Abstract
Persistent infection with hepatitis C virus (HCV) is associated with an increased risk of hepatocellular carcinoma (HCC). Cancer typically develops in a setting of chronic hepatic inflammation and advanced fibrosis or cirrhosis, and such tissue represents a pre-neoplastic “cancer field”. However, not all persistent infections progress to HCC and a combination of viral and host immune factors likely to contribute to carcinogenesis. HCV may disrupt cellular pathways involved in detecting and responding to DNA damage, potentially adding to the risk of cancer. Efforts to unravel how HCV promotes HCC are hindered by lack of a robust small animal model, but a better understanding of molecular mechanisms could identify novel biomarkers for early detection and allow for development of improved therapies.
INTRODUCTION
Approximately 3.5 million persons in the USA are persistently infected with hepatitis C virus (HCV) [1]. Chronic hepatitis C is a slowly progressing disease and many infected persons remain asymptomatic for decades after initial infection. However, the long-term complications of infection are substantial, and include hepatic fibrosis, cirrhosis, and hepatocellular carcinoma (HCC). Many currently asymptomatic individuals acquired infection prior to the identification of HCV as the etiologic agent of chronic ‘non-A non-B’ hepatitis in 1989 [2]. Because the likelihood of disease increases with the length of infection, the incidence of HCV-associated cirrhosis and HCC has been increasing as this cohort ages and is predicted to reach a peak within the next decade [3]. In addition, a recent surge in injection drug use among young, largely white, non-urban Americans is leading to worrisome increases in the incidence of new HCV infections, and is likely predictive of future increases in liver cancer. Compounding the impact of such increases in HCC incidence, the five-year survival rate of patients with liver cancer has remained low (~15%) within the U.S.
As a single-stranded, positive-sense RNA virus (Figure 1), replicating primarily if not exclusively within hepatocytes, HCV is unique among cancer-causing viruses. Its intracellular replication cycle is entirely cytoplasmic and without potential for integration of viral genome into host chromosomal DNA. Similar to infection with other oncogenic viruses, only a minority of chronically infected persons develop cancer, suggesting that viral elements co-operate with host and environmental factors to promote tumorigenesis. Transgenic mice with liver-specific expression of HCV proteins are at increased risk for hepatocellular carcinoma [4,5]. However, the absence of a tractable small animal model of HCV infection and hepatocellular carcinogenesis has slowed progress in this field, such that many questions concerning how HCV causes liver cancer remain unresolved.
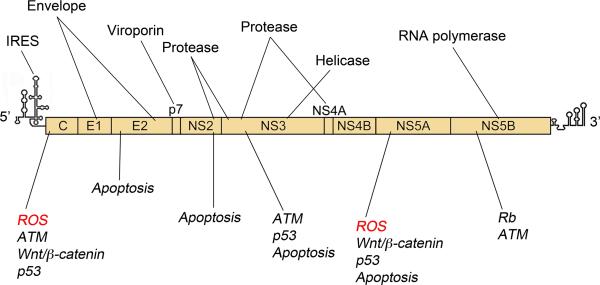
Figure 1. Organization of the 9.7 kb positive-sense HCV RNA genome.
5’ and 3’ untranslated regions (UTRs) contain cis-acting elements essential for virus replication, including a 5’ internal ribosome entry site (IRES) that drives cap-independent expression of a polyprotein (box) that is processed into 10 mature viral proteins by a combination of host and viral proteases. Their functions are highlighted above the genome. The core (C) protein and envelope glycoproteins E1 and E2 are structural components of the virion. Nonstructural (NS) proteins possess functions necessary for replication, including helicase (NS3), protease (NS2 and NS3/4A), and RNA-dependent RNA polymerase (NS5B) activities. NS4B and NS5A drive formation of the ‘membranous web’, a cytoplasmic structure where these proteins accumulate to direct viral RNA synthesis. NS2 and NS5A also function in virion assembly, whereas p7 is essential for egress. Virus-host interactions that may contribute to HCC development are highlighted below the genome. Core and NS5A expression have been linked to the generation of ROS that may contribute to host DNA damage. Multiple HCV proteins interact with and modulate host pathways to facilitate virus replication and may in theory promote carcinogenesis.
Indirect effects of chronic inflammation: a pro-carcinogenic environment
Within the infected liver, double-stranded viral RNA replication intermediates are sensed by host pathogen-associated molecular pattern (PAMP) receptors resulting in the activation of the transcription factors IRF3/7 and NF-κB, and the induction of interferons and related interferon-stimulated genes (ISGs). HCV has evolved several mechanisms that antagonize these responses (reviewed in [6]), and these may contribute to viral persistence by attenuating cell-intrinsic innate immune responses. Despite this virus antagonism, antiviral signaling pathways are activated during persistent HCV infection resulting in increased intrahepatic ISG transcription [7]. Subsequent adaptive immune responses, in particular virus-specific CD8+ cytotoxic T lymphocytes [8,9], while critical for resolving the infection, succeed in clearing the virus only in a minority (~30-40%) of cases [10]. Thus, most infected persons develop lifelong, persistent infection. Such individuals are at risk for protracted but ultimately ineffective immune responses leading to chronic immune-mediated inflammation with repeated cycles of hepatocyte destruction and regeneration.
What are the effects of chronic inflammation?
Persistent immune-mediated hepatic inflammation and associated fibrogenic wound-healing responses are likely to be important drivers of liver cancer in chronic hepatitis C (Figure 2). Activated inflammatory cells promote a pro-carcinogenic microenvironment by releasing reactive oxygen (ROS) and nitrogen (RNS) species and induce lipid peroxidation [11]. The expression of some HCV proteins, in particular core and NS5A, may also add directly to the induction of oxidative stress (see below). The HCV RNA replicase complex possesses a unique ‘sensor’ that responds to lipid peroxidation by down-regulating viral RNA synthesis, thereby maintaining replication at low levels and minimizing oxidative damage [12]. While this may facilitate HCV persistence, hepatic oxidative DNA damage is nonetheless common in chronic hepatitis C [13-15]. Such DNA damage is likely not heritable in terminally differentiated cells. However, in the context of inflammatory liver disease and hepatocellular necrosis associated with chronic hepatitis C, regenerative pathways may be activated allowing dedifferentiation and proliferation to replace damaged tissue.
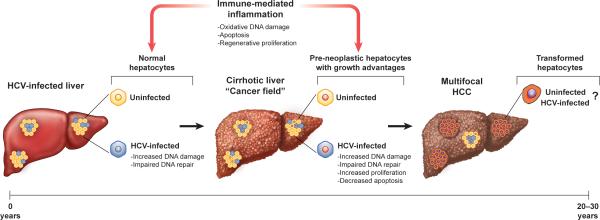
Figure 2. Model for HCV-associated carcinogenesis.
Hepatocellular carcinoma (HCC) likely results from a combination of indirect host- and direct HCV-mediated mechanisms. Persistent immune-mediated inflammation, coupled with expression of core and NS5A, generates ROS that trigger oxidative DNA damage. HCV infection further compromises host genome stability by impairing DNA repair pathways. Repeated cycles of hepatocellular destruction with regenerative proliferation and progressive fibrosis within this pro-mutagenic environment result in a “cancer field” comprised of pre-neoplastic but genetically-altered hepatocytes. Continued hepatocellular turnover may, in turn, select for aberrant hepatocytes with growth advantages. Whether HCC arises directly from HCV-infected hepatocytes remains unclear, although HCV infection may enhance survival of abnormal hepatocytes by promoting cell proliferation and inhibiting apoptosis. Ultimately, these combined mechanisms may select for transformed cells, culminating in development of HCC over 2-3 decades of persistent HCV infection.
Without treatment, approximately 20% of persons with chronic hepatitis C develop severe scarring of the liver, or cirrhosis. Most (perhaps 90%) HCV-associated cancers arise in a background of advanced fibrosis or cirrhosis. A progression of lesions (large regenerative nodules, low-grade and high-grade dysplastic nodules) have been identified in the cirrhotic liver that likely represent HCC precursors [16]. Thus, during the development of hepatocellular cancer, the cirrhotic liver may be considered a pre-neoplastic “cancer field” comprised of genetically abnormal but non-neoplastic tissue that is at high risk for malignant transformation (Figure 2) [17]. Accordingly, HCC is often multifocal in origin in chronic hepatitis C [18]. Cells within this precancerous field already contain mutations that predispose to the cancerous phenotype (discussed below).
What is the impact of chronic hepatocyte turnover?
Over decades of chronic HCV infection, apoptosis of infected hepatocytes (either immune- or virus-mediated) with compensatory hepatocellular proliferation may be strongly pro-carcinogenic. Tumor-promoting effects of apoptosis have been demonstrated in transgenic mouse models where liver-specific knockout of anti-apoptotic proteins such as Mcl-1 promote cancer [19]. Conversely, knockout of the proapoptotic p53 up-regulated modulator of apoptosis (PUMA, or Bbc3) inhibits tumor development following exposure to the chemical carcinogen, diethylnitrosamine [20].
HCV-associated HCC may be considered, like other cancers, a disease of somatic evolution where individual cells are the unit of reproduction [21]. In chronic hepatitis C, the generation of ROS by either viral or immune-mediated mechanisms could create a pro-mutagenic environment, while chronic hepatocellular turnover could select for cells with genetic or epigenetic changes that confer growth advantages allowing clonal expansion (Figure 2).
Direct effects of HCV on the infected hepatocyte
Surprisingly, only a minority of hepatocytes contain detectable viral RNA or proteins within the HCV-infected liver [22,23], suggesting that most cells remain uninfected. The factors restricting replication are not well understood, and whether cancer arises from an infected hepatocyte or an uninfected bystander cell remains an open question.
Does HCV infection render a cell prone to accumulate genetic alterations?
HCCs exhibit a high degree of genetic heterogeneity indicative of a fundamental loss of genomic stability during hepatocellular carcinogenesis [24]. HCV infection is likely to contribute to this genetic instability directly by inducing the generation of reactive oxygen species (ROS) with consequent DNA damage (Figure 2). Although immune-mediated inflammation is at least partly responsible, as discussed above, expression of either the HCV core protein or NS5A, a component of the viral replicase, increases ROS levels and promotes oxidative stress in transgenic mice as well as cultured cells [25-29]. These effects have been attributed to mitochondrial and ER stress initiated by core and NS5A, respectively [25,27,28], although it is uncertain whether these proteins are expressed in sufficient abundance for such direct effects to occur within the infected liver.
In addition to promoting DNA damage, some evidence suggests HCV infection compromises the ability of host cells to detect and repair damaged DNA. The ataxia telangiectasia mutated (ATM) kinase plays a central role in initiating cellular responses to double-strand DNA breaks [30]. Overexpression studies suggest that core, NS3/4A (the major viral protease), and NS5B (RNA-dependent RNA polymerase) are capable of interacting with components of the ATM-driven response, thereby interfering with host DNA repair [31-33]. Although these interactions await confirmation in the context of viral replication, HCV infection impairs phosphorylation of an ATM substrate, histone 2A.X (H2AX), in Huh-7.5 cells [34]. Phosphorylated H2AX (γ-H2AX) normally acts as a platform for the recruitment of DNA repair factors to the sites of DNA damage [35]. As such, inhibition of γ-H2AX might disrupt DNA repair and contribute to genomic instability during HCV infection. Unfortunately, the small proportion of hepatocytes infected with HCV in vivo coupled with low-level expression of viral proteins makes it extraordinarily difficult to confirm or refute such effects in primary human tissues.
The p53 protein is a critical tumor suppressor that coordinates cell-cycle arrest, senescence, and apoptosis in response to DNA damage and other stresses [36]. Mutations that disrupt p53 function are associated with the vast majority of human cancers, including HCC [37,38]. Numerous studies have indicated that HCV proteins, including core, NS3, and NS5A, can interact with p53 when overexpressed in cell culture (for an extensive review see [39]). However, these studies have yielded often-conflicting results as to the impact of HCV proteins on p53 activity, and interactions between HCV proteins and p53 have yet to be reported in the context of infection. The overall impact of HCV infection on p53 function remains undefined, largely because the cell-lines that are most permissive for HCV (Huh-7 hepatoma cells and their derivatives) express a mutated, inactive form of p53 [40,41].
Does HCV infection promote hepatocellular proliferation?
A common feature of oncogenic viruses is their ability to increase cell proliferation, particularly through the inactivation of host tumor suppressors [42]. The retinoblastoma (Rb) protein restricts cell proliferation by repressing the activation of E2F transcription factors necessary for S-phase entry in the cell cycle [43]. HCV infection negatively regulates Rb abundance in cell culture [44-46]. This effect is mediated by the NS5B polymerase, which binds Rb via an LxCxE motif bearing sequence homology to the Rb-binding motifs of DNA virus oncoproteins [46]. NS5B mediates Rb relocalization, promotes proteasomal degradation of Rb in association with the host ubiquitin ligase E6AP, and activates E2F-responsive promoters [45,46]. Rb is essential for optimal innate immune responses in hepatocytes [47], explaining why an RNA virus such as HCV (that does not depend upon DNA synthesis for replication) would have evolved such a mechanism.
Despite E2F activation, however, HCV infection reduces cell proliferation and triggers cell-cycle arrest in cultured cells [48,49]. These findings point towards opposing pro- and anti-proliferative effects during HCV infection. Consistent with this hypothesis, normally quiescent hepatocytes show increased entry into the cell cycle within HCV-infected livers, but markers of G1 arrest are also elevated [50]. However, it is unclear to what extent cell-cycle perturbations observed within the liver represent direct effects of HCV infection since only a minority of hepatocytes are typically infected in vivo [7,22]. It is possible that G1 arrest may be triggered by endogenous or therapeutically administered interferon [51].
HCV may also deregulate the Wnt/β-catenin signaling pathway. This pathway is normally activated upon binding of Wnt ligands to the Frizzled receptor and ultimately triggers activation of the transcription factor β-catenin and consequent upregulation of cellular pro-survival genes. Mutations within this pathway, including activating mutations within β-catenin, occur frequently in HCC, indicating an important role for this pathway in hepatocellular carcinogenesis [37]. Both core and NS5A are capable of activating β-catenin when overexpressed in cell culture [52-55]. Likewise, β-catenin is activated in HCV transgenic mice, wherein it drives overexpression of the oncogene c-myc and thereby contributes to oxidative DNA damage and impaired cell-cycle arrest [26].
Do HCV-infected cells evade apoptosis?
The role of apoptosis in HCV-related HCC is likely complex. On one hand, apoptosis is expected to exert tumor suppressive effects through the removal of aberrant and infected cells. On the other hand, successive rounds of apoptotic cell death with subsequent regenerative proliferation could also contribute to carcinogenesis as discussed above, particularly in an environment of oxidative stress and genomic instability.
Apoptosis can function as a potent antiviral defense, and many viruses thus suppress this host response [56,57]. Whether HCV similarly restricts apoptosis remains unclear. Individual HCV proteins, including core, E2, NS2, NS3, and NS5A, inhibit apoptosis when overexpressed (reviewed in [39]), whereas HCV infection can induce apoptosis in Huh-7 cells and further sensitize them to death receptor-mediated apoptosis [48,49,58-60]. However, most studies of HCV-induced apoptosis have utilized an atypical, robustly replicating, infectious molecular clone of HCV, JFH1 or its derivatives. JFH1 replication is not suppressed by lipid peroxidation [12], and it may trigger apoptosis through artificially high-levels of replication and viral protein expression that do not reflect HCV infections in vivo [7,22]. An alternate molecular clone, H77S.3, replicates at lower levels in cell culture and elicits little apoptosis [48]. Virus produced in cell culture from a closely related clone, H77S.2, established persistent infection in a chimpanzee, whereas cell culture-derived JFH1 virus infects chimpanzees but does not establish persistence [61]. Moreover, passage of JFH1 in chimpanzees resulted in adaptive mutations that rendered it less pro-apoptotic than wild-type JFH1 [62]. Although only a small number of chimpanzees were involved in these studies, they suggest evasion of apoptosis may be important for establishing persistent infection.
What genetic changes contribute to the “cancer field”?
What can we learn from studying the genetics of HCC?
Although it is possible to identify driver genes in HCC by whole exome sequencing [37,63,64], no unique driver genes have yet been linked specifically to HCV infection [64]. A better understanding of genetic and epigenetic changes that drive HCC progression may ultimately allow for improved, personalized therapies, but the genetic diversity of HCC makes such studies very challenging [24].
What are the earliest genetic changes in HCC?
The stepwise accumulation of mutations in HCC is poorly understood. Nonetheless, mutations identified in abnormal, pre-neoplastic tissue may be predictive of progression to HCC. Mutations in the telomerase reverse transcriptase (TERT) promoter are frequently present in high-grade dysplastic nodules (19%) and early HCC (61%) in cirrhosis of several different etiologies, including HCV infection [65,66]. Thus TERT promoter mutations represent one of the earliest recognized genetic changes in hepatocellular carcinogenesis, and could contribute to the cancer field effect in cirrhosis.
What is the cellular origin of HCV-associated HCC?
The precise nature of the founder cell in HCC is also poorly understood. Proliferation is essential for a mutation arising in a pre-neoplastic cell to spread within a population of such cells. Both mature hepatocytes and hepatic progenitor cells are long-lived cells that are capable of repopulation following liver injury, and either could be the source of HCC founder cells in chronic HCV infection [67]. As discussed above, it is not clear whether HCV-associated HCC arises from infected or uninfected cells [68].
An interesting but unanswered question is whether HCV is able to replicate within abnormal hepatocytes in dysplastic nodules, and thus might act co-operatively with mutations that arise early in the progression to cancer. Expression of the liver-specific micro-RNA, miR-122, is often lost during progression to HCC, but is relatively conserved in HCV-associated versus hepatitis B virus (HBV)-associated cancer [69]. Since miR-122 is a critical pro-viral host factor for HCV, this suggests that HCV infection of founder cells may be important at some stage of this process.
What factors enhance the risk of HCC development in chronic hepatitis C?
Host and environmental factors
While the progression of liver disease is highly variable amongst chronically-infected persons, 15-25% progress to cirrhosis by 25-30 years [70]. HCC occurs primarily in persons with cirrhosis, and approximately 20% of cirrhotic individuals ultimately progress to cancer [71]. Such progression is exacerbated by a number of risk factors, including alcohol consumption, older age, male gender, obesity, diabetes, and co-infection with human immunodeficiency virus (HIV) or HBV [72].
Genetic risk factors for HCC development are less well defined. However, genome-wide association studies (GWAS) have revealed several single nucleotide polymorphisms (SNPs) associated with HCC development in HCV-infected persons [73,74]. A SNP located upstream of the MHC class I polypeptide-related sequence A (MICA) gene was associated with a slightly elevated risk for HCC [73]. This SNP appears to influence MICA expression levels, but it is not clear whether it increases the risk of cancer per se, or acts indirectly by influencing progression to cirrhosis. A second GWAS identified an intronic SNP within the DEP domain containing 5 (DEPDC5) locus that nearly doubled the risk of HCC [74]. Both GWAS studies were conducted within Japanese populations, raising the question of whether genetic determinants of HCC vary between ethnic groups. Indeed, an independent study of a Caucasian cohort revealed a reverse correlation between the MICA SNP and HCC risk, and also identified a SNP within the HLA complex P5 (HCP5) locus upstream of MICA associated with an increased risk of HCC in both Caucasian and Japanese populations [75]. The DEPDC5 SNP was not associated with cancer in the Caucasian cohort, further highlighting differences in genetic determinants of HCC across ethnicities.
Viral factors
There are seven different genotypes of HCV that vary by over 30% at the nucleotide level. Epidemiological studies suggest that HCC risk may differ amongst these genotypes, as genotypes 1b and 3 are associated more commonly with HCC than other genotypes [76,77]. In contrast, genotype 2 may be associated with a lower risk of HCC [77]. The mechanisms underlying these potential genotype-specific differences in HCC risk are not clear, nor is it proven that such differences do not arise from genetic or other differences in the infected host populations.
How will new HCV antivirals impact the incidence of HCC?
Over the last few years, the introduction of potent direct-acting antiviral (DAA) drugs targeting the NS3/4A protease, NS5A, or the NS5B polymerase has revolutionized the treatment of chronic hepatitis C. All oral combinations of these DAAs are well tolerated, and typically achieve a sustained virologic response (“SVR”) in 90-95% of patients with genotype 1 infections after only 12 weeks of therapy [78]. Patients with SVR are truly cured of the infection, as HCV does not archive its genome like HIV or HBV. These new drugs thus have the potential to substantially reduce HCC incidence over the coming decades. Several cohort studies demonstrate that SVR achieved with older interferon-based regimens reduces, but does not completely eliminate the risk of HCC in cirrhotic patients (reviewed in [79]). It is likely, but not yet proven, that successful DAA therapy will match these results. Nonetheless, HCC typically grows slowly and may not become clinically evident for many months [80]. Studies are needed to determine how long and to what extent the risk of HCC will persist in successfully treated patients. Until these questions are answered, screening for HCC will remain important for those patients achieving SVR.
What does the future hold?
A better understanding of the molecular drivers of HCV-associated HCC is likely to result in improved and increasingly personalized therapies for this cancer with attendant increases in five-year survival rates. Improved screening and early diagnostic modalities are also likely, along with reductions in the numbers of individuals reaching advanced stages of HCV-related liver disease due to continued improvements in antiviral therapy. Despite this, however, HCV-associated liver cancer will remain an important public health problem for the foreseeable future. There are several reasons for this. First, many infected persons with chronic hepatitis C have no access to new DAAs, either because of their high cost or lack of medical insurance. Second, because of coexisting conditions, the potential for drug-drug interactions, and/or HCV genotype-specific differences in antiviral response, a substantial proportion of patients remain difficult to treat with DAAs. Additionally, a large fraction of individuals are simply unaware of their infection until they develop symptoms of advanced liver disease [1]. And, not to be overlooked, persons fortunate enough to achieve SVR lack protective immunity and are at risk for re-infection. Thus, the elimination of HCV-associated liver cancer seems a far-off and potentially elusive goal, and one that is unlikely to be achieved without development of an effective HCV vaccine.
HIGHLIGHTS.
The incidence of HCV-associated liver cancer will rise over the next decade.
Chronic inflammation and hepatic fibrosis are important drivers of liver cancer.
HCV-infected cirrhotic liver represents a “field of cancerization”.
Genetic studies highlight a role for host genome instability in carcinogenesis.
Virus disruption of host responses to DNA damage may contribute to HCC.
Acknowledgements
Supported in part by grants from the National Institutes of Health, F32-CA192633 (JKM), R21-AI115207 (DRM), and R01-CA164029 and R01-AI095690 (SML), and the University Cancer Research Fund of the University of North Carolina.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Holmberg SD, Spradling PR, Moorman AC, Denniston MM. Hepatitis C in the United States. N Engl J Med. 2013;368:1859–1861. doi: 10.1056/NEJMp1302973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 3.Davis GL, Alter MJ, El-Serag H, Poynard T, Jennings LW. Aging of hepatitis C virus (HCV)-infected persons in the United States: a multiple cohort model of HCV prevalence and disease progression. Gastroenterology. 2010;138:513–521. 521, e511–516. doi: 10.1053/j.gastro.2009.09.067. [DOI] [PubMed] [Google Scholar]
- 4*.Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat.Med. 1998;4:1065–1067. doi: 10.1038/2053. [First report of HCC in transgenic mice expressing the HCV core protein.] [DOI] [PubMed] [Google Scholar]
- 5**.Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, Okuda M, Gosert R, Xiao SY, Weinman SA, et al. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–365. doi: 10.1053/gast.2002.31001. [Transgenic mice expressing the entire HCV polyprotein develop liver cancer in the absence of antiviral immune responses and inflammation.] [DOI] [PubMed] [Google Scholar]
- 6.Li K, Lemon SM. Innate immune responses in hepatitis C virus infection. Semin Immunopathol. 2013;35:53–72. doi: 10.1007/s00281-012-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wieland S, Makowska Z, Campana B, Calabrese D, Dill MT, Chung J, Chisari FV, Heim MH. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology. 2014;59:2121–2130. doi: 10.1002/hep.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, Chien DY, Houghton M, Parham P, Walker CM. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 9.Weiner A, Erickson AL, Kansopon J, Crawford K, Muchmore E, Hughes AL, Houghton M, Walker CM. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Natl Acad Sci U S A. 1995;92:2755–2759. doi: 10.1073/pnas.92.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10:553–562. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- 11.Bartsch H, Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch Surg. 2006;391:499–510. doi: 10.1007/s00423-006-0073-1. [DOI] [PubMed] [Google Scholar]
- 12.Yamane D, McGivern DR, Wauthier E, Yi M, Madden VJ, Welsch C, Antes I, Wen Y, Chugh PE, McGee CE, et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med. 2014;20:927–935. doi: 10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita N, Sugimoto R, Ma N, Tanaka H, Iwasa M, Kobayashi Y, Kawanishi S, Watanabe S, Kaito M, Takei Y. Comparison of hepatic oxidative DNA damage in patients with chronic hepatitis B and C. J Viral Hepat. 2008;15:498–507. doi: 10.1111/j.1365-2893.2008.00972.x. [DOI] [PubMed] [Google Scholar]
- 14.Shimoda R, Nagashima M, Sakamoto M, Yamaguchi N, Hirohashi S, Yokota J, Kasai H. Increased formation of oxidative DNA damage, 8-hydroxydeoxyguanosine, in human livers with chronic hepatitis. Cancer Res. 1994;54:3171–3172. [PubMed] [Google Scholar]
- 15.Tanaka H, Fujita N, Sugimoto R, Urawa N, Horiike S, Kobayashi Y, Iwasa M, Ma N, Kawanishi S, Watanabe S, et al. Hepatic oxidative DNA damage is associated with increased risk for hepatocellular carcinoma in chronic hepatitis C. Br J Cancer. 2008;98:580–586. doi: 10.1038/sj.bjc.6604204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pathologic diagnosis of early hepatocellular carcinoma: a report of the international consensus group for hepatocellular neoplasia. Hepatology. 2009;49:658–664. doi: 10.1002/hep.22709. [DOI] [PubMed] [Google Scholar]
- 17.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 18.Colombo M. Natural history and pathogenesis of hepatitis C virus related hepatocellular carcinoma. J Hepatol. 1999;31(Suppl 1):25–30. doi: 10.1016/s0168-8278(99)80370-5. [DOI] [PubMed] [Google Scholar]
- 19.Weber A, Boger R, Vick B, Urbanik T, Haybaeck J, Zoller S, Teufel A, Krammer PH, Opferman JT, Galle PR, et al. Hepatocyte-specific deletion of the antiapoptotic protein myeloid cell leukemia-1 triggers proliferation and hepatocarcinogenesis in mice. Hepatology. 2010;51:1226–1236. doi: 10.1002/hep.23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu W, Wang X, Leibowitz B, Yang W, Zhang L, Yu J. PUMA-mediated apoptosis drives chemical hepatocarcinogenesis in mice. Hepatology. 2011;54:1249–1258. doi: 10.1002/hep.24516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: origin and consequences. Annu Rev Pathol. 2010;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, Vargas G, Lemon SM. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 23.Kandathil AJ, Graw F, Quinn J, Hwang HS, Torbenson M, Perelson AS, Ray SC, Thomas DL, Ribeiro RM, Balagopal A. Use of laser capture microdissection to map hepatitis C virus-positive hepatocytes in human liver. Gastroenterology. 2013;145:1404–1413. doi: 10.1053/j.gastro.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 25.Gong G, Waris G, Tanveer R, Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc Natl Acad Sci U S A. 2001;98:9599–9604. doi: 10.1073/pnas.171311298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Higgs MR, Lerat H, Pawlotsky JM. Hepatitis C virus-induced activation of beta-catenin promotes c-Myc expression and a cascade of pro-carcinogenetic events. Oncogene. 2013;32:4683–4693. doi: 10.1038/onc.2012.484. [Activation of β-catenin in transgenic mice expressing the entire HCV polyprotein suggests that this cellular signaling pathway may be directly modulated by viral proteins.] [DOI] [PubMed] [Google Scholar]
- 27**.Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, Weinman SA. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481–37488. doi: 10.1074/jbc.M506412200. [Direct induction of ROS by HCV core protein contributes to oxidative stress and potentially DNA damage in the HCV-infected liver. (See also reference 28).] [DOI] [PubMed] [Google Scholar]
- 28.Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 29.Wang AG, Lee DS, Moon HB, Kim JM, Cho KH, Choi SH, Ha HL, Han YH, Kim DG, Hwang SB, et al. Non-structural 5A protein of hepatitis C virus induces a range of liver pathology in transgenic mice. J Pathol. 2009;219:253–262. doi: 10.1002/path.2592. [DOI] [PubMed] [Google Scholar]
- 30.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 31.Ariumi Y, Kuroki M, Dansako H, Abe K, Ikeda M, Wakita T, Kato N. The DNA damage sensors ataxia-telangiectasia mutated kinase and checkpoint kinase 2 are required for hepatitis C virus RNA replication. J Virol. 2008;82:9639–9646. doi: 10.1128/JVI.00351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai CK, Jeng KS, Machida K, Cheng YS, Lai MM. Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology. 2008;370:295–309. doi: 10.1016/j.virol.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 33.Machida K, McNamara G, Cheng KT, Huang J, Wang CH, Comai L, Ou JH, Lai MM. Hepatitis C virus inhibits DNA damage repair through reactive oxygen and nitrogen species and by interfering with the ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and hepatocytes. J Immunol. 2010;185:6985–6998. doi: 10.4049/jimmunol.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duong FH, Christen V, Lin S, Heim MH. Hepatitis C virus-induced up-regulation of protein phosphatase 2A inhibits histone modification and DNA damage repair. Hepatology. 2010;51:741–751. doi: 10.1002/hep.23388. [DOI] [PubMed] [Google Scholar]
- 35.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 39.McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene. 2011;30:1969–1983. doi: 10.1038/onc.2010.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bressac B, Galvin KM, Liang TJ, Isselbacher KJ, Wands JR, Ozturk M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990;87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu IC, Tokiwa T, Bennett W, Metcalf RA, Welsh JA, Sun T, Harris CC. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis. 1993;14:987–992. doi: 10.1093/carcin/14.5.987. [DOI] [PubMed] [Google Scholar]
- 42.Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 44.McGivern DR, Villanueva RA, Chinnaswamy S, Kao CC, Lemon SM. Impaired replication of hepatitis C virus containing mutations in a conserved NS5B retinoblastoma protein-binding motif. J Virol. 2009;83:7422–7433. doi: 10.1128/JVI.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45**.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, Lemon SM. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–1347. doi: 10.1371/journal.ppat.0030139. [HCV NS5B protein targets the Rb tumor suppressor protein for proteasomal degradation in infected cell cultures, suggesting that disruption of the Rb pathway may be a mechanism by which HCV directly increases hepatocellular proliferation and promotes cancer. (see also reference 46).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:18159–18164. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutcheson J, Bourgo RJ, Balaji U, Ertel A, Witkiewicz AK, Knudsen ES. Retinoblastoma protein potentiates the innate immune response in hepatocytes: significance for hepatocellular carcinoma. Hepatology. 2014;60:1231–1240. doi: 10.1002/hep.27217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kannan RP, Hensley LL, Evers LE, Lemon SM, McGivern DR. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J Virol. 2011;85:7989–8001. doi: 10.1128/JVI.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, Katze MG. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 2009;5:e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marshall A, Rushbrook S, Davies SE, Morris LS, Scott IS, Vowler SL, Coleman N, Alexander G. Relation between hepatocyte G1 arrest, impaired hepatic regeneration, and fibrosis in chronic hepatitis C virus infection. Gastroenterology. 2005;128:33–42. doi: 10.1053/j.gastro.2004.09.076. [DOI] [PubMed] [Google Scholar]
- 51.Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 52.Liu J, Ding X, Tang J, Cao Y, Hu P, Zhou F, Shan X, Cai X, Chen Q, Ling N, et al. Enhancement of canonical Wnt/beta-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS One. 2011;6:e27496. doi: 10.1371/journal.pone.0027496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, Jung G, Choi KY, Hwang SB. Nonstructural 5A protein activates beta-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J Hepatol. 2009;51:853–864. doi: 10.1016/j.jhep.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 54.Street A, Macdonald A, Crowder K, Harris M. The Hepatitis C virus NS5A protein activates a phosphoinositide 3-kinase-dependent survival signaling cascade. J Biol Chem. 2004;279:12232–12241. doi: 10.1074/jbc.M312245200. [DOI] [PubMed] [Google Scholar]
- 55.Street A, Macdonald A, McCormick C, Harris M. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. J Virol. 2005;79:5006–5016. doi: 10.1128/JVI.79.8.5006-5016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Best SM. Viral subversion of apoptotic enzymes: escape from death row. Annu Rev Microbiol. 2008;62:171–192. doi: 10.1146/annurev.micro.62.081307.163009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fuentes-Gonzalez AM, Contreras-Paredes A, Manzo-Merino J, Lizano M. The modulation of apoptosis by oncogenic viruses. Virol J. 2013;10:182. doi: 10.1186/1743-422X-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng L, Adachi T, Kitayama K, Bungyoku Y, Kitazawa S, Ishido S, Shoji I, Hotta H. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J Virol. 2008;82:10375–10385. doi: 10.1128/JVI.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lan L, Gorke S, Rau SJ, Zeisel MB, Hildt E, Himmelsbach K, Carvajal-Yepes M, Huber R, Wakita T, Schmitt-Graeff A, et al. Hepatitis C virus infection sensitizes human hepatocytes to TRAIL-induced apoptosis in a caspase 9-dependent manner. J Immunol. 2008;181:4926–4935. doi: 10.4049/jimmunol.181.7.4926. [DOI] [PubMed] [Google Scholar]
- 60.Mateu G, Donis RO, Wakita T, Bukh J, Grakoui A. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology. 2008;376:397–407. doi: 10.1016/j.virol.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yi M, Hu F, Joyce M, Saxena V, Welsch C, Chavez D, Guerra B, Yamane D, Veselenak R, Pyles R, et al. Evolution of a cell culture-derived genotype 1a hepatitis C virus (H77S.2) during persistent infection with chronic hepatitis in a chimpanzee. J Virol. 2014;88:3678–3694. doi: 10.1128/JVI.03540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saeed M, Shiina M, Date T, Akazawa D, Watanabe N, Murayama A, Suzuki T, Watanabe H, Hiraga N, Imamura M, et al. In vivo adaptation of hepatitis C virus in chimpanzees for efficient virus production and evasion of apoptosis. Hepatology. 2011;54:425–433. doi: 10.1002/hep.24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cleary SP, Jeck WR, Zhao X, Kuichen, Selitsky SR, Savich GL, Tan TX, Wu MC, Getz G, Lawrence MS, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 2013;58:1693–702. doi: 10.1002/hep.26540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Nault JC, Calderaro J, Di Tommaso L, Balabaud C, Zafrani ES, Bioulac-Sage P, Roncalli M, Zucman-Rossi J. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology. 2014;60:1983–1992. doi: 10.1002/hep.27372. [The identification of TERT promoter mutations as an early genetic change in abnormal but pre-neoplastic tissue suggests a genetic basis for increased cancer risk in patients with cirrhosis. (Also see reference 66).] [DOI] [PubMed] [Google Scholar]
- 66.Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, Laurent A, Cherqui D, Balabaud C, Zucman-Rossi J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alison MR. Liver stem cells: implications for hepatocarcinogenesis. Stem Cell Rev. 2005;1:253–260. doi: 10.1385/SCR:1:3:253. [DOI] [PubMed] [Google Scholar]
- 68.Lemon SM, McGivern DR. Is hepatitis C virus carcinogenic? Gastroenterology. 2012;142:1274–1278. doi: 10.1053/j.gastro.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spaniel C, Honda M, Selitsky SR, Yamane D, Shimakami T, Kaneko S, Lanford RE, Lemon SM. microRNA-122 abundance in hepatocellular carcinoma and non-tumor liver tissue from Japanese patients with persistent HCV versus HBV infection. PLoS One. 2013;8:e76867. doi: 10.1371/journal.pone.0076867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freeman AJ, Dore GJ, Law MG, Thorpe M, Von Overbeck J, Lloyd AR, Marinos G, Kaldor JM. Estimating progression to cirrhosis in chronic hepatitis C virus infection. Hepatology. 2001;34:809–816. doi: 10.1053/jhep.2001.27831. [DOI] [PubMed] [Google Scholar]
- 71.Seeff LB. The history of the “natural history” of hepatitis C (1968-2009). Liver Int. 2009;29(Suppl 1):89–99. doi: 10.1111/j.1478-3231.2008.01927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. e1261. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73**.Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, Hosono N, Otsuka M, Tateishi R, Omata M, Nakagawa H, et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet. 2011;43:455–458. doi: 10.1038/ng.809. [GWAS studies may identify host genetic variants that differ between chronic hepatitis C patients who either progress or do not progress to HCC. Although a potentially powerful technique, identification of patients at higher risk remains elusive. (Also see references 74 and 75).] [DOI] [PubMed] [Google Scholar]
- 74.Miki D, Ochi H, Hayes CN, Abe H, Yoshima T, Aikata H, Ikeda K, Kumada H, Toyota J, Morizono T, et al. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet. 2011;43:797–800. doi: 10.1038/ng.876. [DOI] [PubMed] [Google Scholar]
- 75.Lange CM, Bibert S, Dufour JF, Cellerai C, Cerny A, Heim MH, Kaiser L, Malinverni R, Mullhaupt B, Negro F, et al. Comparative genetic analyses point to HCP5 as susceptibility locus for HCV-associated hepatocellular carcinoma. J Hepatol. 2013;59:504–509. doi: 10.1016/j.jhep.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 76.Raimondi S, Bruno S, Mondelli MU, Maisonneuve P. Hepatitis C virus genotype 1b as a risk factor for hepatocellular carcinoma development: a meta-analysis. J Hepatol. 2009;50:1142–1154. doi: 10.1016/j.jhep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 77.Kanwal F, Kramer JR, Ilyas J, Duan Z, El-Serag HB. HCV genotype 3 is associated with an increased risk of cirrhosis and hepatocellular cancer in a national sample of U.S. Veterans with HCV. Hepatology. 2014;60:98–105. doi: 10.1002/hep.27095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kohli A, Shaffer A, Sherman A, Kottilil S. Treatment of hepatitis C: a systematic review. Jama. 2014;312:631–640. doi: 10.1001/jama.2014.7085. [DOI] [PubMed] [Google Scholar]
- 79.Kwon H, Lok AS. Does antiviral therapy prevent hepatocellular carcinoma? Antivir Ther. 2011;16:787–795. doi: 10.3851/IMP1895. [DOI] [PubMed] [Google Scholar]
- 80.Lok AS, Everhart JE, Wright EC, Di Bisceglie AM, Kim HY, Sterling RK, Everson GT, Lindsay KL, Lee WM, Bonkovsky HL, et al. Maintenance peginterferon therapy and other factors associated with hepatocellular carcinoma in patients with advanced hepatitis C. Gastroenterology. 2011;140:840–849. doi: 10.1053/j.gastro.2010.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]