin Res Ed) 1983;286:425–428. doi: 10.1136/bmj.286.6363.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Truedsson L, Bengtsson AA, Sturfelt G. Complement deficiencies and systemic lupus erythematosus. Autoimmunity. 2007;40:560–566. doi: 10.1080/08916930701510673. [DOI] [PubMed] [Google Scholar]
- 66.Davies EJ, Steers G, Ollier WE, Grennan DM, Cooper RG, Hay EM, et al. Relative contributions of HLA-DQA and complement C4A loci in determining susceptibility to systemic lupus erythematosus. Br J Rheumatol. 1995;34:221–225. doi: 10.1093/rheumatology/34.3.221. [DOI] [PubMed] [Google Scholar]
- 67.Munoz LE, van Bavel C, Franz S, Berden J, Herrmann M, van der Vlag J. Apoptosis in the pathogenesis of systemic lupus erythematosus. Lupus. 2008;17:371–375. doi: 10.1177/0961203308089990. [DOI] [PubMed] [Google Scholar]
- 68.Ren Y, Tang J, Mok MY, Chan AW, Wu A, Lau CS. Increased apoptotic neutrophils and macrophages and impaired macrophage phagocytic clearance of apoptotic neutrophils in systemic lupus erythematosus. Arthritis Rheum. 2003;48:2888–2897. doi: 10.1002/art.11237. [DOI] [PubMed] [Google Scholar]
- 69.Fukuyama H, Adachi M, Suematsu S, Miwa K, Suda T, Yoshida N, et al. Requirement of Fas expression in B cells for tolerance induction. Eur J Immunol. 2002;32:223–230. doi: 10.1002/1521-4141(200201)32:1<223::AID-IMMU223>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi T, Tanaka M, Brannan CI, Jenkins NA, Copeland NG, Suda T, et al. Generalized lymphoproliferative disease in mice, caused by a point mutation in the Fas ligand. Cell. 1994;76:969–976. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- 71.Shah S, Wu E, Rao VK, Tarrant TK. Autoimmune lymphoproliferative syndrome: an update and review of the literature. Curr Allergy Asthma Rep. 2014;14:462. doi: 10.1007/s11882-014-0462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiang N, Li XM, Wang GS, Tao JH, Li XP. Association of Fas gene polymorphisms with systemic lupus erythematosus: a meta-analysis. Mol Biol Rep. 2013;40:407–415. doi: 10.1007/s11033-012-2075-0. [DOI] [PubMed] [Google Scholar]
- 73.Budagyan VM, Bulanova EG, Sharova NI, Nikonova MF, Stanislav ML, Yarylin AA. The resistance of activated T-cells from SLE patients to apoptosis induced by human thymic stromal cells. Immunol Lett. 1998;60:1–5. doi: 10.1016/s0165-2478(97)00128-4. [DOI] [PubMed] [Google Scholar]
- 74.Wu J, Metz C, Xu X, Abe R, Gibson AW, Edberg JC, et al. A novel polymorphic CAAT/enhancer-binding protein beta element in the FasL gene promoter alters Fas ligand expression: a candidate background gene in African American systemic lupus erythematosus patients. J Immunol. 2003;170:132–138. doi: 10.4049/jimmunol.170.1.132. [DOI] [PubMed] [Google Scholar]
- 75.Pinti M, Troiano L, Nasi M, Moretti L, Monterastelli E, Mazzacani A, et al. Genetic polymorphisms of Fas (CD95) and FasL (CD178) in human longevity: studies on centenarians. Cell Death Differ. 2002;9:431–438. doi: 10.1038/sj.cdd.4400964. [DOI] [PubMed] [Google Scholar]
- 76.Moudi B, Salimi S, Farajian Mashhadi F, Sandoughi M, Zakeri Z. Association of FAS and FAS ligand genes polymorphism and risk of systemic lupus erythematosus. ScientificWorldJournal. 2013;2013:176741. doi: 10.1155/2013/176741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sallai K, Nagy E, Derfalvy B, Muzes G, Gergely P. Antinucleosome antibodies and decreased deoxyribonuclease activity in sera of patients with systemic lupus erythematosus. Clin Diagn Lab Immunol. 2005;12:56–59. doi: 10.1128/CDLI.12.1.56-59.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 79.Yasutomo K, Horiuchi T, Kagami S, Tsukamoto H, Hashimura C, Urushihara M, et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 80.Al-Mayouf SM, Sunker A, Abdwani R, Abrawi SA, Almurshedi F, Alhashmi N, et al. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat Genet. 2011;43:1186–1188. doi: 10.1038/ng.975. [DOI] [PubMed] [Google Scholar]
- 81.Belot A, Kasher PR, Trotter EW, Foray AP, Debaud AL, Rice GI, et al. Protein kinase cdelta deficiency causes mendelian systemic lupus erythematosus with B cell-defective apoptosis and hyperproliferation. Arthritis Rheum. 2013;65:2161–2171. doi: 10.1002/art.38008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steinberg SF. Distinctive activation mechanisms and functions for protein kinase Cdelta. Biochem J. 2004;384:449–459. doi: 10.1042/BJ20040704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mecklenbrauker I, Saijo K, Zheng NY, Leitges M, Tarakhovsky A. Protein kinase Cdelta controls self-antigen-induced B-cell tolerance. Nature. 2002;416:860–865. doi: 10.1038/416860a. [DOI] [PubMed] [Google Scholar]
- 84.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramantani G, Kohlhase J, Hertzberg C, Innes AM, Engel K, Hunger S, et al. Expanding the phenotypic spectrum of lupus erythematosus in Aicardi-Goutieres syndrome. Arthritis Rheum. 2010;62:1469–1477. doi: 10.1002/art.27367. [DOI] [PubMed] [Google Scholar]
- 86.Namjou B, Kothari PH, Kelly JA, Glenn SB, Ojwang JO, Adler A, et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes and immunity. 2011;12:270–279. doi: 10.1038/gene.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, et al. Mutations in the gene encoding the 3’-5’ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nature genetics. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 88.Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 89.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lo M, Tsokos G. Monogenic lupus. International Journal of Clinical Rheumatology. 2014;9:543–546. [Google Scholar]
- 91.Jeremiah N, Neven B, Gentili M, Callebaut I, Maschalidi S, Stolzenberg MC, et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest. 2014;124:5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, et al. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 2011;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, et al. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem. 2013;288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xin B, Jones S, Puffenberger EG, Hinze C, Bright A, Tan H, et al. Homozygous mutation in SAMHD1 gene causes cerebral vasculopathy and early onset stroke. Proc Natl Acad Sci U S A. 2011;108:5372–5377. doi: 10.1073/pnas.1014265108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chahwan C, Chahwan R. Aicardi-Goutieres syndrome: from patients to genes and beyond. Clin Genet. 2012;81:413–420. doi: 10.1111/j.1399-0004.2011.01825.x. [DOI] [PubMed] [Google Scholar]
- 97.Li N, Zhang W, Cao X. Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol Lett. 2000;74:221–224. doi: 10.1016/s0165-2478(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 98.Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Welbourn S, Miyagi E, White TE, Diaz-Griffero F, Strebel K. Identification and characterization of naturally occurring splice variants of SAMHD1. Retrovirology. 2012;9:86. doi: 10.1186/1742-4690-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Briggs TA, Rice GI, Daly S, Urquhart J, Gornall H, Bader-Meunier B, et al. Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet. 2011;43:127–131. doi: 10.1038/ng.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lausch E, Janecke A, Bros M, Trojandt S, Alanay Y, De Laet C, et al. Genetic deficiency of tartrate-resistant acid phosphatase associated with skeletal dysplasia, cerebral calcifications and autoimmunity. Nat Genet. 2011;43:132–137. doi: 10.1038/ng.749. [DOI] [PubMed] [Google Scholar]
- 104.Andersson G, Ek-Rylander B, Hollberg K, Ljusberg-Sjolander J, Lang P, Norgard M, et al. TRACP as an osteopontin phosphatase. J Bone Miner Res. 2003;18:1912–1915. doi: 10.1359/jbmr.2003.18.10.1912. [DOI] [PubMed] [Google Scholar]
- 105.Han S, Guthridge JM, Harley IT, Sestak AL, Kim-Howard X, Kaufman KM, et al. Osteopontin and systemic lupus erythematosus association: a probable gene-gender interaction. PLoS One. 2008;3:e0001757. doi: 10.1371/journal.pone.0001757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Beyaert R, Heyninck K, Van Huffel S. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem Pharmacol. 2000;60:1143–1151. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 107.Flesher DL, Sun X, Behrens TW, Graham RR, Criswell LA. Recent advances in the genetics of systemic lupus erythematosus. Expert Rev Clin Immunol. 2010;6:461–479. doi: 10.1586/eci.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bates JS, Lessard CJ, Leon JM, Nguyen T, Battiest LJ, Rodgers J, et al. Meta-analysis and imputation identifies a 109 kb risk haplotype spanning TNFAIP3 associated with lupus nephritis and hematologic manifestations. Genes Immun. 2009;10:470–477. doi: 10.1038/gene.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lodolce JP, Kolodziej LE, Rhee L, Kariuki SN, Franek BS, McGreal NM, et al. African-derived genetic polymorphisms in TNFAIP3 mediate risk for autoimmunity. Journal of immunology. 2010;184:7001–7019. doi: 10.4049/jimmunol.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Adrianto I, Wen F, Templeton A, Wiley G, King JB, Lessard CJ, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nature genetics. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang S, Wen F, Wiley GB, Kinter MT, Gaffney PM. An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression. PLoS Genet. 2013;9:e1003750. doi: 10.1371/journal.pgen.1003750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 115.Adrianto I, Wang S, Wiley GB, Lessard CJ, Kelly JA, Adler AJ, et al. Association of two independent functional risk haplotypes in TNIP1 with systemic lupus erythematosus. Arthritis and rheumatism. 2012;64:3695–3705. doi: 10.1002/art.34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Caster DJ, Korte EA, Nanda SK, McLeish KR, Oliver RK, G’Sell TR, et al. ABIN1 dysfunction as a genetic basis for lupus nephritis. Journal of the American Society of Nephrology : JASN. 2013;24:1743–1754. doi: 10.1681/ASN.2013020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 118.Guthridge JM, Lu R, Sun H, Sun C, Wiley GB, Dominguez N, et al. Two functional lupus-associated BLK promoter variants control cell-type- and developmental-stage-specific transcription. American journal of human genetics. 2014;94:586–598. doi: 10.1016/j.ajhg.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol Cell Biol. 2003;23:1946–1960. doi: 10.1128/MCB.23.6.1946-1960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007;204:2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang C, Ahlford A, Jarvinen TM, Nordmark G, Eloranta ML, Gunnarsson I, et al. Genes identified in Asian SLE GWASs are also associated with SLE in Caucasian populations. Eur J Hum Genet. 2013;21:994–999. doi: 10.1038/ejhg.2012.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang D, John SA, Clements JL, Percy DH, Barton KP, Garrett-Sinha LA. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int Immunol. 2005;17:1179–1191. doi: 10.1093/intimm/dxh295. [DOI] [PubMed] [Google Scholar]
- 123.Lu X, Zoller EE, Weirauch MT, Wu Z, Namjou B, Williams AH, et al. Lupus Risk Variant Increases pSTAT1 Binding and Decreases ETS1 Expression. Am J Hum Genet. 2015;96:731–739. doi: 10.1016/j.ajhg.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wojcik H, Griffiths E, Staggs S, Hagman J, Winandy S. Expression of a non-DNA-binding Ikaros isoform exclusively in B cells leads to autoimmunity but not leukemogenesis. Eur J Immunol. 2007;37:1022–1032. doi: 10.1002/eji.200637026. [DOI] [PubMed] [Google Scholar]
- 125.Hu W, Sun L, Gao J, Li Y, Wang P, Cheng Y, et al. Down-regulated expression of IKZF1 mRNA in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Rheumatol Int. 2011;31:819–822. doi: 10.1007/s00296-010-1576-1. [DOI] [PubMed] [Google Scholar]
- 126.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood. 1999;93:2013–2024. [PubMed] [Google Scholar]
- 128.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG, Nath SK, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases--a meta-analysis. Rheumatology (Oxford) 2007;46:49–56. doi: 10.1093/rheumatology/kel170. [DOI] [PubMed] [Google Scholar]
- 129.Orru V, Tsai SJ, Rueda B, Fiorillo E, Stanford SM, Dasgupta J, et al. A loss-of-function variant of PTPN22 is associated with reduced risk of systemic lupus erythematosus. Hum Mol Genet. 2009;18:569–579. doi: 10.1093/hmg/ddn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stanford SM, Bottini N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol. 2014;10:602–611. doi: 10.1038/nrrheum.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kariuki SN, Crow MK, Niewold TB. The PTPN22 C1858T polymorphism is associated with skewing of cytokine profiles toward high interferon-alpha activity and low tumor necrosis factor alpha levels in patients with lupus. Arthritis and rheumatism. 2008;58:2818–2823. doi: 10.1002/art.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Y, Shaked I, Stanford SM, Zhou W, Curtsinger JM, Mikulski Z, et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity. 2013;39:111–122. doi: 10.1016/j.immuni.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Habib T, Funk A, Rieck M, Brahmandam A, Dai X, Panigrahi AK, et al. Altered B cell homeostasis is associated with type I diabetes and carriers of the PTPN22 allelic variant. Journal of immunology. 2012;188:487–496. doi: 10.4049/jimmunol.1102176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Menard L, Saadoun D, Isnardi I, Ng YS, Meyers G, Massad C, et al. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. The Journal of clinical investigation. 2011;121:3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cella M, Dohring C, Samaridis J, Dessing M, Brockhaus M, Lanzavecchia A, et al. A novel inhibitory receptor (ILT3) expressed on monocytes, macrophages, and dendritic cells involved in antigen processing. J Exp Med. 1997;185:1743–1751. doi: 10.1084/jem.185.10.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jensen MA, Yanowitch RN, Reder AT, White DM, Arnason BG. Immunoglobulin-like transcript 3, an inhibitor of T cell activation, is reduced on blood monocytes during multiple sclerosis relapses and is induced by interferon beta-1b. Mult Scler. 2010;16:30–38. doi: 10.1177/1352458509352794. [DOI] [PubMed] [Google Scholar]
- 137.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol Med. 2011;17:714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Javierre BM, Richardson B. A new epigenetic challenge: systemic lupus erythematosus. Adv Exp Med Biol. 2011;711:117–136. doi: 10.1007/978-1-4419-8216-2_9. [DOI] [PubMed] [Google Scholar]
- 140.Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9:e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Koelsch KA, Webb R, Jeffries M, Dozmorov MG, Frank MB, Guthridge JM, et al. Functional characterization of the MECP2/IRAK1 lupus risk haplotype in human T cells and a human MECP2 transgenic mouse. J Autoimmun. 2013;41:168–174. doi: 10.1016/j.jaut.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 144.Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 2010;11:124–133. doi: 10.1038/gene.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dozmorov MG, Wren JD, Alarcon-Riquelme ME. Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes. Epigenetics. 2014;9:276–285. doi: 10.4161/epi.27021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 148.Qu B, Shen N. miRNAs in the Pathogenesis of Systemic Lupus Erythematosus. Int J Mol Sci. 2015;16:9557–9572. doi: 10.3390/ijms16059557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hou J, Wang P, Lin L, Liu X, Ma F, An H, et al. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 151.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 152.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhou H, Huang X, Cui H, Luo X, Tang Y, Chen S, et al. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885–5894. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- 154.Carlsen AL, Schetter AJ, Nielsen CT, Lood C, Knudsen S, Voss A, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013;65:1324–1334. doi: 10.1002/art.37890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hikami K, Kawasaki A, Ito I, Koga M, Ito S, Hayashi T, et al. Association of a functional polymorphism in the 3’-untranslated region of SPI1 with systemic lupus erythematosus. Arthritis Rheum. 2011;63:755–763. doi: 10.1002/art.30188. [DOI] [PubMed] [Google Scholar]
- 157.Deng Y, Zhao J, Sakurai D, Kaufman KM, Edberg JC, Kimberly RP, et al. MicroRNA-3148 modulates allelic expression of toll-like receptor 7 variant associated with systemic lupus erythematosus. PLoS genetics. 2013;9:e1003336. doi: 10.1371/journal.pgen.1003336. [DOI] [PMC free article] [PubMed] [Google Scholar]