Abstract
Recently, inhibition of the SH2-containing inositol 5′-phosphatase 1 (SHIP1) has become an attractive strategy for facilitating engraftment of MHC-I mismatched bone marrow grafts, increasing the number of adult stem cells in vivo, and inducing mobilization of hematopoietic stem cells. Utilizing high-throughput screening, two quinoline small molecules (NSC13480 and NSC305787) that inhibit SHIP1 enzymatic activity were discovered. New syntheses of these inhibitors have been developed which verified the relative stereochemistry of these structures. Utilizing this synthetic route, some analogs of these quinolines have been prepared and tested for their ability to inhibit SHIP. These structure activity studies determined that an amine tethered to the quinoline core is required for SHIP inhibition. SHIP inhibition may explain the antitumor effects of similar quinoline amino alcohols and provides an impetus for further synthetic studies in this class of compounds.
Keywords: quinoline, SHIP, inositol, phosphatase, inhibitor
Graphical Abstract

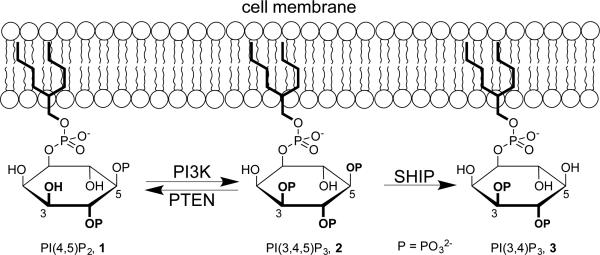
Genetic analysis has indicated the SHIP1 gene is necessary for acute bone marrow graft rejection1 and bone marrow retention of hematopoietic stem cells (HSC) and their homeostasis.2 We have therefore recently focused on the inhibition of the SH2 domain containing inositol-5′-phosphatase-1 (SHIP1) as a potential therapeutic approach to facilitate engraftment of MHC-I mismatched bone marrow grafts,3 increase the number and recovery of adult stem cells in vivo4 and mobilization of HSC.4 In addition, SHIP inhibition has been shown to be effective in killing cancer cells in a number of systems.5 SHIP plays an important role in modulating cell signaling through its phosphatase activity, which modifies the phosphorylation pattern of Phosphatidylinositol Phosphates (PIPs). PIPs play an important role in cell signaling, with molecules like phosphatidylinositol-3,4,5-triphosphate (PI(3,4,5)P3, 2) and phosphatidylinositol-3,4-bisphosphate (PI(3,4)P2, 3) functioning as second messengers that can activate PH domain containing Serine/Threonine kinases like Akt and PDK1.6 In response to extracellular stimuli, enzymes like Phosphatidylinositide 3-Kinase (PI3K), Phosphatase and tensin homolog (PTEN) and SHIP1 modify the phosphorylation patterns on PIPs while they are embedded in the internal leaflet of the plasma membrane lipid bilayer (Figure 1). The concentration of different PIPs significantly influences many downstream effector cascades through what is commonly referred to as the PI3K/Akt signaling pathway, which regulates a number of aspects of cellular physiology.7
Figure 1.
Enzymatic modification of phosphatidylinositol phosphate second messengers in the PI3K/Akt pathway
The two major SHIP paralogs encoded in mammalian genomes are SHIP1 and SHIP2. Expression of SHIP1 is primarily confined to cells of the hematopoietic lineage, typically blood and bone marrow cells, although it is also expressed by embryonic (ES)8 and mesenchymal stem cells (MSC).2b,9 SHIP2 is ubiquitously expressed in all cell and tissue types in rodents and humans, with especially high levels of SHIP2 being found in the heart, skeletal muscle, and the placenta.10 While SHIP1 and SHIP2 share a high rate of amino acid conservation,11 they differ significantly in their cellular expression and receptor recruitment.12 These differences partially explain the different roles these two highly homologous proteins play in cellular pathology. For example, SHIP2 acts as a significant negative regulator of the insulin-signaling pathway.13 Alternatively, SHIP1 functions as an important negative controller in immunoreceptor signaling,14 in hematopoietic progenitor cell proliferation/survival,11 and as an inducer of cellular apoptosis.15 Interestingly, SHIP1 has also been implicated both as a hematopoietic tumor suppressor and activator.6 The SHIP1/2 proteins influence cellular events in two separate ways: through their phosphatase activity and as part of a scaffolding system, which recruits other proteins to the cell membrane utilizing their SH2 domains.6 While genetic knockout studies have been utilized to remove the enzyme entirely,16,6 and thereby deleting both functions of the protein, the use of a small molecule inhibitor to halt the enzymatic function provides the opportunity to evaluate the effects that are primarily associated with the phosphatase activity. In addition, an inhibitor can be introduced and then be allowed to dissipate, so the effects of transient and reversible inhibition on the signaling pathway may be evaluated, in a manner where sequelae associated with long-term SHIP1 deficiency are avoided.
Given the role of SHIP in regulating many aspects of cellular pathology, interest has become focused on influencing the enzymatic activity of the 5′-phosphatase using small molecule inhibitors and agonists.17 Modulation of PIP levels has become a hotly pursued goal, as these molecules play a critical role in signal transduction. Controlling the synthesis of PI(3,4,5)P3 by inhibiting PI3K has been the most heavily pursued strategy.18 However, while several excellent inhibitors have been developed, efforts have been complicated by the requirement of selectively targeting PI3K isoforms to disrupt PI3K signaling in specific cell types. An alternative approach to modify PI3K signaling involves increasing or decreasing the activity of the phosphatases that modulate distal effectors (PI(3,4,5)P3, PI(3,4)P2) of PI3K, such as SHIP1. Intrigued by the possibility of influencing signals in the PI3K/Akt pathway by modulating a phosphatase instead of a kinase, a high-throughput screen utilizing the National Cancer Institute (NCI) diversity set was conducted to find inhibitors of SHIP1.5a This screen successfully identified several compounds, including two quinoline-based amino alcohols, NSC13480 (4•HCl) and NSC305787 (5•HCl) (Figure 2), as SHIP1/2 inhibitors.5b
Figure 2.
Quinoline-based SHIP Inhibitors and similar compounds
Both quinolines 4•HCl and 5•HCl are derived from a program to develop antimalarial agents undertaken by the Walter Reed Army Institute of Research.19 This program developed synthetic analogues of quinine 6 (these products are also similar to quinidine (7), see Figure 2) and culminated in the discovery of mefloquine (8),20 marketed as Larium for prophylactic and therapeutic use against malaria. More recent research has shown that mefloquine possesses anticancer21 properties that are observed at significantly higher concentrations than those required for antimalarial activity. A number of similar quinoline amino alcohols have also shown significant bioactivity in other biological assays, specifically as antibacterials,22 as inhibitors of biofilm formation,22b and as inducers of vacuolization and cell death in glioblastoma cells.23 As the SHIP gene is not present in bacteria, SHIP inhibition is not likely to play a role in these antibacterial activities or in inhibiting biofilm formation. Nevertheless, SHIP inhibition has been shown to be cytotoxic for a number of cancer types both in vitro and in vivo.5,6,17
In order to fully evaluate the biological properties of quinolines 4 and 5, we designed a scalable synthetic route to provide gram-scale quantities of the molecules. Initially, the relative stereochemistry of the amino alcohol portion of quinolines 4 and 5 was indeterminate, as the structure from the NCI library did not specify the stereochemistry, and the original syntheses of the two molecules were not obviously stereoselective. Preliminary 1H NMR analysis of samples from the NCI showed a single diastereomer. Based on the anti (erythro) aminoalcohol relationship seen in mefloquine (8), it was assumed that quinolines 4 and 5 contained the same relative stereochemistry, and structures with this stereochemistry became the desired synthetic targets. As the literature syntheses of these molecules24 did not form the aminoalcohol portion of the molecule in a stereocontrolled manner, efforts were directed towards a route that could selectively provide the parent compounds and that could eventually be adapted to a stereoselective and enantioselective approach. The route defined by the retrosynthetic analysis presented in Figure 3, featuring a stereospecific epoxide opening-cyclization procedure, met these requirements and was therefore investigated further. First used in the synthesis of quinuclidines by Lygo,25 this route appeared to be the most promising means of rapidly accessing the aminoalcohol section of the SHIP1/2 inhibitors, especially since this method has seen widespread application26 in syntheses of quinine, mefloquine, and their analogues, all of which are structurally similar to 4 and 5. The required epoxide 10 may be obtained from the corresponding alkene 11 by way of an E-selective olefination between 12 and 13. Utilization of a Horner-Wadsworth-Emmons (HWE) olefination was anticipated based on precedence established by Kobayashi and co-workers on several similar substrates.26c High selectivity in this olefination was critical, as the olefin stereochemistry defines the desired anti-amino alcohol configuration in the final product.
Figure 3.
Retrosynthetic analysis of quinoline SHIP inhibitors 4 and 5
The synthesis of quinoline 4 commenced with the Doebner condensation of 1-naphthylamine, benzaldehyde, and pyruvic acid which produced carboxylic acid 15 in 26% yield (Scheme 1).24a While not high-yielding, the low cost of the starting materials, the ease with which the product is isolated (simple vacuum filtration provided the product in high purity), and the ease of scale-up made this transformation attractive. Reduction of carboxylic acid 15 to alcohol 16 using BH3•THF was found to be superior to other methods such as sodium borohydride-iodine reduction of the acid, which resulted in incomplete conversion, or lithium aluminum hydride reduction of the corresponding ethyl ester, which resulted in decomposition of the starting material. Conversion of alcohol 16 to chloride 17 using thionyl chloride followed by an Arbuzov reaction provided the desired phosphonate 18.
Scheme 1.
Synthesis of phosphonate 18
With phosphonate 18 in hand, the aldehyde condensation partner 13 was synthesized in two steps from 5-aminopentan-1-ol (19) (Scheme 2). The TEMPO oxidation conditions of De Luca, Giacomelli and Porcheddu27 which utilized trichloroisocyanuric acid (TCCA) as the stoichiometric oxidant proved to be superior to PCC for the oxidation, consistently providing the desired aldehyde in high yields. No chlorination of the phthalimide was observed under these conditions. Sodium hydride was initially used for the HWE olefination; however, this base proved to be unreliable, as the olefination yields varied unpredictably. Masamune and Roush's modified conditions28 for HWE olefinations provided more reproducible yields, with the combination of DBU and lithium chloride providing olefin 20 in 68% yield with >20:1 E-selectivity (as determined by 1H NMR analysis). Subsequent electrophilic epoxidation of the olefin with m-CPBA was predictably reliable, as was removal of the phthalimide protecting group followed by spontaneous cyclization to produce the piperidinylmethanol moiety with anti-stereochemistry. Formation of the mono-HCl salt then provided the desired 4•HCl. Only the mono-HCl salt was observed in the precipitate (the identity of which was confirmed by 1H NMR and combustion analysis), which was attributed to 4•HCl precipitating from the diethyl ether solvent as a white solid before formation of the bis-HCl salt could occur. Comparison by 1H NMR of our synthetic sample of 4•HCl with the NCI sample showed that they were identical. Therefore, the anti-stereochemistry was correctly anticipated. With the structure of quinoline 4•HCl established, we turned our attention to the other quinoline-based SHIP inhibitor, 5•HCl.
Scheme 2.
Synthesis of 4•HCl
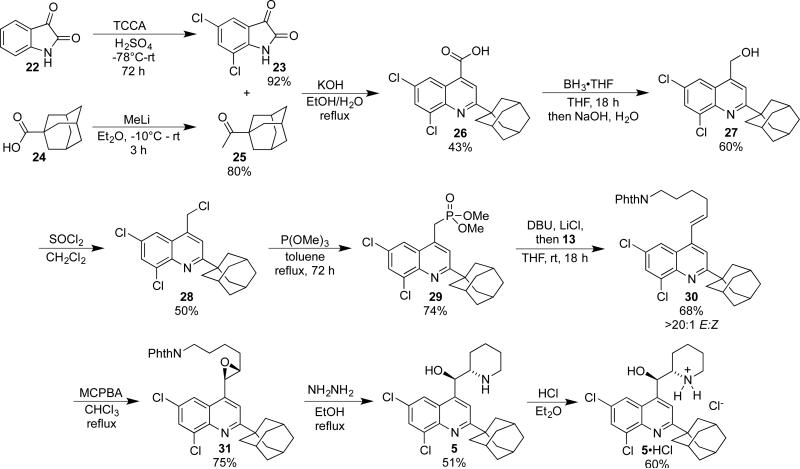
While a scalable synthesis of quinoline 5 has been published,24b it required access to a high-pressure reactor capable to attaining 200 psi of hydrogen on large scale. Instead of pursuing a route that required special equipment, we chose to instead adapt our route for making quinoline 4 to the synthesis of 5 (Scheme 3). Dichlorination of isatin (22) with TCCA, which functions as an effective chlorinating agent when sulfuric acid is utilized as a promoter, provided 5,7-dichloroisatin (23) in good yield as reported by Ribeiro and co-workers.29 On large scale this process resulted in a highly exothermic reaction, so the procedure was modified to begin the reaction as a heterogeneous mixture at −78 °C, which was then allowed to mix and warm slowly to room temperature providing 5,7-dichloroisatin 23 in 75% yield. Adamantyl carboxylic acid 24 was conveniently converted to ketone 25 with methyl lithium and was then used in the Pfitzinger quinoline synthesis to provide the desired quinoline carboxylic acid 26. Refluxing isatin 23 with 1.1 equivalents of ketone 25 in EtOH for 24 h consistently provided 20-25% of quinoline 26 after recrystallization, with complete consumption of the starting isatin 23, but with approximately 50% of starting ketone 25 recovered. This was not surprising, as the Pfitzinger reaction is high-yielding with aryl ketones, but increasing the steric bulk of the starting ketone, as in this example, significantly decreases the efficiency of the process.30 To compensate, the less expensive isatin was used in 2-fold excess, which provided carboxylic acid 26 in 43% yield after recrystallization. As with quinoline 4, reduction of carboxylic acid 26 to alcohol 27 was optimally performed with BH3•THF. The conversion to chloride 28 was unexpectedly difficult. Thionyl chloride at room temperature, though successful in the synthesis of quinoline 17, gave inconsistent results with alcohol 27. Lowering the temperature of the reaction to 0 °C and shortening the reaction time to 1 h gave a moderate, but consistent, yield of 50%, and so these conditions were adopted. Arbuzov reaction with trimethyl phosphate then provided phosphonate 29. The HWE olefination using DBU/LiCl, epoxidation with m-CPBA, cyclization, and acidification steps followed to provide the mono-HCl salt of quinoline 5. A comparison of synthetic 5•HCl with a sample obtained from the NCI showed that they were essentially identical, verifying the structure of NSC305787 as 5•HCl.
Scheme 3.
Synthesis of 5•HCl
With the structures secure, attention was turned to the biological activity of these molecules and their synthetic intermediates (Table 1). The original high-throughput screen was performed utilizing a Fluorescence Polarization assay for phosphatase activity,5a but more recently we have employed a colorimetric Malachite Green assay to evaluate SHIP activity, as this assay is less expensive and less care needs to be taken with compounds that have fluorescent signatures like the quinolines. Unfortunately, attempts to test the activity of quinoline 4•HCl in the Malachite Green assay proved difficult, as the compound formed a precipitate under the assay conditions, which made the assay results unreliable. Utilizing the citrate salt avoided this issue and allowed for evaluation of the compound, but SHIP1 inhibition was somewhat moderate. Quinoline 5 provided more significant inhibition of SHIP1/2, showing an especially strong inhibition of SHIP2. Additionally, mefloquine hydrochloride 8•HCl showed significant inhibition of both SHIP paralogs. This SHIP1/2 inhibition may account for the antitumor activity of mefloquine that has been previously reported.21 Both quinine sulfate 6•(0.5 H2SO4) and quinidine sulfate 7•(0.5 H2SO4) showed little to no activity against SHIP1 or SHIP2. Prior to beginning an enantioselective synthesis of 4 and 5, some synthetic intermediates (carboxylic acids 15 and 26 as well as alcohols 16 and 27) were tested to determine if the aminoalcohol core of the molecule was important to the pharmacophore. Little activity was seen for these molecules, except for moderate activity from carboxylic acid 26 against SHIP1. From these results, it was hypothesized that perhaps the aminoalcohol portion of the molecule was more significant, so (1R, 2S)-(-)-ephedrine 32 and (1S, 2S)-(+)-pseudephedrine 33 were evaluated to determine if the quinoline core was essential, but these also showed no SHIP activity. These results led us to hypothesize that both the amine and the quinoline core were necessary for significant SHIP activity, and this was tested with the synthesis of quinoline 34•HCl (synthesized from alcohol 16 utilizing classical Gabriel conditions, see the supporting information for details). Quinoline 34•HCl gave significantly better activity against both SHIP paralogs than acid 15 or alcohol 16. This result verifies the importance of the quinoline core and the pendant amine, and also indicates that the piperidine ring and the alcohol present in amino alcohols 4, 5 and 8 are not critical for SHIP phosphatase inhibition.
Table 1.
SHIP inhibitory activity of quinolines 4 and 5 and related moleculesa
| Entry | Compound | SHIP1 Inhibition | SHIP2 Inhibition |
|---|---|---|---|
| 1 | 4•HCl | N/Ab | N/Ab |
| 2 | 4•citrate | 38% | 17% |
| 3 | 5•HCl | 38% | 73% |
| 4 | mefloquine hydrochloride (8•HCl) | 54% | 54% |
| 5 | quinine sulfate (6•0.5 H2SO4) | 0% | 8% |
| 6 | quinidine sulfate (7•0.5 H2SO4) | 19% | 5% |
| 7 | carboxylic acid 15 | 7%c | 0%c |
| 8 | carboxylic acid 26 | 44%c | 0%c |
| 9 | alcohol 16 | 2%c | 18%c |
| 10 | alcohol 27 | 0%c | 14%c |
| 11 | (1R, 2S)-(-)-ephedrine 32 | 0% | 0% |
| 12 | (1S, 2S)-(+)-pseudephedrine 33 | 2% | 13% |
| 13 | 34•HCl | 41% | 66% |
Results from the Malachite Green assay performed at 1mM concentration.
Compound precipitated during testing.
Evaluated at 500μM

In summary, new synthetic routes to quinoline-based SHIP inhibitors 4 and 5 have been developed which do not require special equipment for their synthesis. These routes provide the products in a stereoselective manner. The parent quinolines, several intermediates and similar commercially available compounds have been tested for activity against SHIP1 and SHIP2. In addition to the activity demonstrated by the parent compounds, mefloquine hydrochloride 8•HCl proved to be active as a SHIP inhibitor at high concentrations. The SHIP1/2 activity of mefloquine may explain the reports of the antitumor activity of the molecule, as the concentration seen for SHIP inhibition is similar to the concentration observed for cancer cell killing.20 Quinine and quinidine were not active in the assay, and neither were ephedrine or pseudoephedrine. A brief structure activity study showed that both the quinoline core and a pendant amine at the 4-position of the quinoline were required for significant SHIP1/2 inhibition, but the alcohol and the piperidine ring were not critical components for this activity. Future studies exploring the structure-activity relationships in these systems are underway and will be reported in due course.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (RO1 HL72523, R01 HL085580) and the Paige Arnold Butterfly Run.W.G.K. is an Empire Scholar of the State University of New York and a Senior Scholar of the Crohn's & Colitis Foundation of America (CCFA). S.F. is a Research Fellow of the CCFA. Support for the Syracuse University NMR facility was provided by the NSF (CHE-1229345), which is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Copies of 1H NMR and 13C NMR spectra as well as full experimental details. Supplementary data associated with this article can be found, in the online version, at http://XXXXXXXXXXXXX.
References and notes
- 1.a Wang JW, Howson JM, Ghansah T, Desponts C, Ninos JM, May SL, Nguyen KH, Toyama-Sorimachi N, Kerr WG. Science. 2002;295:2094. doi: 10.1126/science.1068438. [DOI] [PubMed] [Google Scholar]; b Gumbleton M, Vivier E, Kerr WG. J. Immunol. 2015;194:2847. doi: 10.4049/jimmunol.1402930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Desponts C, Hazen AL, Paraiso KH, Kerr WG. Blood. 2006;107:4338. doi: 10.1182/blood-2005-12-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hazen AL, Smith MJ, Desponts C, Winter O, Moser K, Kerr WG. Blood. 2009;113:2924. doi: 10.1182/blood-2008-02-138008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandes S, Brooks R, Park M-Y, Srivastava N, Russo CM, Chisholm JD, Kerr WG. EBioMedicine. 2015;2:205. doi: 10.1016/j.ebiom.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks R, Iyer S, Akada H, Neelam S, Russo CM, Chisholm JD, Kerr WG. Stem Cells. 2015;33:848. doi: 10.1002/stem.1902. [DOI] [PubMed] [Google Scholar]
- 5.a Brooks R, Fuhler GM, Iyer S, Smith MJ, Park MY, Paraiso KH, Engelman RW, Kerr WG. J. Immunol. 2010;184:3582. doi: 10.4049/jimmunol.0902844. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Fuhler GM, Brooks R, Toms B, Iyer S, Gengo EA, Park MY, Gumbleton M, Viernes DR, Chisholm JD, Kerr WG. Mol. Med. 2012;18:65. doi: 10.2119/molmed.2011.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Park MY, Srivastava N, Sudan R, Viernes DR, Chisholm JD, Engelman RW, Kerr WG. Mucosal Immunol. 2014;7:1429. doi: 10.1038/mi.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L, Titz B, Graeber TG, Park E, Tan YX, Satterthwaite A, Paietta E, Hunger SP, Willman CL, Melnick A, Loh ML, Jung JU, Coligan JE, Bolland S, Mak TW, Limnander A, Jumaa H, Reth M, Weiss A, Lowell CA, Muschen M. Nature. 2015;521:357. doi: 10.1038/nature14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerr WG. Ann. N. Y. Acad. Sci. 2011;1217:1. doi: 10.1111/j.1749-6632.2010.05869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantley LC. Science. 2002;296:1655. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 8.Tu Z, Ninos JM, Ma Z, Wang JW, Lemos MP, Desponts C, Ghansah T, Howson JM, Kerr WG. Blood. 2001;98:2028. doi: 10.1182/blood.v98.7.2028. [DOI] [PubMed] [Google Scholar]
- 9.Iyer S, Brooks R, Gumbleton M, Kerr WG. Stem Cells Dev. 2015;24:1073. doi: 10.1089/scd.2014.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pesesse X, Deleu S, De Smedt F, Drayer L, Erneux C. Biochem. Bioph. Res. Co. 1997;239:697. doi: 10.1006/bbrc.1997.7538. [DOI] [PubMed] [Google Scholar]
- 11.Krystal G, Damen JE, Helgason CD, Huber M, Hughes MR, Kalesnikoff J, Lam V, Rosten P, Ware MD, Yew S, Humphries RK. Int. J. Biochem. Cell Biol. 1999;31:1007. doi: 10.1016/s1357-2725(99)00072-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Wavreille AS, Kunys AR, Pei D. Biochemistry. 2009;48:11075. doi: 10.1021/bi9012462. [DOI] [PubMed] [Google Scholar]
- 13.Hakim S, Bertucci MC, Conduit SE, Vuong DL, Mitchell CA. Curr. Top. Microbiol. Immunol. 2012;362:247. doi: 10.1007/978-94-007-5025-8_12. [DOI] [PubMed] [Google Scholar]
- 14.Veillette A, Latour S, Davidson D. Annu. Rev. Immunol. 2002;20:669. doi: 10.1146/annurev.immunol.20.081501.130710. [DOI] [PubMed] [Google Scholar]
- 15.Erneux C, Govaerts C, Communi D, Pesesse X. Biochim. Biophys. Acta. 1998;1436:185. doi: 10.1016/s0005-2760(98)00132-5. [DOI] [PubMed] [Google Scholar]
- 16.a Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM, Borowski A, Jirik F, Krystal G, Humphries RK. Genes Dev. 1998;12:1610. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Paraiso KH, Ghansah T, Costello A, Engelman RW, Kerr WG. J. Immunol. 2007;178:2893. doi: 10.4049/jimmunol.178.5.2893. [DOI] [PubMed] [Google Scholar]
- 17.Viernes DR, Choi LB, Kerr WG, Chisholm JD. Med. Res. Rev. 2014;34:795. doi: 10.1002/med.21305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a Bauer TM, Patel MR, Infante JR. Pharmacol. Ther. 2015;146:53. doi: 10.1016/j.pharmthera.2014.09.006. [DOI] [PubMed] [Google Scholar]; b Vanhaesebroeck B, Stephens L, Hawkins P. Nat. Rev. Mol. Cell Biol. 2012;13:195. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]; c Wong K-K, Engelman JA, Cantley LC. Curr. Opin. Genet. Dev. 2010;20:87. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Garcia-Echeverria C, Sellers WR. Oncogene. 2008;27:5511. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]; e Blunt MD, Ward SG. Curr. Opin. Pharmacol. 2012;12:444. doi: 10.1016/j.coph.2012.02.015. [DOI] [PubMed] [Google Scholar]; f Wu P, Hu Y. MedChemComm. 2012;3:1337. [Google Scholar]; g Courtney KD, Corcoran RB, Engelman JA. J. Clin. Oncol. 2010;28:1075. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croft AM. J. R. Soc. Med. 2007;100:170. doi: 10.1258/jrsm.100.4.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trenholme CM, Williams RL, Desjardins RE, Frischer H, Carson PE, Rieckmann KH, Canfield CJ. Science. 1975;190:792. doi: 10.1126/science.1105787. [DOI] [PubMed] [Google Scholar]
- 21.Carson DA, Leoni LM, Cottam HB. U.S. Patent Appl. 20050154010:2005. [Google Scholar]
- 22.a Mao J, Wang Y, Wan B, Kozikowski AP, Franzblau SG. ChemMedChem. 2007;2:1624. doi: 10.1002/cmdc.200700112. [DOI] [PubMed] [Google Scholar]; b Leon B, Fong JCN, Peach KC, Wong WR, Yildiz FH, Linington RG. Org. Lett. 2013;15:1234. doi: 10.1021/ol400150z. [DOI] [PubMed] [Google Scholar]; c Kunin CM, Ellis WY. Antimicrob. Agents Chemother. 2000;44:848. doi: 10.1128/aac.44.4.848-852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitambi SS, Toledo EM, Usoskin D, Wee S, Harisankar A, Svensson R, Sigmundsson K, Kalderén C, Niklasson M, Kundu S, Aranda S, Westermark B, Uhrbom L, Andäng M, Damberg P, Nelander S, Arenas E, Artursson P, Walfridsson J, Forsberg Nilsson K, Hammarström LGJ, Ernfors P. Cell. 2014;157:313. doi: 10.1016/j.cell.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 24.a Buchman ER, Howton DR. J. Org. Chem. 1949;14:895. doi: 10.1021/jo01153a001. [DOI] [PubMed] [Google Scholar]; b Novotny J, Collins CH, Starks FW. J. Pharm. Sci. 1974;63:1264. doi: 10.1002/jps.2600630821. [DOI] [PubMed] [Google Scholar]
- 25.a Lygo B, Crosby J, Lowdon TR, Wainwright PG. Tetrahedron. 1999;55:2795. [Google Scholar]; b Lygo B, Crosby J, Lowdon TR, Wainwright PG. Tetrahedron Lett. 1997;38:2343. [Google Scholar]
- 26.a Adam S. Tetrahedron. 1994;50:3327. [Google Scholar]; b Adam S. Bioorg. Med. Chem. Lett. 1992;2:53. [Google Scholar]; c Igarashi J, Katsukawa M, Wang YG, Acharya HP, Kobayashi H. Tetrahedron Lett. 2004;45:3783. [Google Scholar]; d Raheem IT, Goodman SN, Jacobsen EN. J. Am. Chem. Soc. 2004;126:706. doi: 10.1021/ja039550y. [DOI] [PubMed] [Google Scholar]; e Barlin GB, Ireland SJ, Jiravinyu C, Nguyen TMT, Kotecka B, Rieckmann KH. Aust. J. Chem. 1993;46:1695. [Google Scholar]
- 27.De Luca L, Giacomelli G, Porcheddu A. Org. Lett. 2001;3:3041. doi: 10.1021/ol016501m. [DOI] [PubMed] [Google Scholar]
- 28.Blanchette MA, Choy W, Davis JT, Essenfeld AP, Masamune S, Roush WR, Sakai T. Tetrahedron Lett. 1984;25:2183. [Google Scholar]
- 29.Ribeiro NM, Da Silva BV, de Almeida Violante F, Rezende CM, Pinto AC. Org. Prep. Proc. Int. 2005;37:265. [Google Scholar]
- 30.Fugitt RB, Roberts RM. J. Med. Chem. 1973;16:875. doi: 10.1021/jm00266a002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.