Abstract
Benzophenone is a popular photophore for photoaffinity-labeling. It is also an important framework for drug development; many drugs contain benzophenone or analogous frameworks. The current work reports that benzophenone and its analogs bind to human glyoxalase 1. The binding, however, has little effect on the catalytic activity of this enzyme. The implications of the finding in terms of both drug development and photoaffinity-labeling are discussed.
Keywords: GLO1, Photoaffinity-labeling, Off-target, Ketoprofen, Fenofibrate, Doxorubicin
Graphical abstract
Benzophenone is a popular photophore for photoaffinity-labeling.1,2 It is widely used to study ligand-receptor interactions.3 More recently, benzophenone is employed in chemical proteomics and drug target identification.4-6
Benzophenone is also an important framework in medicinal chemistry. The framework of benzophenone is seen in many drugs, including ketoprofen (an analgesic targeting COX1 and COX2),7 fenofibrate (a lipid lowering agent targeting PPARα),8 and doxorubicin (an anticancer agent targeting DNA).9 According to the SciFinder database, there are more than 300 benzophenone-containing molecules with diverse pharmacological activities.10 As such, benzophenone is a versatile framework that can target a wide variety of macromolecules. The versatility of benzophenone, however, also raises a concern about its selectivity. Do these benzophenone-containing drugs really bind only to their intended targets? At present, little is known about the off-target activity of benzophenone derivatives.
Here, we report our fortuitous finding that benzophenone binds to human glyoxalase 1 (GLO1). Further analysis revealed that several clinical drugs containing the benzophenone framework also bind to human GLO1. The binding of these compounds does not affect the catalytic activity of GLO1. Implications of this finding in terms of both drug development and photoaffinity-labeling are discussed (vide infra).
The finding was made during our study of biotinylated benzophenone-adenine photoprobe 1 (Fig. 1a), which was originally designed to enrich adenine-binding proteins, such as kinases and ATPases, from complex proteomes.11-14 1 was prepared by standard solid phase synthesis using the Fmoc chemistry.14,15 A preliminary photolabeling analysis of five different cell lines (Hela, MCF7, T24, Calu1, THP-1) revealed that THP-1 (human monocytic leukemia cells) expresses a large amount of a 20 kDa protein that can be readily enriched by 1 (See Supplementary Figure S1). As such we decided to use THP-1 to identify the binding targets of 1.
Figure 1.
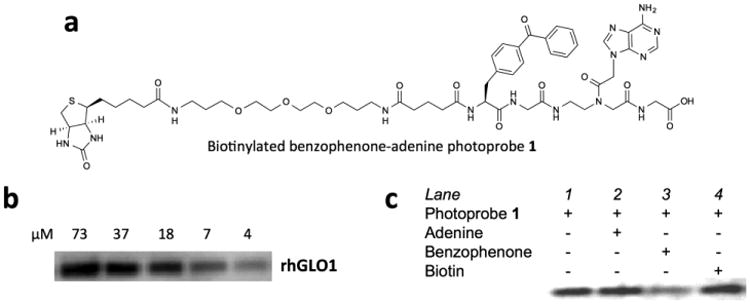
Photoaffinity-labeling of human GLO1 by biotinylated benzophenone-adenine photoprobe 1. (a) The structure of 1. (b) Photolabeling of recombinant human GLO1 (rhGLO1) with different concentrations of 1. 30 μL aliquots of rhGLO1 (0.05 μg/μl) in 1× Tris-buffered saline (TBS: 20 mM Tris-HCl, pH 7.4) were photolabeled with 1 at the final photoprobe concentrations of 73, 37, 18, 7 and 4 μM. Photocrosslinking was carried out under UV light (λmax 350 nm) for 2 h. See Supporting Information for more experimental details. (c) Photolabeling of rhGLO1 by 1 (7 μM) in the presence of excess (20×, 140 μM) adenine, benzophenone (4-methylbenzophenone), and biotin.
To obtain sufficient amounts of target proteins, THP-1 was cultured in a large scale and subjected to the photoaffinity-labeling by 1 (Supplementary Scheme S1). Photochemically biotinylated proteins were purified by streptavidin affinity chromatography, and separated by SDS PAGE. Coomassie blue staining visualized an intense band at 20 kDa as seen in the preliminary analysis. In addition, a weaker band was also observed at 40 kDa. Both of these bands were excised and subjected to mass spectrometric (MS) analysis. Table 1 summarizes the proteins identified by MS. The 40 kDa band contained expected targets of 1, namely, α, β, γ-isoforms of actin proteins, which are well-known ATPases.
Table 1. Target proteins of 1 in THP-1 (human monocytic leukemia cells).
| Banda | Gene Identifier | Gene | MASCOT Scoreb |
|---|---|---|---|
| 20 kDa | 2292338 | Glyoxalase 1 | 1387 |
| 40 kDa | 16359158 | Actin, beta | 963 |
| 40 kDa | 4885049 | Actin, alpha cardiac muscle 1 | 791 |
| 40 kDa | 4501889 | Actin, gamma 2 | 764 |
Coomassie blue stained bands that were subjected to the MS analysis.
The table shows the proteins with the MASCOT score of 500 or higher.
The 20 kDa band, on the other hand, turned out to be GLO1 (Table 1), which is a homodimeric zinc metalloenzyme for the conversion of methylglyoxal (a toxic by-product of glycolytic pathway) into S-lactoylglutathione. The finding of GLO1 was surprising because it was not known as an adenine-binding protein. To verify this finding, recombinant human GLO1 (rhGLO1) was obtained and subjected to the photoaffinity-labeling with 1. Western blot of the photolabeled samples showed that rhGLO1 was indeed photolabeled by 1 in a dose-dependent manner with an EC50 value16 of 5 μM (Fig. 1b).
Next, we subjected rhGLO1 to the photolabeling in the presence of excess adenine. The working-hypothesis was that, if GLO1 recognizes the adenine moiety of 1, the excess adenine would compete for the binding pocket and block the photolabeling. A 20-fold molar excess of adenine over 1, however, did not affect the band intensity (Fig. 1c), suggesting that GLO1 did not recognize adenine. Similar experiments were carried out in the presence of excess benzophenone and biotin. While excess biotin did not affect the band intensity, excess benzophenone clearly blocked the photolabeling (Fig. 1c).
There were two possible mechanisms by which benzophenone blocked the photolabeling. One possibility was that excess benzophenone and 1 competed for a binding pocket on GLO1, which resulted in the diminished band intensity. Alternatively, it was also possible that excess benzophenone photochemically quenched the photolabeling reaction without binding to GLO1. A clue to distinguish the two mechanisms was obtained with the subsequent study, in which the photolabeling reaction was conducted in the presence of excess ketoprofen, a clinical drug containing the benzophenone framework. Excess ketoprofen blocked the photolabeling more efficiently than excess benzophenone (4-methylbenzophenone) (Fig. 2), even though both blockers have the same benzophenone chromophore. Since the concentration of the benzophenone chromophore was same in both cases, the photochemical quenching mechanism could not account for the observed difference between the two blockers. It was more likely that the difference in binding affinity to GLO1 resulted in the different blocking efficiency. Thus the result suggested that human GLO1 had a hydrophobic binding-pocket for benzophenone analogs.
Figure 2.
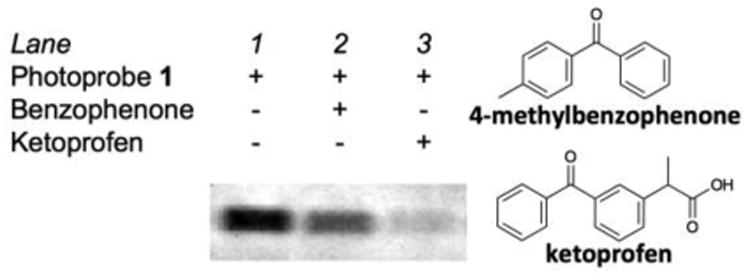
Photoaffnity-labeling of rhGLO1 by 1 (7 μM) in the presence of excess (20×, 140 μM) benzophenone (4-methylbenzophenone) and ketoprofen. See the caption for Fig. 1b and Supporting Information for more experimental details.
The finding opened a possibility that human GLO1 might also recognize various drugs containing the benzophenone framework. To test this, similar blocking experiments were carried out with a series of benzophenone analogs (Supplementary Fig. S2). It turned out that, in addition to ketoprofen, fenofibrate and doxorubicin blocked the photolabeling. No blocking was observed with carmine, which contains a highly substituted benzophenone framework, whereas resveratrol, which has a hydrophobic framework with two benzene rings, was also an efficient blocker of photolabeling. Taken together, these results suggested that various structural analogs of benzophenone bind to human GLO1.
Next, we examined whether drug binding affects the catalytic activity of human GLO1. Kinetic analyses were carried out for rhGLO1 treated with quercetin (a known inhibitor of GLO1) and ketoprofen at different substrate concentrations. The Dixon plot analysis showed that quercetin (a positive control) potently inhibited GLO1 with a Ki value of 1.69 ± 0.14 μM (Fig. 3a). Ketoprofen, on the other hand, did not inhibit the catalytic activity of GLO1 (Fig. 3b).
Figure 3.
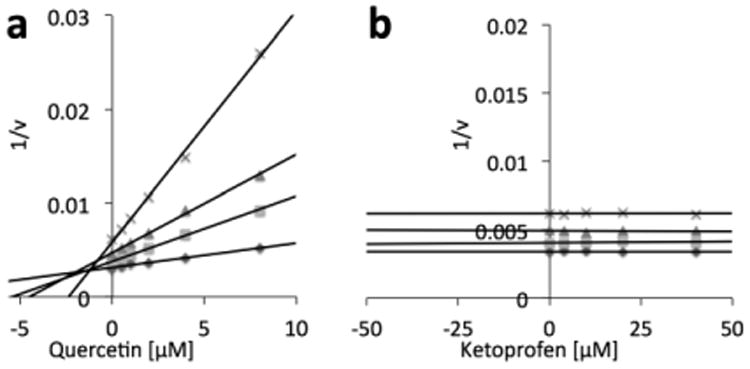
The effect of ketoprofen binding on the catalytic activity of rhGLO1. (a) The Dixon plot of quercetin, a known inhibitor of GLO1 (positive control). Quercetin gave a Ki value of 1.69 ± 0.14 μM in this assay. (b) The Dixon plot of ketoprofen. The initial substrate concentrations were (◆) 400 μM, (■) 200 μM, (▲) 100 μM, and (×) 50 μM.
The current work indicates the presence of a hydrophobic binding pocket for benzophenone analogs in human GLO1. Binding of ketoprofen did not have any effect on the catalytic activity, suggesting that the binding pocket is located outside of the active site. Similar binding of small molecules to GLO1 has been observed previously by using affinity chromatography. In one study, nonsteroidal antiinflammatory drugs (NSAIDS), including ketoprofen, were found to bind to mouse GLO1 (Supplementary Fig. S3).17 Another study found that methylgerfelin, an inhibitor of osteoclastogenesis, binds to mouse GLO1 (Supplementary Fig. S3).18 As such, GLO1 may recognize a wide variety of hydrophobic molecules. It is known that GLO1 expression is up-regulated in many human cancers with high glycolytic rates, and the overexpression of GLO1 has been associated with multidrug resistance.19 Our finding of the drug-binding pocket on human GLO1 provides a new clue to understand the mechanism of multidrug resistance associated with this enzyme.
The benzophenone-binding site on GLO1 is also important in photoaffinity-labeling studies. In chemical proteomics and drug-target identification, benzophenone photophores above micromolar concentration might identify GLO1 especially when studies use cells or tissues overexpressing this enzyme. In such cases, careful control experiments are needed to decipher whether GLO1 is indeed the target of the drug of interest.
In conclusion, the current work suggests that GLO1 is an intracellular “magnet” for benzophenone analogs and possibly other hydrophobic drugs. The finding provides new clues to understand multidrug resistance as well as to improve photoaffinity-labeling studies.
Supplementary Material
Acknowledgments
This research was supported in part by PSC CUNY grant (PSCREG-37-813). NIMHD/NIH G12 MD007599-27, which supports the research infrastructure of Hunter College, is also acknowledged.
Footnotes
Supplementary Material: Supplementary data associated with this article can be found in the online version at xxxxx
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Dorman G, Prestwich GD. Biochemistry. 1994;33:5661. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 2.Prestwich GD, Dorman G, Elliott JT, Marecak DM, Chaudhary A. Photochem Photobiol. 1997;65:222. doi: 10.1111/j.1751-1097.1997.tb08548.x. [DOI] [PubMed] [Google Scholar]
- 3.Wittelsberger A, Mierke DF, Rosenblatt M. Chem Biol Drug Des. 2008;71:380. doi: 10.1111/j.1747-0285.2008.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salisbury CM, Cravatt BF. Proc Natl Acad Sci U S A. 2007;104:1171. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salisbury CM, Cravatt BF. J Am Chem Soc. 2008;130:2184. doi: 10.1021/ja074138u. [DOI] [PubMed] [Google Scholar]
- 6.Tantama M, Lin WC, Licht S. J Am Chem Soc. 2008;130:15766. doi: 10.1021/ja805868x. [DOI] [PubMed] [Google Scholar]
- 7.Kantor TG. Pharmacotherapy. 1986;6:93. doi: 10.1002/j.1875-9114.1986.tb03459.x. [DOI] [PubMed] [Google Scholar]
- 8.Yang LP, Keating GM. Am J Cardiovasc Drugs. 2009;9:401. doi: 10.2165/11203920-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Tacar O, Sriamornsak P, Dass CR. J Pharm Pharmacol. 2013;65:157. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 10.As of July 2015.
- 11.Hindi S, Deng H, James L, Kawamura A. Bioorg Med Chem Lett. 2006;16:5625. doi: 10.1016/j.bmcl.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 12.Kawamura A, Hindi S. Chirality. 2005;17:332. doi: 10.1002/chir.20169. [DOI] [PubMed] [Google Scholar]
- 13.Kawamura A, Hindi S, Mihai DM, James L, Aminova O. Bioorg Med Chem. 2008;16:8824. doi: 10.1016/j.bmc.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamura A, Mihai DM. Methods Mol Biol. 2012;803:65. doi: 10.1007/978-1-61779-364-6_6. [DOI] [PubMed] [Google Scholar]
- 15.Fields GB, Noble RL. Int J Pept Protein Res. 1990;35:161. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 16.EC50 refers to the photoprobe concentration at which 50% of the maximum photolabeling is observed.
- 17.Sato S, Kwon Y, Kamisuki S, Srivastava N, Mao Q, Kawazoe Y, Uesugi M. J Am Chem Soc. 2007;129:873. doi: 10.1021/ja0655643. [DOI] [PubMed] [Google Scholar]
- 18.Kawatani M, Okumura H, Honda K, Kanoh N, Muroi M, Dohmae N, Takami M, Kitagawa M, Futamura Y, Imoto M, Osada H. Proc Natl Acad Sci U S A. 2008;105:11691. doi: 10.1073/pnas.0712239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thornalley PJ, Rabbani N. Semin Cell Dev Biol. 2011;22:318. doi: 10.1016/j.semcdb.2011.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


