Abstract
The Nrf2-Keap1-ARE pathway is a redox and xenobiotic sensitive signaling axis that functions to protect cells against oxidative stress, environmental toxicants, and harmful chemicals through the induction of cytoprotective genes. To enforce strict regulation, cells invest a great deal of energy into the maintenance of the Nrf2 pathway to ensure rapid induction upon cellular insult and rapid return to basal levels once the insult is mitigated. Because of the protective role of Nrf2 transcriptional programs, controlled activation of the pathway has been recognized as a means for chemoprevention. On the other hand, constitutive activation of Nrf2, due to somatic mutations of genes that control Nrf2 degradation, promotes carcinogenesis and imparts chemoresistance to cancer cells. Autophagy, a bulk protein degradation process, is another tightly regulated complex cellular process that functions as a cellular quality control system to remove damaged proteins or organelles. Low cellular nutrient levels can also activate autophagy, which acts to restore metabolic homeostasis through the degradation of macromolecules to provide nutrients. Recently, these two cellular pathways were shown to intersect through the direct interaction between p62 (an autophagy adaptor protein) and Keap1 (the Nrf2 substrate adaptor for the Cul3 E3 ubiquitin ligase). Dysregulation of autophagy was shown to result in prolonged Nrf2 activation in a p62-dependent manner. In this review, we will discuss the progress that has been made in dissecting the intersection of these two pathways and the potential tumor-promoting role of prolonged Nrf2 activation.
1. Introduction
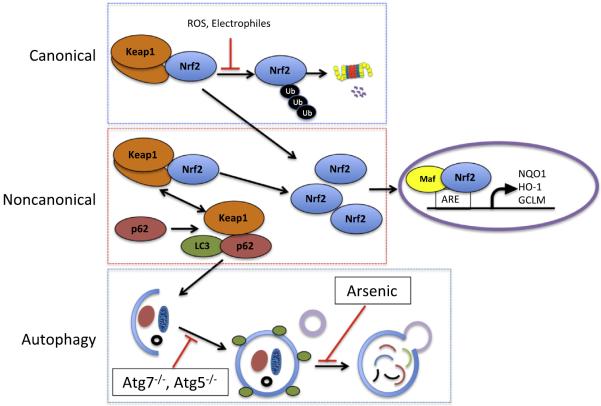
The nuclear factor erytheroid-derived-2-like 2 (Nrf2)-Kelch-like ECH-associated protein 1 (Keap1)-antioxidant response element (ARE) field has expanded at an extraordinary rate since the cloning of Nrf2 [1,2]. Over two decades of research, it has been firmly established that the Nrf2-Keap1-ARE pathway is an adaptive cellular response conferring protection against oxidative and xenobiotic stress. Modification of Keap1 cysteine residues leads to inhibition of Nrf2 ubiquitylation and stabilization of Nrf2, allowing Nrf2 to accumulate in the cytosol and then to translocate into the nucleus where it binds to a small Maf protein and activates transcription of genes containing antioxidant response elements (AREs) in their regulatory regions [3–5] (Fig. 1, canonical pathway).
Fig. 1.
The canonical and noncanonical regulatory pathways of Nrf2 signaling. (1) Canonical pathway: under normal conditions, Nrf2 is bound to the E3 ubiquitin ligase adaptor protein Keap1, which leads to its ubiquitylation and proteasomal degradation. When Keap1 is challenged with ROS or electrophiles, critical cysteines are modified, blocking Nrf2 ubiquitylation, increasing the level of Nrf2 and activating the ARE-mediated transcription. (2) Noncanonical pathway: when autophagic flux is compromised and p62 accumulates, Keap1 is sequestered by p62 and can no longer bind Nrf2, leading to increased Nrf2 signaling. (3) Autophagy pathway: the pathway can be dysregulated by the blockage of autophagosome maturation (e.g. deletion of Atg7 or Atg5) or the fusion of autophagosome-lysosome (e.g. arsenic treatment).
In addition to Keap1-mediated regulation, recently, two other E3 ubiquitin ligases have been found to regulate the protein level of Nrf2. In 2010, the Neh6 domain of Nrf2 was shown to be phosphorylated by GSK-3β, creating a phosphodegron, which is recognized by the Skp1-Cul1-Rbx1-βTrCP E3 ubiquitin ligase complex [6,7]. Furthermore, our group has found that Hrd1, an integral endoplasmic reticulum (ER) membrane E3 ligase, negatively regulates Nrf2 in patients with liver cirrhosis [8]. These Keap1-independent mechanisms of Nrf2 regulation highlight the functional importance of Nrf2 beyond redox sensing.
Macroautophagy (henceforth “autophagy”) is an important catabolic process that degrades cytoplasmic components in bulk (Fig. 1, autophagy) [9–11]. Autophagy is responsible for the clearance of damaged organelles, long-lived proteins, or misfolded proteins. This degradation process acts to preserve cellular homeostasis and to defend against oxidative or proteotoxic stress. Both intracellular and extracellular signals can induce autophagy. These signals include oxidative stress, ER stress, or nutrient deprivation [12]. Autophagy also plays an important role in eliminating intracellular pathogens, in antigen presentation, and in maintaining cellular longevity [13,14]. Given its many critical roles, it is not surprising that impaired autophagic function contributes to a diverse collection of diseases such as cancer, neurodegeneration, cardiomyopathy, Crohn's disease, diabetes, and fatty liver disease [10,12]. On the other hand, increased autophagic activity allows cancer cells to cope with high metabolic and proteotoxic stress, which is essential to their survival [15]. Thus, similar to Nrf2, autophagy plays seemingly contradictory roles in cancer [9,16,17], and the roles of Nrf2 and autophagy in cancer are context-dependent.
The crosstalk between the Nrf2-Keap1-ARE axis and autophagy was revealed in 2010 when five separate groups independently confirmed the association between p62/sequestosome 1(SQSTM1), an autophagy adaptor, and Keap1 [18–22]. Following this finding, great progress has been made to understand the mechanistic underpinnings linking the Nrf2 and autophagy pathways, the functional consequence of autophagy dysregulation, and prolonged Nrf2 signaling.
1.1. p62-dependent Nrf2 activation – the noncanonical pathway
p62 is an autophagy adaptor protein, initially identified as a tyrosine kinase p56lck binding protein, that gained more attention when mutations in p62 were found to be associated with Paget's disease of bone [23–25]. However, the functional importance of p62 did not emerge until Komatsu et al. reported an association between p62 and LC3, indicating a role for p62 in autophagy [26]. In this study, p62 was found to mediate the formation of protein aggregates destined for autophagic turnover, which suggested that p62 facilitated selective degradation of protein cargo via autophagy [26]. It is now understood that p62 works as an adaptor, binding ubiquitylated protein aggregates and delivering them to the autophagosomes.
Although the p62-mediated induction of Nrf2 with subsequent nuclear translocation and activation of ARE-driven genes was first reported in 2007 [27], the molecular mechanism was not fully elucidated until the discovery of the association between p62 and Keap1 [18–22]. Three of the five groups confirmed that the 349-DPSTGE-354 motif in the Keap1-interacting region (KIR) domain of p62, which resembles the Keap1-interating ETGE motif in the Neh2 domain of Nrf2, accounts for the direct interaction between p62 and Keap1 [18–20]. This interaction allows p62 to sequester Keap1 into the autophagosomes, which impairs the ubiquitylation of Nrf2, leading to activation of the Nrf2 signaling pathway (Fig. 1, noncanonical pathway). However, the details of the association between p62, Keap1, and LC3 remain controversial. For example, the LC3-interacting region (LIR) domain in p62 is close to the KIR domain in p62. This adjacency was shown to cause competitive binding between LC3 and Keap1 to p62 [20]. In contrast, another study showed that Keap1-p62-LC3 complex formation was required for the clearance of ubiquitin aggregates in response to oxidative stress [21]. This controversy remains to be solved.
In addition to the sequestration of Keap1 by p62 into aggregates or autophagosomes, Keap1 was found to be a p62-regulated substrate for autophagy-mediated degradation. Therefore, p62 plays a key role in controlling Keap1 turnover, as indicated by the results showing that overexpression of p62 significantly decreased the half-life of Keap1 whereas knockdown of p62 increased it [22]. It was shown that the protein level of Keap1 in the liver was much lower in wild type mice compared to liver-specific Atg7-deficient or p62-deficient mice [28]. These results indicate that p62-mediated Keap1 turnover, in addition to physical sequestration of Keap1, also contributes to the noncanonical activation of Nrf2 (Fig. 1, canonical pathway).
Recent structural and functional studies have further illuminated the details of the noncanonical mechanism of Nrf2 activation. Previous structural studies demonstrated that Keap1 interacts through its Kelch domain with either the DLG or the ETGE motif of Nrf2 in a 2:1 ratio. According to the hinge and latch model, the ETGE motif has a higher affinity for Keap1 than the DLG motif, which causes the latter to dynamically associate and dissociate from Keap1 to generate oscillating “closed” (associated) and “open” (dissociated) conformations [29–33]. Interestingly, p62 contains an STGE motif that binds to the Kelch domain of Keap1 and it has been speculated that this binding occurs during the open conformation of the Keap1-Nrf2 complex [33]. Another study demonstrated that phosphorylation of p62 S351 within this motif significantly increased the affinity between p62 and Keap1, which resulted in prolonged accumulation of Nrf2 and transcriptional upregulation of its target genes [34]. To date, however, the kinase responsible for p62 phosphorylation at S351 has not been identi-fied [34]. Recently, Sestrins, especially Sestrin2 was found to be in complex with p62, Keap1, and Rbx1, and the association of these proteins facilitated p62-dependent autophagic degradation of Keap1 and subsequent activation of Nrf2 [35]. Another study suggests that Sestrin2 interacted with Unc-51-like kinase 1 (ULK1) and p62 to promote p62 phosphorylation by ULK1 at S403, which facilitated p62-mediated degradation of cargo proteins including Keap1 [36].
2. Prolonged Nrf2 activation results from the noncanonical mechanism – a novel mechanism of arsenic action
It is reasonable to envision that this noncanonical mechanism of Nrf2 induction results in prolonged Nrf2 signaling, relative to the canonical pathway, because protein aggregates have to be resolved and new Keap1 protein has to be made in order to restore the functional, canonical Nrf2-Keap1-ARE axis. This was experimentally confirmed during mechanistic investigations of toxic and carcinogenic effects of arsenic. Inorganic arsenic, an environmentally ubiquitous toxic metalloid that pollutes drinking water, soil, and air all over the world, has been implicated in many human diseases including cancer, cardiovascular diseases, neurological diseases, respiratory diseases, and potentially diabetes [37–41]. It seems reasonable that there would be different cellular signaling events or outcomes elicited by acute high doses vs. chronic low doses of arsenic exposure. Some detrimental outcomes of arsenic toxicity resulting from acute exposure, typically higher than environmentally relevant doses, include ROS production, metabolic stress due to an alteration of enzyme function, DNA damage, inhibition of DNA repair mechanisms, and activation of cell death pathways such as necrosis and apoptosis [42–46]. In contrast, chronic exposure to environmentally relevant low doses of arsenic results in tumorigenesis without provoking a measurable amount of the aforementioned cellular responses. Recently, our group determined that autophagic flux was blocked after exposure to low doses of arsenic, which may represent one of the underlying mechanisms of arsenic carcinogenicity [47,48]. In this study, we used low doses of arsenic to understand a long standing dilemma: why arsenic, a carcinogenic metal, is also able to induce the chemopreventive Nrf2 signaling pathway and whether there is a distinction in the way Nrf2 is activated by arsenic vs. chemo-preventive compounds such as sulforaphane. Through detailed investigations, we found that sodium arsenite employs a unique mechanism to activate Nrf2, without modification of Keap1 cysteines, but with diminished Nrf2 ubiquitylation [49]. Subsequently, we determined that arsenic activated Nrf2 in a p62-dependent (noncanonical) manner, through inhibition of autophagic flux [47,48]. Arsenic was shown to increase the accumulation of autophagosomes and the colocalization of p62, Keap1, and LC3, which in turn led to chronic and sustained activation of Nrf2 [47,48]. Generally, autophagy activation coincides with diminished p62 levels, so the accumulation of p62 would suggest blockage of autophagy rather than induction. It remains to be determined how arsenic blocks autophagic flux.
In addition to arsenic, another metal was found to work by a similar mechanism to activate the Nrf2 signaling pathway. In cadmium-transformed human lung bronchial epithelial BEAS-2B cells, an acquired autophagy deficiency was observed, which led to constitutive accumulation of p62 and Nrf2, giving rise to apoptosis-resistant cells [50]. More interestingly, cadmium behaves differently in BEAS-2B parental cells vs. cadmium-transformed BEAS-2B cells. In non cadmium-transformed BEAS-2B cells, cadmium served as an autophagy inducer, but it lost the ability to induce autophagy in cadmium-transformed cells, as indicated by a lack of GFP-LC3 puncta in response to short term cadmium treatment [50]. Therefore, long-term exposure to cadmium might affect the initiation or elongation steps of autophagy in these transformed cells.
In addition to metals, the central nervous system stimulant methamphetamine and the skin sensitizer 1-fluoro-2,4-dinitrobenzene were reported to induce the Nrf2 signaling pathway through impaired autophagic flux and p62 accumulation in a mouse atrial cardiac cell line and in a human monocytic cell line, respectively [51,52]. These studies provide a critical link between autophagy dysregulation and prolonged Nrf2 signaling, which provides important environmental health implications.
3. The pathological consequences of prolonged Nrf2 activation through the p62-mediated noncanonical mechanism
Starting in 2006, the “dark side” of Nrf2 has gradually emerged. Somatic mutations in Nrf2 or its negative regulators (Keap1 and Cul3) have been shown to be responsible for the high levels of Nrf2 in certain tumors, such as lung cancer, melanoma, renal clear cell carcinoma, and hepatocellular carcinoma [53–57]. High levels of Nrf2 in these cancer cells provide a cellular environment conducive to growth and survival under detrimental conditions. Furthermore, Nrf2 has proven to contribute to chemoresistance [17,58]. More importantly, adjuvant therapy using the Nrf2 inhibitor brusatol imparted drug sensitivity to chemoresistant cancer cells, and enhanced the survival of mice bearing KRas-induced lung tumors [59,60]. Therefore, persistent and constitutive Nrf2 activation due to genetic mutations has been recognized to be pathogenic, especially in cancer. This “dark side” role of Nrf2 in cancer due to persistent Nrf2 activation led us to postulate that prolonged Nrf2 activation through the p62-mediated non-canonical mechanism during low dose, chronic arsenic exposure may promote cell transformation.
Recently, studies using autophagy-defective mouse models provided strong evidence that prolonged Nrf2 activation, due to autophagy dysregulation, leads to tissue injuries and cancer. For instane, when autophagy is ablated due to deletion of Atg5, Atg7, or Beclin-1, p62-Keap1 aggregates accumulate in the cytosol, resulting in prolonged Nrf2 activation [61–64]. Atg7 serves as an E1 ubiquitin-activating enzyme in the initiation of autophagosome synthesis. It plays an important role in the elongation step of the phagophore by activating Atg12 and LC3-I mediated conjugation systems [65]. Mice deficient in the Atg7 gene have impaired autophagosome formation [66]. In these mice, phagophore elongation is blocked and accumulation of protein aggregates rich in p62 and Keap1 is observed [18]. Therefore, Atg7-deficient mice provided a valuable in vivo model for mechanistic studies aimed at dissecting the crosstalk between the autophagy and Nrf2 pathways.
A positive correlation between p62 and Nrf2 was initially observed in a liver-specific Atg7 knockout mouse [67]. Subsequently, several reports confirmed the accumulation of Nrf2 in the liver as being due to p62-mediated Keap1 inactivation, which in turn triggered liver damage, inflammation, fibrosis, and tumorigenesis [18,28,35,61,63,68]. For example, hepatocellular adenoma was detected in liver-specific Atg7-deficient mice at the age of 7 months. In these mice, p62- and Keap1-containing protein aggregates accumulated and Nrf2 was persistently activated [61]. Interestingly, similar aggregates were detected in human hepatocellular carcinomas (HCC), with activation of the Nrf2 pathway. In these HCC cell lines, deletion of p62 using a zinc finger nuclease system blocked aggregate formation and Nrf2 activation, and suppressed anchorage-independent growth [61]. Another study using the same liver specific Atg7-deficient mice confirmed aberrant p62 and Keap1 accumulation in aggregates and prolonged Nrf2 activation. Liver damage was observed in these mice, as displayed by swelling of hepatocytes, infiltration of inflammatory cells, and collapse of the hepatic lobule [28]. In support of Nrf2 activation as a crucial factor for liver injury, concurrent p62 or Nrf2 ablation in Atg7-deficient mice corrected the pathological effects observed in Atg7-deficient mice [18,26,28].
In addition to liver injury and tumorigenesis, similar non-canonical activation of Nrf2 was observed in lung, epidermal keratinocytes and melanocytes in cell type-specific Atg7-deficient mice. In these studies, autophagy deficiency was shown to disturb redox homeostasis through persistent accumulation of Nrf2, which contributed to airway hyper-responsiveness, impaired removal of oxidized phospholipids and protein aggregates and hypo-pigmentation of the skin [69–71]. In another study, latent Kaposi's sarcoma-associated herpesvirus infection was reported to induce aberrant Nrf2 accumulation due to p62-mediated Keap1 inactivation, which was associated with the initiation and development of Kaposi's sarcoma [72].
The impaired autophagic pathway due to Atg7 deficiency caused not only p62 accumulation and prolonged Nrf2 activation, but also increased the level of polyubiquitylated protein aggregates and inclusion bodies. Interestingly, increases in protein aggregates or inclusion bodies in liver and brain from Atg7-deficient mice were completely suppressed by simultaneous loss of either p62 or Nrf2, thus the accumulation of poly-ubiquitylated proteins is thought to be a consequence of prolonged Nrf2 activation [68]. Further studies are warranted to understand the requirement of poly-ubiquitylated protein aggregates in the pathogenesis resulting from prolonged Nrf2 activation and the molecular mechanism driving protein aggregate formation.
Intriguingly, in BRafV600E-driven lung tumors, although Atg7 deficiency led to robust tumor induction at an early stage (40% more at 5 weeks post-BRaf activation), tumor growth was eventually retarded and the morphology of the tumors switched from adenoma or adenocarcinoma to oncocytoma, a benign tumor with accumulation of defective mitochondria, due to limited glutamine supply. Therefore, the mice bearing Atg7-deficient tumors had better survival rates [73,74]. Furthermore, in BRafV600E-driven tumors, Nrf2 deficiency and Atg7 deficiency share similar phenotypes in that deletion of either promotes early tumorigenesis but extended survival of mice by promoting oncocytomas [73,74]. In accordance with this finding that basal levels of autophagy are necessary for cancer cell growth, a basal amount of autophagy is also required for normal functions of melanocytes since Atg7 deficiency caused premature senescence and impaired pigment production in these cells [71]. Therefore, it may be concluded that the roles of autophagy and Nrf2 in cancer are context-dependent. A comprehensive understanding of the context-dependent mechanistic details is crucial before optimal therapeutic modulation of Nrf2 and autophagy can be achieved.
The understanding of the prolonged Nrf2 activation through this noncanonical mechanism explains why high levels of Nrf2 were observed in certain cancer cells that do not bear mutations in genes controlling the expression of Nrf2. For example, persistent activation of Nrf2 in human hepatocellular carcinoma cells was due to Keap1-p62 aggregate formation, implicating the involvement of the noncanonical mechanism of Nrf2 activation, instead of somatic mutations in Nrf2, Keap1, or Cul3[61]. In hepatocellular carcinoma patients, the co-localization of p62 and Keap1 in aggregates is as high as 25.5% [61], compared to the 8% mutation rate of Keap1[55]. Therefore, the impaired production of autophagosomes (e.g. Atg7-deficient mice) or blocked autophagic flux (e.g. in response to low dose arsenic exposure) may be a contributing factor to achieve high Nrf2 levels in the absence of somatic mutations in genes controlling Nrf2 expression levels, leading to tumor development and progression.
4. Conclusion
Both the autophagy and Nrf2-Keap1 pathways antagonize cellular stress by upregulating a battery of antioxidant and cellular defense genes. Typically, intermittent activation of Nrf2 through the canonical pathway confers cellular protection and functional integrity. Conversely, constitutive activation of Nrf2, due to somatic mutations in genes regulating Nrf2, leads to tumor promotion and contributes to chemoresistance [75]. Similarly, prolonged activation of Nrf2 through the noncanonical mechanism seems to be detrimental, resulting in tissue injuries, inflammation, and tumorigenesis, the “dark side” of Nrf2. Autophagy, another cellular stress response, is crucial to proteostasis and organelle health in cells. Recent findings indicate that these two pathways are intimately linked by the autophagy adaptor protein, p62. The observation of this interconnectedness has revealed the mechanistic underpinnings of prolonged Nrf2 signaling. When autophagy is compromised, p62 sequesters Keap1 into aggregates, Nrf2 is then stabilized and ARE-regulated genes are upregulated. Because this system lacks the normal, facile means of deactivating Nrf2 signaling, this leads to sustained activation of the Nrf2-ARE system. Although some details remain to be illuminated, these studies have provided new paradigms for the treatment of disease. Conceivably, Nrf2 inhibitors can be used to suppress the prolonged Nrf2 activation resulting from autophagy defects. The detailed mechanistic understanding of the interaction between the Nrf2-Keap1-ARE axis and autophagy is a clear demonstration of the ongoing need for detailed mechanistic pictures of biological and pathological phenomena to facilitate the discovery of new therapies.
References
- [1].Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol. 1995;15:4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Itoh K, Ishii T, Wakabayashi N, Yamamoto M. Regulatory mechanisms of cellular response to oxidative stress. Free Radic Res. 1999;31:319–324. doi: 10.1080/10715769900300881. [DOI] [PubMed] [Google Scholar]
- [4].Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu T, Zhao F, Gao B, Tan C, Yagishita N, Nakajima T, Wong PK, Chapman E, Fang D, Zhang DD. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014;28:708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- [10].Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].White E. The role for autophagy in cancer. The Journal of clinical investigation. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- [13].Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Munz C. Enhancing immunity through autophagy. Annual review of immunology. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- [15].Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fulda S, Kogel D. Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene. 2015 doi: 10.1038/onc.2014.458. [DOI] [PubMed] [Google Scholar]
- [17].Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- [19].Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. The Journal of biological chemistry. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, Zhu M, Zhong Q. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6:614–621. doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, Park BK. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. The Journal of biological chemistry. 2010;285:16782–16788. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Joung I, Strominger JL, Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc Natl Acad Sci U S A. 1996;93:5991–5995. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Laurin N, Brown JP, Morissette J, Raymond V. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am J Hum Genet. 2002;70:1582–1588. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hocking LJ, Lucas GJ, Daroszewska A, Mangion J, Olavesen M, Cundy T, Nicholson GC, Ward L, Bennett ST, Wuyts W, Van Hul W, Ralston SH. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget's disease. Hum Mol Genet. 2002;11:2735–2739. doi: 10.1093/hmg/11.22.2735. [DOI] [PubMed] [Google Scholar]
- [26].Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- [27].Liu Y, Kern JT, Walker JR, Johnson JA, Schultz PG, Luesch H. A genomic screen for activators of the antioxidant response element. Proc Natl Acad Sci U S A. 2007;104:5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- [30].Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. The Journal of biological chemistry. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- [33].Baird L, Lleres D, Swift S, Dinkova-Kostova AT. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc Natl Acad Sci U S A. 2013;110:15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- [35].Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, Lee HE, Kang D, Rhee SG. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013;17:73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- [36].Ro SH, Semple IA, Park H, Park H, Park HW, Kim M, Kim JS, Lee JH. Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1. FEBS J. 2014;281:3816–3827. doi: 10.1111/febs.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ray PD, Yosim A, Fry RC. Incorporating epigenetic data into the risk assessment process for the toxic metals arsenic, cadmium, chromium, lead, and mercury: strategies and challenges. Frontiers in genetics. 2014;5:201. doi: 10.3389/fgene.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Farzan SF, Karagas MR, Chen Y. In utero and early life arsenic exposure in relation to long-term health and disease. Toxicol Appl Pharmacol. 2013;272:384–390. doi: 10.1016/j.taap.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang CH, Hsiao CK, Chen CL, Hsu LI, Chiou HY, Chen SY, Hsueh YM, Wu MM, Chen CJ. A review of the epidemiologic literature on the role of environmental arsenic exposure and cardiovascular diseases. Toxicol Appl Pharmacol. 2007;222:315–326. doi: 10.1016/j.taap.2006.12.022. [DOI] [PubMed] [Google Scholar]
- [40].Argos M, Ahsan H, Graziano JH. Arsenic and human health: epidemiologic progress and public health implications. Rev Environ Health. 2012;27:191–195. doi: 10.1515/reveh-2012-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Maull EA, Ahsan H, Edwards J, Longnecker MP, Navas-Acien A, Pi J, Silbergeld EK, Styblo M, Tseng CH, Thayer KA, Loomis D. Evaluation of the association between arsenic and diabetes: a National Toxicology Program workshop review. Environ Health Perspect. 2012;120:1658–1670. doi: 10.1289/ehp.1104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Andrew AS, Burgess JL, Meza MM, Demidenko E, Waugh MG, Hamilton JW, Karagas MR. Arsenic exposure is associated with decreased DNA repair in vitro and in individuals exposed to drinking water arsenic. Environ Health Perspect. 2006;114:1193–1198. doi: 10.1289/ehp.9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Andrew AS, Karagas MR, Hamilton JW. Decreased DNA repair gene expression among individuals exposed to arsenic in United States drinking water. International journal of cancer, Journal international du cancer. 2003;104:263–268. doi: 10.1002/ijc.10968. [DOI] [PubMed] [Google Scholar]
- [44].Ebert F, Leffers L, Weber T, Berndt S, Mangerich A, Beneke S, Burkle A, Schwerdtle T. Toxicological properties of the thiolated inorganic arsenic and arsenosugar metabolite thio-dimethylarsinic acid in human bladder cells. J Trace Elem Med Biol. 2014;28:138–146. doi: 10.1016/j.jtemb.2013.06.004. [DOI] [PubMed] [Google Scholar]
- [45].Jiang X, Chen C, Liu Y, Zhang P, Zhang Z. Critical role of cellular glutathione homeostasis for trivalent inorganic arsenite-induced oxidative damage in human bronchial epithelial cells. Mutat Res Genet Toxicol Environ Mutagen. 2014;770:35–45. doi: 10.1016/j.mrgentox.2014.04.016. [DOI] [PubMed] [Google Scholar]
- [46].Tokumoto M, Lee JY, Fujiwara Y, Uchiyama M, Satoh M. Inorganic arsenic induces apoptosis through downregulation of Ube2d genes and p53 accumulation in rat proximal tubular cells. J Toxicol Sci. 2013;38:815–820. doi: 10.2131/jts.38.815. [DOI] [PubMed] [Google Scholar]
- [47].Lau A, Whitman SA, Jaramillo MC, Zhang DD. Arsenic-mediated activation of the Nrf2-Keap1 antioxidant pathway. J Biochem Mol Toxicol. 2013;27:99–105. doi: 10.1002/jbt.21463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, Zhang DD. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol. 2013;33:2436–2446. doi: 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang XJ, Sun Z, Chen W, Li Y, Villeneuve NF, Zhang DD. Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction. Toxicol Appl Pharmacol. 2008;230:383–389. doi: 10.1016/j.taap.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Son YO, Pratheeshkumar P, Roy RV, Hitron JA, Wang L, Zhang Z, Shi X. Nrf2/p62 signaling in apoptosis resistance and its role in cadmium-induced carcinogenesis. The Journal of biological chemistry. 2014;289:28660–28675. doi: 10.1074/jbc.M114.595496. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [51].Funakoshi-Hirose I, Aki T, Unuma K, Funakoshi T, Noritake K, Uemura K. Distinct effects of methamphetamine on autophagy-lysosome and ubiquitin-proteasome systems in HL-1 cultured mouse atrial cardiomyocytes. Toxicology. 2013;312:74–82. doi: 10.1016/j.tox.2013.07.016. [DOI] [PubMed] [Google Scholar]
- [52].Luis A, Martins JD, Silva A, Ferreira I, Cruz MT, Neves BM. Oxidative stress-dependent activation of the eIF2alpha-ATFr unfolded protein response branch by skin sensitizer 1-fluoro-2,4-dinitrobenzene modulates dendritic-like cell maturation and inflammatory status in a biphasic manner. Free Radic Biol Med. 2014;77:217–229. doi: 10.1016/j.freeradbiomed.2014.09.008. [DOI] [PubMed] [Google Scholar]
- [53].Miura S, Shibazaki M, Kasai S, Yasuhira S, Watanabe A, Inoue T, Kageshita Y, Tsunoda K, Takahashi K, Akasaka T, Masuda T, Maesawa C. A somatic mutation of the KEAP1 gene in malignant melanoma is involved in aberrant NRF2 activation and an increase in intrinsic drug resistance. J Invest Dermatol. 2014;134:553–556. doi: 10.1038/jid.2013.343. [DOI] [PubMed] [Google Scholar]
- [54].Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- [55].Cleary SP, Jeck WR, Zhao X, Chen K, Selitsky SR, Savich GL, Tan TX, Wu MC, Getz G, Lawrence MS, Parker JS, Li J, Powers S, Kim H, Fischer S, Guindi M, Ghanekar A, Chiang DY. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 2013;58:1693–1702. doi: 10.1002/hep.26540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer research. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- [58].Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tao S, Wang S, Moghaddam SJ, Ooi A, Chapman E, Wong PK, Zhang DD. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer research. 2014;74:7430–7441. doi: 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ni HM, Boggess N, McGill MR, Lebofsky M, Borude P, Apte U, Jaeschke H, Ding WX. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–450. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ni HM, Woolbright BL, Williams J, Copple B, Cui W, Luyendyk JP, Jaeschke H, Ding WX. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, Cervo M, Yao H, Chung AL, Mizumura K, An CH, Shan B, Franks JM, Haley KJ, Owen CA, Tesfaigzi Y, Washko GR, Quackenbush J, Silverman EK, Rahman I, Kim HP, Mahmood A, Biswal SS, Ryter SW, Choi AM. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. The Journal of clinical investigation. 2013;123:5212–5230. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- [66].Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Matsumoto N, Ezaki J, Komatsu M, Takahashi K, Mineki R, Taka H, Kikkawa M, Fujimura T, Takeda-Ezaki M, Ueno T, Tanaka K, Kominami E. Comprehensive proteomics analysis of autophagy-deficient mouse liver. Biochem Biophys Res Commun. 2008;368:643–649. doi: 10.1016/j.bbrc.2008.01.112. [DOI] [PubMed] [Google Scholar]
- [68].Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, Yamamoto M, Tanaka K, Mizushima N, Komatsu M, Kopito RR. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol. 2010;191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhao Y, Zhang CF, Rossiter H, Eckhart L, Konig U, Karner S, Mildner M, Bochkov VN, Tschachler E, Gruber F. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. J Invest Dermatol. 2013;133:1629–1637. doi: 10.1038/jid.2013.26. [DOI] [PubMed] [Google Scholar]
- [70].Inoue D, Kubo H, Taguchi K, Suzuki T, Komatsu M, Motohashi H, Yamamoto M. Inducible disruption of autophagy in the lung causes airway hyper-responsiveness. Biochem Biophys Res Commun. 2011;405:13–18. doi: 10.1016/j.bbrc.2010.12.092. [DOI] [PubMed] [Google Scholar]
- [71].Zhang CF, Gruber F, Ni C, Mildner M, Koenig U, Karner S, Barresi C, Rossiter H, Narzt MS, Nagelreiter IM, Larue L, Tobin DJ, Eckhart L, Tschachler E. Suppression of Autophagy Dysregulates the Antioxidant Response and Causes Premature Senescence of Melanocytes. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.439. [DOI] [PubMed] [Google Scholar]
- [72].Gjyshi O, Flaherty S, Veettil MV, Johnson KE, Chandran B, Bottero V. Kaposi's Sarcoma-Associated Herpesvirus Induces Nrf2 Activation in Latently Infected Endothelial Cells through SQSTM1 Phosphorylation and Interaction with Polyubiquitinated Keap1. J Virol. 2015;89:2268–2286. doi: 10.1128/JVI.02742-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Strohecker AM, White E. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy. 2014;10:384–385. doi: 10.4161/auto.27320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer discovery. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]