Abstract
Interaction between the Notch receptor and Delta-Serrate-Lag2 (DSL) ligands is generally deemed to be the starting point of the Notch signaling cascade, after which, Notch is cleaved and the intracellular domain acts as a transcriptional co-activator. By contrast, Notch protein can become activated independent of ligand stimulus through recently identified endosomal trafficking routes as well as through aberrant regulation of Notch components during Notch trafficking, ubiquitination, and degradation. In this review, we summarize genes implicated in ligand-independent Notch activity and remark on the mechanisms by which this process could occur.
Keywords: Developmental biology, Notch signaling, non-canonical cell signaling, Drosophila
The many ways to activate Notch signaling
The Notch pathway has emerged as one of the major signaling cascades activated throughout development and its misregulation has been associated with many diseases. Canonical Notch signaling begins upon DSL ligand (Delta (Dl) or Serrate (Ser) in Drosophila; Delta or Jagged orthologues in mammals) binding to the extracellular domain of Notch (NECD), allowing subsequent proteolytic cleavage of the Notch receptor (reviewed in [1]). This cleavage releases the intracellular fragment of the Notch receptor (NICD), which can translocate to the nucleus and form a complex with a CSL transcription factor (CBF-1 in mammals, Suppressor of Hairless in Drosophila, and Lag-2 in Caenorhabditis), resulting in expression of downstream target loci [2–5]. However, recent work has shown that the Notch pathway can be utilized noncanonically, including signaling independent of CSL transcription factors through the Wnt pathway (reviewed in [6]) or in a DSL ligand-independent manner. This ligand-independent activation of the Notch receptor is primarily caused by the disruption of genes that control endosomal sorting and ubiquitination, resulting in accidental and often detrimental pathway activity (reviewed in [7,8]). Conversely, mechanisms are emerging by which ligand-independent Notch activity is controlled endogenously.
When Notch is not required to signal in a cell, the receptor must be tightly regulated in order to make certain no aberrant signal is produced. Furthermore, there is a constant turnover of Notch in the cell, as pulse-chase experiments show the disappearance of labeled Notch within hours [e.g. 9]. Notch is translated, processed into its heterodimer form, and transported to the membrane, where it awaits ligand presentation from a neighboring cell. If ligand is not presented, Notch will be marked for degradation. This occurs by the addition of a monoubiquitin signal to the Notch intracellular domain by the E3 ubiquitin ligase, Deltex (Dx), spurring its internalization [10]. Another E3 ubiquitin ligase, Kurtz (Krz) has been shown to complex with Dx and Notch, promoting polyubiquitination of the receptor [11, 12]. The Endosomal Complex Required for Transport (ESCRT) is able to recognize polyubiqutinated proteins, and has been implicated in the regulation of many membrane bound receptors [13, 14]. There are four distinct ESCRT complexes, which work sequentially to sort Notch into intraluminal vesicles (ILVs) of the multivesicular body (MVB). Eventually, the cargo in the lumen of the MVB is transferred to the lumen of the lysosome for degradation. If malfunctions in this process leave Notch ensnared on the outer endosome membrane, then NICD can be accidentally removed from NECD, mimicking ligand-dependent Notch activation. This model is primarily supported by genetic loss- and gain-of-function studies in Drosophila, although some genes have been characterised in zebrafish as well. Among the genes identified are E3 ligases, endosomal sorting proteins, metalloproteases, basal body proteins (in zebrafish) [15], small GTPases [16,17], hif1-alpha, and Notch’s own ligands (Table 1). Note that some of these genes are also involved in ligand-dependent Notch signaling. Most of these factors have been implicated in endosomal regulation [18–22] and appear to act in the same overarching process. For example, the ESCRT-III component, Shrub, serves as a link between the ligand-independent Notch activation observed in ESCRT complex mutants and that witnessed in both lethal giant discs (lgd) mutants and dx-expressing cells [7, 11, 17].
Table 1.
Genes implicated in positive or negative regulation of ligand-independent Notch activity.
| Gene(s) | Function | Regulation | Mechanism | Refs |
|---|---|---|---|---|
| Lethal(2) Giant Discs (lgd) | Phospholipid binding | Negative | Trafficking defect in lgd mutant cells where ubiquitinated Notch accumulates on lysosomal limiting membrane | 41–43, 81, 82 |
| Deltex (Dx) | E3 ligase | Positive | Promotoesmonoubiquitination, endocytosis, and stabilization of Notch on lysosomal limiting membrane | 23–28, 35 |
| Su(Dx) and dNedd4 | E3 ligase | Both | Promotes internalization of Notch, resulting in either inhibition (via poly-ubiquitination) or promotion (via Dx-mediated route) of ligand-independent Notch activity depending on temperature. | 30–35 |
| ESCRT I-III complexes | Sorting of ubiquitinated proteins | Negative | MVB biogenesis and sorting of ubiqutinated proteins is disrupted in ESCRT mutants (e.g. Vps25, tsg101, shrub) leading to high levels of Notch accumulating in malformed endosomes. | 7, 9–11, 44–49, 52 |
| Kuz and Tace | ADAM proteases | Positive | Removes Notch ectodomain, leaving as a viable substrate for presenlin-mediated S3 cleavage. | 64, 71 |
| Ras-like Protein A | Small GTPase | Negative | Mechanism unclear, although has been implicated in exocysts and endocytosis. | 16–19 |
| BBS1/BBS4 | BBSome components | Negative | Shifts subcellular location of Notch from membrane, recycling endosome, and lysosome towards the MVB and late endosome. | 15 |
| Hif1-alpha | Hypoxic stress response | Positive | Stabilizes Notch in hrs positive endosomes | 22 |
| Cis-DSL ligands | Notch ligands | Negative | Ligands in same cell as receptor buffer against accidental activation. | 37 |
While the endogenous regulation of Notch requires more investigation, a clearer picture is emerging of the mechanism of ligand-independent activation that occurs in mutant tissue. In this review, we address the role and mechanisms of endogenous ligand-independent Notch activation in development. We focus on studies in Drosophila as the fruit fly has the least number of Notch ligands of any model organism and therefore serves as an ideal model for studying ligand-independent Notch activity.
An endogenous buffering system for ligand-independent Notch activity
Notch protein is under a constant state of turnover, characterized by simultaneous production and degradation of Notch. As a result, some Notch may become “accidentally” activated en route to lysosomes, and development must buffer against this activation, use it to its advantage, or risk phenotypic consequences. It seems that a dynamic interplay between trafficking, ubiquitination, temperature, and ligand concentration aids in the formation of these buffers (Figure 1).
Figure 1.
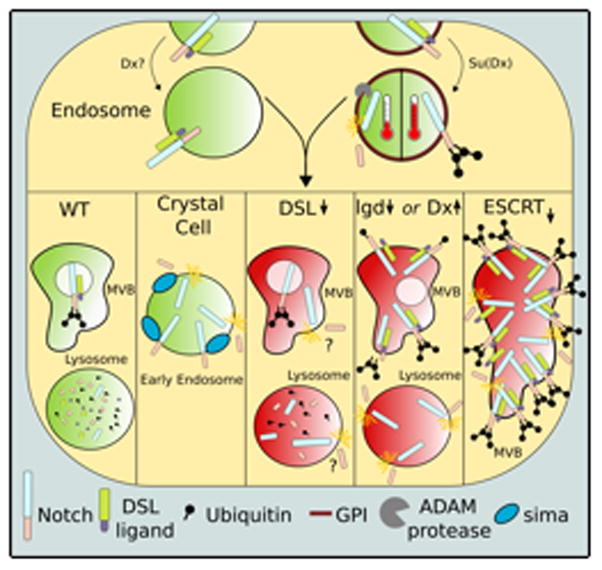
The flow of Notch through endosomes endogenously and in mutant cells.
In wild-type cells (marked as green endosomes), Notch can be internalized into either the GPI positive or negative endosomal routes and will be activated or degraded depending on the presence of E3 ligases and temperature. How these routes integrate into the other illustrated scenarios of ligand-independent activity is unknown. In crystal cells, sima-promoted ligand independent Notch activation occurs in Hrs-positive early endosomes. Ligand-independent Notch activity occurs in many mutant tissues as well (marked by red endosomes). Upon loss of DSL ligands, Notch will become activated, however the specific endosomal compartment where this activation occurs is unknown. In lgd mutants and Dx-overexpressing cells, Notch will remain on the lysosomal limiting membrane where activation occurs. In ESCRT mutants, Notch will gather in large irregular endosomes where it will be activated.
Specifically, the E3 ubiquitin ligases, Dx and Suppressor of Deltex (Su(dx)), and the Notch ligands, Dl and Ser, in Drosophila play integral roles in regulating this process. Dx is a RING domain containing E3 ubiquitin ligase that promotes Notch activation by interacting with the ankyrin repeats of the Notch intracellular domain, facilitating its monoubiquitination, and helping it evade destruction through the trafficking pathway [23–28]. Overexpression of dx causes activation of Notch downstream genes and reporters in the Drosophila wing imaginal disc, as well as loss of Notch from the adherens junction, indicating that Dx actively promotes Notch internalization leading to its activation [27,28]. Localization of Notch to the late endosomal limiting membrane is required for Dx-dependent Notch activation, as blocking Rab5, Rab7, HOPS complex, or AP-3 complex components attenuates Notch signaling in dx-expressing cells [27,28] (Figure 1). This requirement for the localization of Notch on the late endosomal limiting membrane is also observed in Notch activation occurring in lgd mutants (Table 1, Figure 1), and localization to the limiting membrane is directed by a conserved dileucine motif in mammals [29]. In addition, the E3 ubiquitin protein ligase family of Nedd4 proteins, dNedd4 and Su(dx), antagonize Notch signaling through ubiquitination [30–34].
A recent study revealed that Dx and Su(dx) can induce the internalization of Notch into two different endosomal compartments to promote ligand-independent Notch activity [35]. Su(dx) promotes Notch endocytosis through a glycophosphatidylinositol (GPI)-positive, sterol-dependent endosomal route, whereas Dx promotes Notch internalization through a GPI-negative, sterol-independent route [35] (Figure 1). Su(dx) has traditionally been considered a negative regulator of Notch signaling, but at low temperatures, Su(dx) can promote Notch signaling [35]. Indeed, the HECT domain acts in a temperature-dependent manner, and Su(dx)-induced Notch internalization is independent of the HECT domain. Therefore, at low temperatures, both routes (i.e. Dx- or Su(dx)-dependent) lead to Notch activation in different endosomal compartments. However, at moderate temperatures, Su(dx) acts as a negative regulator of Notch signaling by inducing Notch degradation via the HECT domain, and therefore Dx and Su(dx) are in competition for Notch to enter their respective endosomal route. This competition results in canalization of Notch signaling against temperature fluctuations, because Notch is internalized at an increased rate at higher temperatures. Furthermore, this competition may serve to either increase or decrease ligand-independent Notch signaling depending on temperature, which could account for the temperature sensitivity inherent in genetic interactions between Notch and its ligands [35]. Consistently, the embryonic defects observed in HOPS, AP-3, and dx null embryos are temperature-sensitive, and their phenotypes worsen at higher temperatures [28].
Ligand-independent Notch activity may also be buffered against by DSL ligands (Figure 1). DSL ligand-expressing cells adjacent to Notch-expressing cells (trans-ligand) activate Notch signaling, while DSL ligands present in the same cell as the Notch receptor (cis-ligand) repress ligand-dependent Notch signaling, which is referred to as cis-inhibition [reviewed in 36]. cis-ligands also endogenously repress the accidental activation of ligand-independent Notch [37]. Upon removal of both cis- and trans-ligands, Notch is activated cell-autonomously in both the ovarian follicle cells and the imaginal discs in Drosophila [37]. Furthermore, Notch expressed alone in S2 cells results in ligand-independent Notch activity [35,37], but co-expression of a form of Ser that only has cis-inhibitory potential [38] almost completely blocks this activation, indicating that cis-ligands can act as an efficient buffer against ligand-independent Notch activity. Also, increased cis-ligand expression can be used to decrease the ligand-independent signaling found in various mutant tissues including lgd, shrub, and dx, as well as that endogenously occurring in crystal cells [37]. The mechanism by which DSL ligands offer protection against ligand-independent Notch activation has yet to be explored, although they possibly exert their function through the stabilization of the Notch heterodimer, increased receptor degradation, or through binding competition for activating factors such as Dx or Su(dx). How the buffering activity of E3 ligases and cis-ligands may interact is also unknown, and the predicted signaling output (NICD production) could vary with the degree of dominance or synergism between the two systems. Another important point is that these two buffering systems are buffering against different phenomena – whereby the E3 ligases hypothetically ensure that a sufficient amount of Notch is activated in times of directed trans-activation, while the cis-ligand system ensures that Notch remains inactive during periods without trans-activation. Modeling suggests that maintaining translated Notch can keep the pathway in a state of readiness [39], and it is plausible that the cis-ligand buffering system aids in strict temporal regulation of Notch-dependent developmental events.
The aptitude for ligand-independent activation of Notch in endosomes can also be exploited by developing cells. The first case of ligand independent Notch activation occurring endogenously during development was described in Drosophila crystal cells, which are of the hematopoetic lineage and involved in the immune response [22,40]. In crystal cells, the Notch pathway is coopted to promote cell survival, and silencing Notch with RNAi in crystal cells leads to cell bursting and a decrease in crystal cell number [22]. Furthermore, reduction of Drosophila Hif1-alpha homologue, similar (sima), from crystal cells causes an increase in cell bursting and a decrease in Notch reporter activity, indicating Sima promotes Notch activation [22]. This Notch activation is endosomal in nature because live trafficking assays show Notch is stabilized in Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hrs)-positive endosomes specifically in the crystal cells, and overexpression of Rab5 decreased the ability of Notch to signal [22]. Inhibition of Dl or Ser by RNAi after crystal cell fate determination has no effect on cell survival. Similarly the ubiquitin ligases Mindbomb and Neuralized, which are required for ligand-dependent signaling, have no role in mature crystal cell survival [22]. Together, these data indicate that crystal cells undergo ligand-independent Notch activation by Sima-dependent endosomal stabilization. Overexpression of cis-ligand was found to lessen this ligand-independent activation [37]; however, it remains to be investigated if this is utilized as a developmental or immunological strategy. A tempting explanation for the usefulness of a ligand-independent activation mechanism in crystal cells is that as circulating blood cells, there is no guaranteed ligand source, so the ability to regulate the Notch signaling pathway cell-autonomously would be advantageous.
Together, these studies indicate that E3 ligases, temperature, specific trafficking compartments, the presence of DSL ligand, and endosomal half-life all have an effect on buffering for or against endogenous ligand-independent Notch activity.
An emerging picture of ligand independent Notch activation
Notch localization on endosomes
Although ligand-independent activity occurs endogenously, most of our knowledge about its regulation and mechanism comes from studying the behavior of Notch in mutant tissue. This insight is especially important in understanding how misregulation of Notch trafficking could relate to human disease, but also provides hints as to how Notch ligand-independent regulation occurs naturally. As mechanisms have been described for the lgd, dx, Su(dx)/dNedd4, ESCRT, and sima mutant tissues, we focus on these (see Figure 1 for illustrative summary). Interestingly, each case may not have the same trafficking requirements, as the only shared similarity is prolonged endosomal half-life, whether it be through increased stability in endosomes, as promoted by Sima and Dx (and Su(dx) at lower temperatures), or degradation malfunctions, such as in mutants for lgd, Su(dx)/dNedd4, or ESCRT components. The endosomal compartments wherein Notch becomes activated may also differ between mutant tissues, and could imply different mechanisms.
It is apparent that in the case of lgd mutant tissue and dx over-expressing tissue, Notch must be transported to the lysosomal limiting membrane in order for activation to occur, and in these cells Notch can be found colocalized with late endosomal and lysosomal markers [27,28,35,41–43]. However, in crystal cells or ESCRT mutant cells, Notch seems to accumulate in an earlier stage of the trafficking pathway, and mainly colocalizes with Hrs [22, 44]. Furthermore, in the Su(dx)-mediated, sterol-dependent endosomal route to activation (or inactivation, depending on temperature), Notch is trafficked through GPI-positive endosomes to an unknown endosomal compartment [35]. We propose two explanations that are not entirely mutually exclusive, for the activation of Notch in different endosomal compartments. First, the three known locations of Notch activation have different mechanisms or factors that promote NICD formation. In lgd and dx tissues, a lysosome-specific environmental factor produces the membrane tethered Notch fragment. In ESCRT mutant tissue, Notch does not reach the lysosome, but becomes trapped in high quantities in large irregular endosomes [44–49]. Therefore, it could be imagined that a high quantity of closely contained membrane proteins might alter intra-protein kinetics resulting in auto-activation. As unbound Notch also has a steady-state level of endogenous ligand-independent activation [37], more time and receptors could simply mean a higher probability of reaching the threshold of NICD production required for downstream transcriptional activation. The second possibility is that in ESCRT mutant cells, the endosomal compartment in which it is trapped becomes lysosome-like. In ESCRT mutants, the enlarged endosomes that trap Notch do not fuse with lysosomes [50], whereas the factors leading to endosomal acidification may still be present. If these factors contribute to Notch auto-activation on the lysosomal limiting membrane, then the same mechanism might facilitate NICD production in different stages of endosomal trafficking. However, there is no colocalization of lysotracker with late endosomal markers upon expression of a dominant negative form of ESCRT-III component vps4 [51], arguing that this hypothetical shared component between the two compartments would not be pH-dependent. Further, this would not explain Notch activation in crystal cells, unless their trafficking steps are temporally altered. Alternatively, keeping high early endosomal levels of Notch in crystal cells might redirect the flow of a significant portion of the receptor, causing more to be directed to the lysosomal limiting membrane (or through the sterol-dependent trafficking route), as the effect of late trafficking steps on crystal-cell Notch activation have yet to be explored.
Either explanation could be extended to Notch activation that is dependent on Su(dx)-mediated endocytosis and cholesterol, as these Notch-containing endosomes may have proteins or environmental factors that overlap with those of the lysosome. It is also possible that Notch is present on the endosomal limiting membrane at an unusually high concentration. However, given the different requirements for S2 cleavage between Dx- and Su(dx)-mediated endosomal routes, it is likely that the mechanism of Su(dx)-induced Notch activation differs drastically from the Dx-dependent route. It is worth noting that the basal level of ligand-independent Notch activity that occurs when cis-ligand is absent seems to mainly use the Su(dx)-mediated activation route in S2 cells [35]. Whether this is the case in vivo upon relief of cis-inhibition has yet to be explored.
Although these above situations connect accumulated Notch with resultant activation, this is not always the case. Accumulation at the cell surface does not promote ligand-independent Notch activation as shown by shibire (Drosophila dynamin), Rab5, or avalanche deficient cells [9]. Mutants that accumulate Notch after ILV formation do not exhibit aberrant Notch signaling, such as seen in mutants for the lysosomal component, fab1 [9]. Although all ESCRT-II component mutants show high levels of ubiquitinated Notch accumulation and enlarged endosomes, only vps22 and vps25 mutants show Notch activation [52]. The presence of mutant-specific activation implies there may be a hierarchy to the importance of ESCRT-II component involvement in Notch regulation, or that these subunits may have alternative functions. Perhaps most interestingly, ESCRT-0 mutants accumulate Notch similarly to other ESCRT mutants, except without Notch activation [9,53,54]. Therefore, endosome-ensnared Notch alone is insufficient to promote ligand-independent Notch activity. Since the ESCRT-0 complex functions to cluster ubiquitinated cargo on clathrin-rich areas of the endosomes and subsequently recruit the other ESCRT complexes, this dense grouping of Notch receptors may be required for ligand-independent Notch activity [54]. This observation would be consistent with the mechanism of ligand-independent Notch activation, whereby a high concentration of endosomal Notch alters protein kinetics, thus resulting in accidental NICD release. A possible prediction arising from this hypothesis would be the requirement of Notch clustering with clathrin in crystal cell endosomes and a requirement for ESCRT-0 in that process. Alternatively, studies have identified other genes that might function in a similar manner as ESCRT-0 [e.g. 55, 56], indicating there may be other routes to degradation in ESCRT-0 mutants. Therefore, ESCRT-0 would not act as a complete block in degradation as seen with the other ESCRT mutants, rather as a “traffic jam” in earlier endosomal compartments. Notably, hrs lgd double mutant cells do not show ectopic Notch signaling and Dx-mediated Notch signaling requires Hrs [41, 57], which is consistent with a model where specifically ESCRT-0 function (i.e. gathering Notch on clathrin coats) is required for ligand-independent signal production. Therefore, in a variety of mutant tissues, Notch held in endosomal stages after the early endosome may be particularly susceptible to ligand-independent activation.
Imitation of Notch cleavage events
Notch undergoes several cleavage events en route to ligand-dependent activation. S1 cleavage occurs before transportation to the cell membrane and cleaves full length Notch, which is subsequently reassembled into a heterodimer consisting of an extracellular domain and a transmembrane fragment bound by noncovalent interactions [58, 59]. Upon ligand binding, the next two cleavage events, S2 and S3, create the NICD fragment, with S2 cleavage as the major rate-limiting step in this process and S3 cleavage occurring seemingly constitutively [60–64]. Severing of Notch at the S2 site is performed by an ADAM metalloprotease, primarily Kuzbanian (Kuz) in Drosophila and TNF-alpha converting enzyme (TACE) in mammals [63, 64]. S2 cleavage is inhibited by a LIN-12-Notch (LNR) repeat containing negative regulatory region (NRR) of the Notch protein located N-terminal to the sites of S2 and S1 cleavage, which folds around and blocks access of ADAM to the S2 cleavage site [65]. Ligand binding and endocytosis exerts a pulling force on the Notch receptor, causing a conformational change in the NRR region and leaving the S2 site unprotected [66–68]. After the extracellular domain is removed, the transmembrane fragment becomes a suitable substrate for the presenilin-containing gamma-secretase complex, which performs S3 cleavage C-terminal to the transmembrane domain, releasing NICD [61]. As S2 cleavage is directly dependent on ligand stimulus, the entire process must be emulated by a different mechanism in ligand-independent Notch activation.
An endosomal factor could induce ectodomain shedding either by inducing a conformational change in the NRR region or facilitating ectodomain removal itself (Figure 2). In the former, NICD production would still be dependent on a protease for S2 cleavage. Although kuz is required in the Su(dx)-mediated route to Notch activation [35], it is not required In lgd mutant cells and in dx-overexpressing cells for the observed ligand-independent Notch activity [35, 43]. However, there are five ADAM metalloproteases including kuz in Drosophila, many of which are poorly characterized, and there may be a co-option of one or many of these during ligand-independent Notch activation. ADAM co-option during ligand-independent Notch1 activity seems to occur in mammals, as ADAM17 is not required for ligand-dependent signaling but can promote S2 cleavage independent of ligand [69]. Interestingly, the co-option of ADAM17 varies between species and Notch paralogues, exemplified by its ability to process Notch1 and Notch2 in mice, but only Notch1 in humans [70]. Also consistent with this idea, overexpression of dTACE is a viable substitute for S2 cleavage in Drosophila cultured cells [71]. Furthermore, S2 cleavage of mammalian Notch1 still occurs upon treatment with metalloprotease inhibitors, indicating that other families of proteases may be able to cleave Notch [72]. Therefore, it is possible that each ADAM protease has different trafficking preferences, which may be reflected as different requirements specifically for Kuz. It is also possible that complete auto-dissociation at the S1 site occurs in endosomes (Figure 2). This would create a Notch isoform that requires an ADAM protease for maximal activity but may still be a viable target for S3 cleavage directly, as metalloprotease inhibitors against the dissociated Notch1 heterodimer transmembrane segment only partially reduce signaling [73]. Similar mechanisms could explain both NRR unfolding or complete heterodimer dissociation. Calcium chelation, such as that caused by EDTA, can dissociate and activate Notch in an ADAM dependent manner, as each of the LNR repeats uses a calcium ion to form a “calcium bridge”, thus bringing stability to the NRR region [74]. Calcium efflux from the vesicle lumen into the cytosol is required for the fusion between the endosome and lysosome [75; 76], indicating that Notch may be unstable during some trafficking stages due to the reduction of luminal calcium levels. As the integrity of the NRR is maintained through a complex web of hydrogen bonding, an endosomal factor disrupting these bonds might also be to blame [77, 78]. Another explanation might be the complete degradation of the extracellular domain and the intraluminal portion of the transmembrane domain by the lysosomal acid hydrolases (Figure 2). In this case, the S2 cleavage would not be required, as this portion of the transmembrane domain would be degraded. Consistent with this explanation is the requirement of late endosomal to lysosomal fusion for Dx-mediated Notch activation and in lgd mutant cells, and the requirement of the vacuolar ATPase (vATPase) in lgd and ESCRT mutant cells [28, 43, 79] (Figure 1). However vATPase mutants have previously been shown to affect earlier trafficking steps and inhibit ligand-dependent Notch activation in other tissues [80], and therefore more work must be done in order to distinguish between these possibilities. Finally, Presenilin (Psn) is more active in acidic compartments [81], and therefore another possibility is that ectodomain removal is bypassed completely.
Figure 2.

Hypotheses regarding S2 cleavage emulators.
Simplified schematic of the Notch protein (top left) in a WT endosome. S2 cleavage could be imitated by autodissociation at the S1 site (top right), accidental unfolding of the NRR domain (bottom left), or degradation of the extracellular domain (bottom right).
Ligand-independent Notch activity can also be promoted by overexpression of ADAM proteases [71]. However, this mechanism may require special tissue-specific conditions, as overexpression of Kuz in Drosophila ovarian follicle cells and wing imaginal discs shows no aberrant Notch activation (unpublished observations). These special conditions might include a requirement for both high Kuz and high Notch concentrations in the same cell or reduced cis-inhibition of Notch. Alternatively there could exist specific trafficking requirements, as Kuz is a membrane protein [82] and may also travel through endosomes with Notch.
Although the requirement for S2 cleavage remains unclear, there is evidence that S3 cleavage by Psn and downstream events occurs in a similar manner as in ligand-dependent Notch activity. Psn is required in Dx-induced Notch activation, crystal cell Notch activity, and in mutant clones for lgd or shrub [10, 22, 53, 83, 84]. Furthermore, Su(H) is required in dx overexpressing cells, lgd and shrub mutant cells, crystal cells, and ligand-independent Notch activity observed upon the removal of cis-inhibiting ligands [10, 22, 37, 83]. Therefore, it seems that the mechanistic differences between ligand-independent and ligand-dependent Notch activation remain almost solely dependent on how the ectodomain is removed. No attempt has been made to detect differences in transcriptional responses between ligand-dependent and ligand-independent Notch activation.
Concluding Remarks
Studies from Drosophila tell us that many genes are involved in the negative regulation of aberrant signaling, a handful of genes are involved in the positive regulation of ligand-independent signaling, and that this process plays a role endogenously during development. Based on the evidence discussed, ligand-independent Notch activity seems to occur in different endosomal compartments and with differing mechanisms of ectodomain removal, although how these may correlate is unknown (see Outstanding Questions Box). Although increased endosomal half-life seems to be necessary for ligand-independent Notch activity in mutant tissues thus far, it is not sufficient, implying the existence of unknown factors affecting this process. Whether common factors can explain this observed mechanistic variation, or whether completely distinct processes exist is also still an open question. Future studies focusing on detailed mechanistic differences between, for example, Su(dx)- and Dx-dependent endosomal routes may help in resolving these discrepancies.
Outstanding Questions Box.
How might cis-ligand and the differing endosomal routes to Notch activation relate to one another? These processes both modulate ligand-independent Notch activity; however, whether there is a hierarchical or synergistic relationship between them is unknown.
What is the mechanism of separation between the Notch intracellular and extracellular domain, and is this mechanism the same across different endosomal locations of ligand-independent Notch activity?
Do the transcriptional responses between ligand-independent and ligand-dependent signaling differ in a fundamental way? If so, can this be explained by differences in Notch intracellular domain concentration?
Is ligand-independent Notch activity also prevalent in humans? If so, what role might ligand-independent Notch activity play in disease? Mammals have many different Notch receptors and ligands, and complex relationships and regulation mechanisms could exist that cloud true cases of ligand-independent Notch activity.
The regulation and misregulation of Notch endosomal trafficking and the resulting ligand-independent pathway activation has great importance for human disease. Non-canonical Notch signaling has been observed in cancers including melanoma [85] and T-cell acute lymphoblastic leukaemia [86, 87]. Recently, a study also found that breast cancer stem cell expansion depends on a ligand-independent Notch activation mechanism [88]. Furthermore, many of the ESCRT components have been implicated in cancer [reviewed in 89], although this finding has yet to be functionally linked to ligand-independent Notch activity in vertebrates. To our knowledge, no study has shown that ligand-independent Notch activation plays an endogenous role in mammalian development. However, the multitude of Notch ligands present in mammals hinders our ability to claim complete ligand independence with certainty, and the dearth of ligand-independent Notch involvement in mammalian disease and development could be a reflection of this complication. Therefore, studies in Drosophila may continue to shed light on the intricacies of Notch pathway regulation, and to guide future studies in mammalian systems.
Trends Box.
Notch, a transmembrane receptor and transcriptional co-activator, can be activated independent of ligand through a trafficking-dependent route.
Multiple endosomal routes exist that can lead to ligand-independent Notch activation, which are dependent on temperature and competing E3 ligases.
Notch ligands expressed in the same cell as Notch help to buffer against ligand-independent Notch activity.
Crystal cells utilize ligand-independent Notch activity endogenously for survival.
Defects in trafficking and ubiquitination of Notch cause Notch accumulation and activation, possibly through different mechanisms.
Acknowledgments
We thank G. Calvin and four anonymous reviewers whose comments significantly improved the final manuscript. W.-M. D. is supported by National Science Foundation grant (IOS-1052333) and National Institutes of Health grant (R01 GM072562).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kopan R, Ilagan MXG. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 3.Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 4.Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 5.Kidd S, Lieber T, Young MW. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen P, Uosaki H, Shenje L, Kwon C. Non-Canonical Notch Signaling: Emerging Role and Mechanism. Trends Cell Biol. 2012;22:257–265. doi: 10.1016/j.tcb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Regulation of ligand-independent Notch signal through intracellular trafficking. Commun Integr Biol. 2012;5:374–376. doi: 10.4161/cib.19995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baron M. Endocytic routes to Notch activation. Seminars in Cell & Developmental Biology. 2012;23:437–442. doi: 10.1016/j.semcdb.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol. 2008;180:755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J Cell Biol. 2011;195:1005–1015. doi: 10.1083/jcb.201104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troost T, Jaeckel S, Ohlenhard N, Klein T. The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. J Cell Sci. 2012;125:763–776. doi: 10.1242/jcs.097261. [DOI] [PubMed] [Google Scholar]
- 12.Mukherjee A, et al. Regulation of Notch signalling by non-visual beta-arrestin. Nature Cell Biology. 2005;7(12):1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- 13.Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. The EMBO Journal. 2010;29(6):1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 15.Leitch CC, Lodh S, Prieto-Echagüe V, Badano JL, Zaghloul NA. Basal body proteins regulate Notch signaling via endosomal trafficking. J Cell Sci. 2014 doi: 10.1242/jcs.130344. jcs.130344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho B, Fischer JA. RalGTPase promotes asymmetric Notch activation in the Drosophila eye in response to Frizzled/PCP signaling by repressing ligand-independent receptor activation. Development. 2011;138:1349–1359. doi: 10.1242/dev.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho B, Fischer JA. Ral inhibits ligand-independent Notch signaling in Drosophila. Small GTPases. 2012;3:186–191. doi: 10.4161/sgtp.19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jullien-Flores V, et al. RLIP76, an effector of the GTPaseRal, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. Journal of Cell Science. 2000;113:2837–2844. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 19.Moskalenko S, et al. The exocyst is a Ral effector complex. Nature Cell Biology. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 20.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 21.Westlake CJ, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. PNAS. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction Between Notch and Hif-α in Development and Survival of Drosophila Blood Cells. Science. 2011;332:1210–1213. doi: 10.1126/science.1199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu T, Artavanis-Tsakonas S. deltex, a locus interacting with the neurogenic genes, Notch, Delta and mastermind in Drosophila melanogaster. Genetics. 1990;126:665–677. doi: 10.1093/genetics/126.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorman MJ, Girton JR. A genetic analysis of deltex and its interaction with the Notch locus in Drosophila melanogaster. Genetics. 1992;131:99–112. doi: 10.1093/genetics/131.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diederich RJ, Matsuno K, Hing H, Artavanis-Tsakonas S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development. 1994;120:473–481. doi: 10.1242/dev.120.3.473. [DOI] [PubMed] [Google Scholar]
- 26.Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- 27.Hori K, et al. Drosophila Deltex mediates Suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- 28.Wilkin M, et al. Drosophila HOPS and AP-3 Complex Genes Are Required for a Deltex-Regulated Activation of Notch in the Endosomal Trafficking Pathway. Developmental Cell. 2008;15:762–772. doi: 10.1016/j.devcel.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Zheng L, Saunders Ca, Sorensen EB, Waxmonsky NC, Conner SD. Notch signaling from the endosome requires a conserved dileucine motif. Molecular Biology of the Cell. 2013;24(3):297–307. doi: 10.1091/mbc.E12-02-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fostier M, Evans DA, Artavanis-Tsakonas S, Baron M. Genetic characterization of the Drosophila melanogaster suppressor of deltex gene: a regulator of Notch signaling. Genetics. 1998;150:1477–1485. doi: 10.1093/genetics/150.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornell M, et al. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics. 1999;152:567–576. doi: 10.1093/genetics/152.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazaleyrat SL, et al. Down-regulation of notch target gene expression by suppressor of deltex. Developmental Biology. 2003;255:363–372. doi: 10.1016/s0012-1606(02)00086-6. [DOI] [PubMed] [Google Scholar]
- 33.Sakata T, et al. Drosophila Nedd4 Regulates Endocytosis of Notch and Suppresses Its Ligand-Independent Activation. Current Biology. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 34.Wilkin MB, et al. Regulation of Notch Endosomal Sorting and Signaling by Drosophila Nedd4 Family Proteins. Current Biology. 2004;14:2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu H, et al. Compensatory Flux Changes within an Endocytic Trafficking Network Maintain Thermal Robustness of Notch Signaling. Cell. 2014;157:1160–1174. doi: 10.1016/j.cell.2014.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.del Álamo D, Rouault H, Schweisguth F. Mechanism and Significance of cis-Inhibition in Notch Signalling. Current Biology. 2011;21:R40–R47. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 37.Palmer WH, Jia D, Deng W-M. Cis-interactions between Notch and its ligands block ligand-independent Notch activity. eLife Sciences. 2014:e04415. doi: 10.7554/eLife.04415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleming RJ, et al. An extracellular region of Serrate is essential for ligand-induced cis-inhibition of Notch signaling. Development. 2013;140:2039–2049. doi: 10.1242/dev.087916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meister M, Lagueux M. Drosophila blood cells. Cellular Microbiology. 2003;5:573–580. doi: 10.1046/j.1462-5822.2003.00302.x. [DOI] [PubMed] [Google Scholar]
- 41.Childress JL, Acar M, Tao C, Halder G. Lethal Giant Discs, a Novel C2-Domain Protein, Restricts Notch Activation during Endocytosis. Current Biology. 2006;16:2228–2233. doi: 10.1016/j.cub.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaekel R, Klein T. The Drosophila Notch Inhibitor and Tumor Suppressor Gene lethal (2) giant discs Encodes a Conserved Regulator of Endosomal Trafficking. Developmental Cell. 2006;11:655–669. doi: 10.1016/j.devcel.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 43.Schneider M, Troost T, Grawe F, Martinez-Arias A, Klein T. Activation of Notch in lgd mutant cells requires the fusion of late endosomes with the lysosome. J Cell Sci. 2013;126:645–656. doi: 10.1242/jcs.116590. [DOI] [PubMed] [Google Scholar]
- 44.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 45.Thompson BJ, et al. Tumor Suppressor Properties of the ESCRT-II Complex Component Vps25 in Drosophila. Developmental Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 46.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila Ortholog of Mammalian Tumor Susceptibility Gene 101, Elicit Non-Cell-Autonomous Overgrowth. Developmental Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 47.Herz H-M, et al. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaccari T, et al. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J Cell Sci. 2009;122:2413–2423. doi: 10.1242/jcs.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoyama N, et al. Loss- and gain-of-function analyses of vacuolar protein sorting 2 in Notch signaling of Drosophila melanogaster. Genes Genet Syst. 2013;88:45–57. doi: 10.1266/ggs.88.45. [DOI] [PubMed] [Google Scholar]
- 50.Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, Wang Y, Wong JJL, Lim K-L, Liou Y-C, Wang H, Yu F. Endocytic Pathways Downregulate the L1-type Cell Adhesion Molecule Neuroglian to Promote Dendrite Pruning in Drosophila. Developmental Cell. 2014;30(4):463–478. doi: 10.1016/j.devcel.2014.06.014. http://doi.org/10.1016/j.devcel.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 52.Herz H-M, Woodfield SE, Chen Z, Bolduc C, Bergmann A. Common and Distinct Genetic Properties of ESCRT-II Components in Drosophila. PLoS ONE. 2009;4:e4165. doi: 10.1371/journal.pone.0004165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jékely G, Rørth P. Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 2003;4:1163–1168. doi: 10.1038/sj.embor.7400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tognon E, Wollscheid N, Cortese K, Tacchetti C, Vaccari T. ESCRT-0 Is Not Required for Ectopic Notch Activation and Tumor Suppression in Drosophila. PLoS ONE. 2014;9:e93987. doi: 10.1371/journal.pone.0093987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh Y, et al. Tollip and Tom1 Form a Complex and Recruit Ubiquitin-conjugated Proteins onto Early Endosomes. J Biol Chem. 2004;279:24435–24443. doi: 10.1074/jbc.M400059200. [DOI] [PubMed] [Google Scholar]
- 56.Blanc C, et al. Dictyostelium Tom1 Participates to an Ancestral ESCRT-0 Complex. Traffic. 2009;10:161–171. doi: 10.1111/j.1600-0854.2008.00855.x. [DOI] [PubMed] [Google Scholar]
- 57.Yamada K, et al. Roles of Drosophila Deltex in Notch receptor endocytic trafficking and activation. Genes to cells : devoted to molecular & cellular mechanisms. 2011;16:261–272. doi: 10.1111/j.1365-2443.2011.01488.x. [DOI] [PubMed] [Google Scholar]
- 58.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular Cleavage of Notch Leads to a Heterodimeric Receptor on the Plasma Membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 59.Lake RJ, Grimm LM, Veraksa A, Banos A, Artavanis-Tsakonas S. In Vivo Analysis of the Notch Receptor S1 Cleavage. PLoS ONE. 2009;4:e6728. doi: 10.1371/journal.pone.0006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 61.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 62.Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 63.Brou C, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 64.Lieber T, Kidd S, Young MW. kuzbanian-mediated cleavage of Drosophila Notch. Genes & Development. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanchez-Irizarry C, et al. Notch Subunit Heterodimerization and Prevention of Ligand-Independent Proteolytic Activation Depend, Respectively, on a Novel Domain and the LNR Repeats. Mol Cell Biol. 2004;24:9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 67.Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling: a structural and biochemical perspective. J Cell Sci. 2008;121:3109–3119. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stephenson NL, Avis JM. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc Natl Acad Sci USA. 2012;109:E2757–2765. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Molecular and Cellular Biology. 2009;29(21):5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Habets RA, et al. Human NOTCH2 is resistant to ligand independent activation by metalloprotease Adam17. Journal of Biological Chemistry. 2015;290(23) doi: 10.1074/jbc.M115.643676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delwig A, Rand MD. Kuz and TACE can activate Notch independent of ligand. Cell Mol Life Sci. 2008;65:2232–2243. doi: 10.1007/s00018-008-8127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Tetering G, et al. Metalloprotease ADAM10 Is Required for Notch1 Site 2 Cleavage. J Biol Chem. 2009;284:31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nichols JT, et al. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rand MD, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott CC, Gruenberg J. Ion flux and the function of endosomes and lysosomes: pH is just the start: the flux of ions across endosomal membranes influences endosome function not only through regulation of the luminal pH. Bioessays. 2011;33:103–110. doi: 10.1002/bies.201000108. [DOI] [PubMed] [Google Scholar]
- 76.Tian X, et al. A Voltage-Gated Calcium Channel Regulates Lysosomal Fusion with Endosomes and Autophagosomes and Is Required for Neuronal Homeostasis. PLOS Biology. 2015;13(3):e1002103. doi: 10.1371/journal.pbio.1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 78.Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113(18):4381–90. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vaccari T, Duchi S, Cortese K, Tacchetti C, Bilder D. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development. 2010;137(11):1825–1832. doi: 10.1242/dev.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan Y, Denef N, Schüpbach T. The Vacuolar Proton Pump, V-ATPase, Is Required for Notch Signaling and Endosomal Trafficking in Drosophila. Developmental Cell. 2009;17:387–402. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pasternak SH, et al. Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J Biol Chem. 2003;278:26687–26694. doi: 10.1074/jbc.m304009200. [DOI] [PubMed] [Google Scholar]
- 82.Rooke J, Pan D, Xu T, Rubin GM. KUZ, a Conserved Metalloprotease-Disintegrin Protein with Two Roles in Drosophila Neurogenesis. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- 83.Klein T. The tumour suppressor gene l(2)giant discs is required to restrict the activity of Notch to the dorsoventral boundary during Drosophila wing development. Dev Biol. 2003;255:313–333. doi: 10.1016/s0012-1606(02)00052-0. [DOI] [PubMed] [Google Scholar]
- 84.Gallagher CM, Knoblich JA. The Conserved C2 Domain Protein Lethal (2) Giant Discs Regulates Protein Trafficking in Drosophila. Developmental Cell. 2006;11:641–653. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 85.Ma J, et al. Noncanonical Activation of Notch1 Protein by Membrane Type 1 Matrix Metalloproteinase (MT1-MMP) Controls Melanoma Cell Proliferation. J Biol Chem. 2014;289:8442–8449. doi: 10.1074/jbc.M113.516039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 87.Malecki MJ, et al. Leukemia-Associated Mutations within the NOTCH1 Heterodimerization Domain Fall into at Least Two Distinct Mechanistic Classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hirata N, et al. Sphingosine-1-phosphate promotes expansion of cancer stem cells via S1PR3 by a ligand-independent Notch activation. Nature Communications. 2014;5:4806. doi: 10.1038/ncomms5806. [DOI] [PubMed] [Google Scholar]
- 89.Saksena S, Emr SD. ESCRTs and human disease. Biochem Soc Trans. 2009;37:167–172. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]


