Abstract
NRF2 is a transcription factor that promotes antioxidant and drug-metabolizing gene expression. It also regulates the transcription of genes involved in carbohydrate and lipid metabolism, NADPH regeneration, heme and iron metabolism, as well as proteasome metabolism. Emerging research has identified NRF2 as a critical factor for promoting survival of mammalian cells subjected to ionizing radiation. At a mechanistic level, NRF2 promotes the repair of DNA damage and drives detoxification of superoxide that is generated hours to days after irradiation. This review summarizes research in these areas and discusses targeting of NRF2 in radiation resistant cancer and NRF2’s role in mitigating Acute Radiation Syndrome.
Keywords: Nrf2, ionizing radiation, reactive oxygen species, DNA repair, cell and tissue damage
Introduction
Electrophiles and reactive oxygen intermediates, as well as reactive nitrogen species, can contribute significantly to the etiology of many chronic human diseases. This knowledge has driven a major research effort that focused on providing mechanistic insight and guidance for the development of redox-based therapeutic strategies. The effort, however, was hampered by the structural diversity of electrophilic and oxidative compounds. It took the pioneering work of Talalay and colleagues [1-3] and Pickett and associates [4, 5], as well as research from Violet Daniel’s laboratory [6, 7], to provide a fundamental molecular framework that ultimately was used to explain how a thiol-based protein sensor distinguished between different types of chemistries and translated the information into physiological responses. The sensor is kelch-like ECH-associated protein 1 (Keap1), originally characterized by Itoh et al. [8]. The murine protein contains 25 and the human contains 27 cysteine residues that function as redox sensors capable of integrating diverse chemistries [9], including radiolytically-generated hydroxyl radical (•OH) and hydrogen peroxide [10], into a common signal: activation of nuclear factor erythroid 2-like 2, or NRF2 (reviewed in [11].
NRF2 (HGNC:7782), encoded by NFE2L2, is a member of the cap ‘n’ collar (CnC) family of basic leucine zipper transcription factors [12] that are conserved in mammals [13], birds [8], fish [14], insects [15], and worms [16], but not expressed in plants and fungi [13]. The family is composed of the transcription factors SKN-1, NRF1, NRF2, NFE2, NRF3, CncC, BACH1 and BACH2 [13]. These proteins are characterized by a leucine zipper protein-protein dimerization domain as well as CnC and abasic domains that confer DNA binding activity [17]. An NMR solution structure of the DNA binding domain may be found at http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=105542.
Nrf2 was originally identified as a key regulator of canonical antioxidant and drug-metabolizing gene expression [18, 19]. Nrf2 heterodimerization with MAF-G [20] or the JUN CnC-bZIP factor [21] licenses binding to cis antioxidant response elements (AREs) located in proximal promoters of Nrf2 target genes that now number over 500 [20], including those involved in carbohydrate and lipid metabolism, NADPH regeneration, heme and iron metabolism, as well as proteasome metabolism [19, 22]. The functional ARE has recently been defined as TMANNRTGACTCAGCRWWWW, where M = A or C, R= A or G, and W = A or T [20]. Emerging research now shows that Nrf2-mediated transcription can protect cells and tissues from the pathogenic consequences of hydroxyl radicals that are directly generated by ionizing radiation as well as the hydrogen peroxide and superoxide that are generated as a secondary consequence of irradiation.
NRF2 promotes survival of irradiated cells
Preclinical cell culture models have been used to address the question of whether Nrf2 impacts survival of irradiated cells. Keap1−/− mouse embryo fibroblasts (MEFs) constitutively overexpress Nrf2 and Nrf2 target genes and are characterized by low levels of intracellular reactive oxygen species (ROS) and a radiation-resistant phenotype compared to wild-type MEFs [23]. The generalized term ROS is used in this review when the initial oxidizing species were not identified in the cited papers [24] and has been defined by C Winterbourn as “those initial species generated by oxygen reduction (eg, superoxide) as well as all secondary reactive products. The definition includes overlapping reactive nitrogen species” [25]. Relative to wild type MEFs, Nrf2−/− MEFs express high levels of intracellular ROS and are intrinsically radiosensitive [23, 26]. Activation of Nrf2/NRF2 signaling due to electrophilic adduction of Keap1 or a deficiency in the expression/function of Keap1 has been shown to lower intracellular ROS and confer radioresistance in fibroblasts [27], bronchial and breast epithelial cells [28], DU145 prostate cells [29], squamous cell lung cancer [30], and glioblastoma cells [31]. RNA interference (RNAi) or pharmacological targeting of NRF2 in DU145 prostate cancer cells [29, 32], non-small cell lung cancer A549, H460, or H1299 cells [23, 33], or gliobastoma cells [31] elevates ROS and produces a corresponding radiosensitive phenotype. Taken all together, these investigations support a hypothesis that NRF2 promotes a pro-survival response in irradiated cells.
Molecular effects of ionizing radiation
Initial events
The term ionizing radiation describes a photon or particle with sufficient energy to displace orbital electrons from atoms, thereby yielding ions and ionized electrons [34]. Coulomb interactions occur between ionized charged particles (e.g., an electron) moving through a medium such as a cell and the orbital electrons of the constituent atoms. These interactions result in a transfer of kinetic energy from the ionized charged particles to the electrons in the medium [35] and are quantified as absorbed dose (D), which is defined as the absorption of energy in a medium of known mass by ionizing particles. [35]The units of D are Gy (the SI unit; 1 Gy = 1J/kg) or rad, which is equal to 0.01 Gy. In the case of X- or γ-irradiation, 70% of photons traversing a cell interact with water molecules that ultimately decompose into hydroxyl radicals (•OH), hydrogen radicals (•H), hydrogen peroxide, superoxide, and solvated electrons (eaq−) [36]. The hydroxyl radical can react at diffusion controlled rates with all four purine and pyrimidine bases, as well as 2-deoxyribose. However, neither superoxide nor hydrogen peroxide reacts significantly with DNA bases or 2-deoxyribose [37] and as discussed below, radiation-induced damage to DNA is a critical event. Thus, the initial reactions relevant to this review can be described as follows [38-40].
| (1) |
| (2) |
| (3) |
| (4) |
The radical cation in eq 1 (H2O+•) can donate a proton to a nearby water molecule within 10−14 seconds to yield H3O and the hydroxyl radical (•OH), eq 3 while H2O* (eq 2) can decompose into •OH + •H (eq 4). These reactions are complete on a time scale of milliseconds.
G-value is a term used to quantify the chemical effects of ionizing radiation. The term was originally defined as the number of molecules transformed, produced, destroyed, or changed per 100 eV of energy absorbed. In SI units the G-value is assigned a value of mol/J. G-values are energy-dependent. For example, Cobalt 60 emits two monoenergetic photons of 1.17 and 1.34 MeV. The G-value for the hydroxyl radical was calculated to be 2.74 molecules formed per 100 eV absorbed [38-40]. The LD50/60 is a term to describe the mean lethal dose that will produce 50% mortality in a population over 60 days. For humans the LD50/60 is approximately 4 Gy [34]. The G-values discussed above allow one to calculate the concentration of hydroxyl radicals generated by a given dose of radiation. For example, 4 Gy will generate approximately 20.4 μmol of hydroxyl radicals in a person who weighs 190 pounds.
Radiation-induced damage to DNA
While all cellular macromolecules are susceptible to attack by •OH, damage to DNA represents a critical injury with pluripotent consequences: cell death, tissue injury, and disease. Hydroxyl radicals can abstract a hydrogen atom from the methyl group of thymine and from each carbon atom of the 2-deoxyribose moiety [41]. [41][42]Hydroxyl radicals are able to add to the double bonds in DNA bases to generate hydroxyl DNA base radicals [41]. These and other reactions can result in apurinic/apyrimidinic (AP) sites, single strand breaks [42], double strand breaks (DSBs), and protein DNA crosslinks, as well as other types of DNA damage. It is estimated that 4 Gy will damage more than 2000 base pairs and generate 2000 single strand breaks per cell nucleus, as well as approximately 80 DNA DSBs per nucleus. In addition to hydroxyl-mediated attack on chromatin, 30% of DNA damage is a consequence of direct interaction with ionizing particles.
NRF2 promotes repair of damaged DNA
Base excision repair
Damaged bases, abasic sites, and single strand breaks are repaired by the base excision repair pathway. This pathway can be briefly summarized as follows: a) base recognition and excision by lesion-specific DNA glycosylases, b) incision by endonucleases [43] is followed by formation of an abasic site, c) replacement of the excised base, and d) appropriate end-terminal processing and ligation [44]. These steps are performed with the aid of enzymes such as DNA Polymerase β, poly(ADP-ribose) polymerase 1 (PARP1), and XRCC1/LIG3 [42].
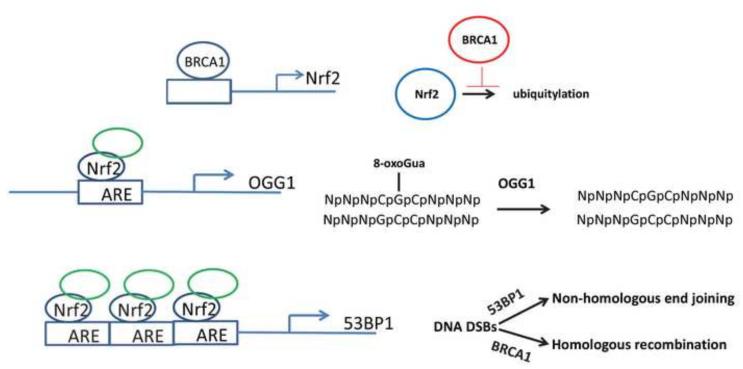
8—oxo-7,8-dihydroguanine (8-oxoGua) is a highly mutagenic DNA lesion that is detected in x-irradiated cells [45{Chen, 2001 #108][45, 46] . It can cause GC to TA transversion mutations [47]. 8-oxoGua lesions are repaired by the enzyme 8-oxoguanine DNA glycosylase (OGG1). Human OGG1 is located on chromosome 3p25 [48]. Alternative splicing of OGG1 results in expression of mitochondrial and nuclear proteins [48]. Dhenaut et al. [49] have shown that the human OGG1 promoter harbors an ARE 29 base pairs from the transcriptional start site. Analysis of 2 kb of the OGG1 promoter region using a luciferase fusion reporter provided evidence of a functional ARE [49]. Chromatin immunoprecipitation (ChIP) assays confirmed that Nrf2 binds to the OGG1 promoter[50] and RNAi experiments demonstrated that NRF2 deficiencies suppressed OGG1 expression [50]. The research of Hyun et al. [51] has shown that OGG1 deficiencies increase the radiation sensitivity of human cells, thus supporting a hypothesis that posits a mechanistic link between Nrf2, repair of DNA base damage, and radiation sensitivity.
Repair of DNA double strand breaks
Radiation-induced lethal damage is a consequence, in large part, to a DNA DSBs. The majority of DNA DSBs are generated by 2 independent hydroxyl radicals that interact in close proximity [34]. DSBs are repaired by either non-homologous end joining (NHEJ) or homologous recombination (HR). In brief, canonical NHEJ, which operates in all phases of the somatic mammalian cell cycle but represents the major pathway in G0/G1 cells [52-54], is activated by DNA DSBs which the Ku70/Ku80 complex binds to and stabilizes [55]. DNA-dependent protein kinase is then recruited to the breaks, becomes activated, and phosphorylates target proteins, including the endonuclease Artemis that processes DNA ends with overhangs. Finally, the XRCC4-DNA ligase 4 (LIG4) complex is recruited and ligates the DNA strand with the help of the XRCC4-like factor (XLF). In addition to XRCC4-LIG4–dependent C-NHEJ, at least one alternative end-joining (A-EJ) pathway exists. This pathway involves microhomology and is mainly used in cells with defects in C-NHEJ [52-54].
HR, which is restricted to late S and G2 cells, begins with resection of the 5’-strands within the DNA DSB, yielding 3’-single stranded DNA overhands that are coated with replication protein A (RPA) [55, 56]. RAD51 then displaces RPA, forming a nucleoprotein filament (one RAD51 monomer for every 3 nucleotides). The filament aligns with homologous DNA sequences on a sister chromatid, forming a structure termed a D-loop. Strand invasion, followed by the capture of a second 3’-single strand DNA overhang that was created by the resection process, forms a Holliday junction. Finally, the junction is dissolved (the major pathway) or cleaved to generate double strand DNA. Cleavage can produce non-crossover and crossover products. Experimental evidence acquired to date indicates that non-crossover products predominate. In addition, there is an alternative HR pathway termed synthesis-dependent strand annealing that is independent of Holliday junction formation.
The choice of repair pathway is impacted by whether or not the broken DNA ends undergo extensive 5’ to 3’ nucleolytic resection that generates 3’ DNA overhangs [57]. Resection promotes initiation of the HR pathway. The question of whether resection will be regulated by the interactions of 53BP1 and breast cancer type 1 susceptibility protein (BRCA1) was reviewed in [58]. A model has been proposed in which 53BP1 is phosphorylated in G1 cells by ataxia telangiectasia mutated (ATM), localizes to sites of DNA breaks, and attenuates HR. In S and G2 cells the protein CtIP is phosphorylated by CDK, inducing the formation of a complex with BRCA1 and Mre11/Rad50/Nbs1. This complex displaces 53BP1 and initiates resection [58].
Emerging research has shown that transcriptional regulation of 53BP1 is regulated in part by Nrf2. Kim et al. [59] examined 53BP1’s proximal promoter region for cis regulatory elements and found 3 putative AREs in normal human colonic epithelial cells. ChIP assays confirmed Nrf2 binding to all 3 cis acting AREs. RNAi-mediated suppression of Nrf2 prevented the electrophilic triterpenoid bardoxolone methyl from protecting human colonic epithelial cells from radiation-induced cytotoxicity [59]. These results are consistent with the work of others who have shown that 53BP1 deficiencies increase radiation sensitivity [60-62].
BRCA1 has several significant roles in the DNA DSB repair process, including regulation of CtIP-mediated DNA end resection [55]. BRCA1 deficiencies result in elevation of ROS and a corresponding radiation hypersensitivity phenotype [63]. Thus, BRCA1 promotes high fidelity DNA repair while suppressing genotoxic ROS. The mechanism involves BRCA1’s ability to bind to Nrf2’s proximal promoter and positively regulate Nrf2 mRNA expression [64, 65]. In addition, BRCA1 physically interacts with and stabilizes Nrf2, thereby promoting Nrf2-mediated promoter transactivation [65]. These concepts are summarized in Figure 1.
Figure 1.
Role of Nrf2 in repair of DNA damage. BRCA1 can promote Nrf2 expression by both increasing transcription and inhibiting Nrf2 degradation. Nrf2 has been shown to bind to AREs located in the proximal promoters of the OGG1 gene and 53PB1, thereby increasing expression of these DNA repair proteins.
Loss of BRCA1 increases the risk of breast, ovary, and fallopian tube cancers (see reference [66] for review). Loss of BRCA1 (or BRCA2) in cancer can be exploited by targeting PARP1, a facilitator of DNA repair [67]. Recently, Wu et al. [68] found that PARP1 interacts directly with small Maf proteins, NRF2’s heterodimeric partners, and that this complex augments Nrf2’s ARE-specific DNA binding, thereby enhancing Nrf2-dependent gene transcription. Thus, one may hypothesize that the therapeutic efficacy obtained by molecular targeting of PARP1 in cancer could be due in part to suppression of Nrf2-mediated DNA repair.
At a biochemical level, acquisition of DNA damage in mitotically active mammalian cells activates the Mre11/Rad50/NBS1 sensor complex whose binding and processing of DNA damage initiates ATM and/or ATR activation, which are followed by transduction of signals to downstream effector pathways [69]. One of these effector proteins, TP53 (p53), is an important regulator of radiation-induced cell cycle checkpoint signaling and apoptosis [70]. ATM-mediated activation of p53 has been shown to suppress Nrf2-mediated target gene expression and contribute to an enhanced apoptotic response [71].
Nrf2 and induction of apoptosis following X-irradiation
Although it is well established that hydroxyl radical-induced nuclear DNA damage is responsible for a significant proportion of cell death following exposure to ionizing radiation, it appears that superoxide generated hours to days after irradiation can also impact radiation sensitivity. Gao et al [72]found that radiation resistance was increased in human glioma cells over expressing SOD1. Similarly, Petkau [73] found that SOD1 activity increased the recovery of hematopoietic myeloid progenitor cell recovery in X-irradiated mice. The mechanisms appears to be a consequence of suppression of superoxide-mediated induction of apoptosis [72]. Although radiation-induced apoptosis is not considered to play a large role in cell death sub-routine execution, computerized video analysis of cell death by Dewey and colleagues demonstrated that apoptosis during interphase was the primary mode of cell death initiated by mitotic catastrophe [74, 75]. SOD1 is a Nrf2 target gene [76]. Thus, one may hypothesize that elevated levels of Nrf2 would lead to increased SOD1 expression and radioprotection.
In total, these studies show that NRF2 contributes to a pro-survival response due to an enhanced detoxification of pathogenic superoxide and promotion of •OH-mediated DNA damage repair. As many human cancers are characterized by elevated NRF2 and NRF2-mediated gene expression [23, 77, 78], the question of targeting NRF2 for treatment of radioresistant disease gains importance. However, this question needs to be addressed in the context of the radiation response of normal tissue.
The radiation response of normal tissue is NRF2-dependent
Stem and progenitor cells are defined by their self-renewal capacity and ability to differentiate. The intrinsic radiosensitivity of mammalian hematopoietic stem and progenitor cells (defined as LIN− SCA1+ c-KIT+) dictates the LD50 dose. Irradiated stem cells can undergo apoptosis, cell cycle arrest, or senescence, all initiated by DNA damage [79, 80], and these responses can negatively impact cell survival and homeostasis, which are critical for prevention/mitigation of the life-threatening Acute Radiation Syndrome (ARS). Recently it has been shown that the Nrf2−/− mouse exhibits a greater radiation sensitivity compared to the wild-type mouse and that the severity of ARS can be partially mitigated by pharmacological activation of NRF2 [59, 79].
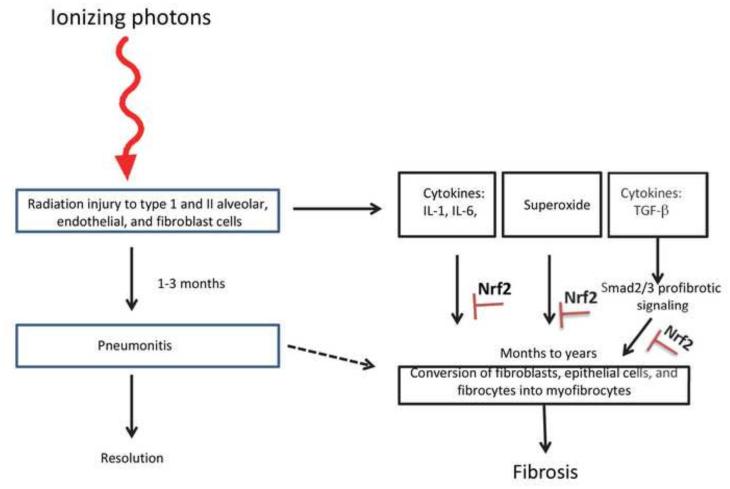
Life-threatening radiation-induced pulmonary fibrosis, characterized by the loss of parenchyma, the progressive accumulation of myofibroblast progenitors, the development of fibrosis, and the subsequent remodeling of lung interstitium, presents 6 to 24 months after absorption of ionizing energy ([81] and references therein, Figure 2). Transforming growth factor (TGF)-β and ROS contribute significantly to the pathogenesis of radiation-induced pulmonary fibrosis [82, 83]. Travis et al. [81] found that Nrf2 binds to CAGA elements in the proximal promoter of the TGF-β1 target gene plasminogen activator inhibitor (PAI)-1 and suppresses its expression. Others have found that transduction of TGF-β1/Smad2/3 target genes collagen 1A1, fibronectin-1, tissue inhibitor of matrix metalloproteinase 1, and PAI-1 were suppressed by elevated levels of Nrf2, a consequence of genetic targeting of Keap1 or electrophilic isothiocyanate inactivation of Keap1 [84, 85]. Consistent with these molecular studies, Travis et al. [81] found that PAI-1 expression in lung was elevated in thoracic irradiated Nrf2−/− C57BL mice compared to irradiated wild-type mice. Irradiated Nrf2−/− mice had fewer alveoli compared to their wild-type counterparts and these were more distended. Most importantly, the life span of Nrf2−/− mice was shortened by thoracic irradiation [81].
Figure 2.
Brief overview of the development of radiation-induced pulmonary fibrosis, summarized from refs [89, 90]. Low linear energy transfer ionizing radiation injures epithelial and stroma cells. One to 3 months later pneumonitis can develop, which can either resolve, convert to chronic inflammation or convert to fibrosis. The relationship, if any, between pneumonitis and fibrosis is not well characterized. Irradiation results in cytokine production, exemplified by IL-1, IL-6 and TGF-β. Additionally, chromic superoxide generation is observed. Cytokines and superoxide contribute to myofibroblast generation, which promotes collagen deposition and the development of fibrosis. Nrf2 can inhibit fibrosis due to its anti-inflammatory activity, it ability to induce antioxidant enzymes and its suppression of R-Smad-dependent expression of pro-fibrotic gene expression.
Rana et al. [86] found that Nrf2 deficiency was associated with a drastic overall decrease in bone volume after irradiation, as quantified by microCT analysis. Loss of bone volume in Nrf2−/− mice was associated with a decrease in osteoblast mineralization and an increase in osteoclasts. RT-PCR analysis of calvarial osteoblasts revealed that in the absence of Nrf2, expression of RANKL was increased after irradiation but could be suppressed by treatment with N-acetyl cysteine, implicating a role for ROS in radiation-induced bone injury.
Inflammatory cytokines are critical to the radiation response of normal tissue (reviewed in [87]) and Nrf2’s role in regulating cytokine expression in inflammatory disease is well characterized ([88] and references therein). Although not well studied, one may hypothesize that part of Nrf2’s role in promoting cell survival following irradiation is linked to modulation of the cytokine response.
Summary
Nrf2 promotes cell survival in irradiated cells and tissues through ROS detoxification, supporting DNA repair and potentially modulating cytokine responses. Although overexpression of NRF2 in human cancer cells may produce a radioresistance phenotype, there is a strong possibility that targeting NRF2 will result in significant normal tissue toxicity. However, administration of NRF2 activators has the potential for ARS mitigation following deployment of a radiological dispersal device.
Acknowledgments
This work was supported in part by NIH/NHLBI grant RO1HL112286. We wish to thank Melissa Stauffer, PhD for her careful reading of the manuscript.
Abbreviations
- ARE
antioxidant response element
- ARS
Acute radiation syndrome
- ATM
Ataxia telangiectasia mutated
- BRCA1
Breast cancer type 1 susceptibility protein
- ChIP
Chromatin immunoprecipitation
- CnC
Cap ‘n’ collar family of basic leucine zipper transcription factors
- DSB
Double strand break
- HR
Homologous recombination
- Keap1
Kelch-like ECH-associated protein 1
- LD50
Lethal dose for 50% of a population
- MEF
Mouse embryo fibroblast
- NADPH
Nicotinamide adenine dinucleotide phosphate
- NHEJ
Non-homologous end joining
- NRF2
Human nuclear factor (erythroid-derived 2)-like 2 protein
- Nrf2
Mouse nuclear factor (erythroid-derived 2)-like 2 protein
- OGG1
8-Oxoguanine DNA glycosylase
- •OH
Hydroxyl radical
- PARP1
Poly(ADP-ribose) polymerase 1
- RNAi
RNA interference
- ROS
Reactive oxygen species
- RPA
Replication protein A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Talalay P, De Long MJ, Prochaska HJ. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc Natl Acad Sci U S A. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Holtzclaw WD, Dinkova-Kostova AT, Talalay P. Protection against electrophile and oxidative stress by induction of phase 2 genes: the quest for the elusive sensor that responds to inducers. Adv Enzyme Regul. 2004;44:335–367. doi: 10.1016/j.advenzreg.2003.11.013. [DOI] [PubMed] [Google Scholar]
- [3].Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nguyen T, Rushmore TH, Pickett CB. Transcriptional regulation of a rat liver glutathione S-transferase Ya subunit gene. Analysis of the antioxidant response element and its activation by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate. J Biol Chem. 1994;269:13656–13662. [PubMed] [Google Scholar]
- [5].Nguyen T, Huang HC, Pickett CB. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- [6].Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc Natl Acad Sci U S A. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Friling RS, Bergelson S, Daniel V. Two adjacent AP-1-like binding sites form the electrophile-responsive element of the murine glutathione S-transferase Ya subunit gene. Proc Natl Acad Sci U S A. 1992;89:668–672. doi: 10.1073/pnas.89.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Holland R, Hawkins AE, Eggler AL, Mesecar AD, Fabris D, Fishbein JC. Prospective type 1 and type 2 disulfides of Keap1 protein. Chem Res Toxicol. 2008;21:2051–2060. doi: 10.1021/tx800226m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Di Simplicio P, Cheeseman KH, Slater TF. The reactivity of the SH group of bovine serum albumin with free radicals. Free Radic Res Commun. 1991;14:253–262. doi: 10.3109/10715769109088954. [DOI] [PubMed] [Google Scholar]
- [11].Sekhar KR, Rachakonda G, Freeman ML. Cysteine-based regulation of the CUL3 adaptor protein Keap1. Toxicol Appl Pharmacol. 2010;244:21–26. doi: 10.1016/j.taap.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- [15].Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- [17].Chevillard G, Blank V. NFE2L3 (NRF3): the Cinderella of the Cap'n'Collar transcription factors. Cell Mol Life Sci. 2011;68:3337–3348. doi: 10.1007/s00018-011-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- [19].Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- [20].Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Singh A, Bodas M, Wakabayashi N, Bunz F, Biswal S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid Redox Signal. 2010;13:1627–1637. doi: 10.1089/ars.2010.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Forman HJ, Augusto O, Brigelius-Flohe R, Dennery PA, Kalyanaraman B, Ischiropoulos H, Mann GE, Radi R, Roberts LJ, 2nd, Vina J, Davies KJ. Even free radicals should follow some rules: a guide to free radical research terminology and methodology. Free Radic Biol Med. 2015;78:233–235. doi: 10.1016/j.freeradbiomed.2014.10.504. [DOI] [PubMed] [Google Scholar]
- [25].Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- [26].McDonald JT, Kim K, Norris AJ, Vlashi E, Phillips TM, Lagadec C, Della Donna L, Ratikan J, Szelag H, Hlatky L, McBride WH. Ionizing radiation activates the Nrf2 antioxidant response. Cancer Res. 2010;70:8886–8895. doi: 10.1158/0008-5472.CAN-10-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mathew ST, Bergstrom P, Hammarsten O. Repeated Nrf2 stimulation using sulforaphane protects fibroblasts from ionizing radiation. Toxicol Appl Pharmacol. 2014;276:188–194. doi: 10.1016/j.taap.2014.02.013. [DOI] [PubMed] [Google Scholar]
- [28].El-Ashmawy M, Delgado O, Cardentey A, Wright WE, Shay JW. CDDO-Me Protects Normal Lung and Breast Epithelial Cells but Not Cancer Cells from Radiation. PLoS One. 9 doi: 10.1371/journal.pone.0115600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Abazeed ME, Adams DJ, Hurov KE, Tamayo P, Creighton CJ, Sonkin D, Giacomelli AO, Du C, Fries DF, Wong KK, Mesirov JP, Loeffler JS, Schreiber SL, Hammerman PS, Meyerson M. Integrative radiogenomic profiling of squamous cell lung cancer. Cancer Res. 2013;73:6289–6298. doi: 10.1158/0008-5472.CAN-13-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cong ZX, Wang HD, Zhou Y, Wang JW, Pan H, Zhang DD, Zhang L, Zhu L. Temozolomide and irradiation combined treatment-induced Nrf2 activation increases chemoradiation sensitivity in human glioblastoma cells. J Neurooncol. 2014;116:41–48. doi: 10.1007/s11060-013-1260-x. [DOI] [PubMed] [Google Scholar]
- [32].Jayakumar S, Kunwar A, Sandur SK, Pandey BN, Chaubey RC. Differential response of DU145 and PC3 prostate cancer cells to ionizing radiation: role of reactive oxygen species, GSH and Nrf2 in radiosensitivity. Biochim Biophys Acta. 2014;1840:485–494. doi: 10.1016/j.bbagen.2013.10.006. [DOI] [PubMed] [Google Scholar]
- [33].Lee S, Lim MJ, Kim MH, Yu CH, Yun YS, Ahn J, Song JY. An effective strategy for increasing the radiosensitivity of Human lung Cancer cells by blocking Nrf2-dependent antioxidant responses. Free Radic Biol Med. 2012;53:807–816. doi: 10.1016/j.freeradbiomed.2012.05.038. [DOI] [PubMed] [Google Scholar]
- [34].Hall EJ. Radiobiology for the Radiologist. Lippincott Williams & Wilkines; NY,NY: 2000. [Google Scholar]
- [35].Tubiana Maurice DJ, Wambersie Andre . Introduction to Radiobiology. Philadelphia Taylor & Francis; London, New York: 1990. [Google Scholar]
- [36].Collinson E, Dainton FS, Kroh J. Effects of linear energy transfer on the radiolysis of water and heavy water. Nature. 1960;187:475–477. doi: 10.1038/187475a0. [DOI] [PubMed] [Google Scholar]
- [37].Cadet J, Douki T, Ravanat JL. Oxidatively generated base damage to cellular DNA. Free Radic Biol Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- [38].Laszlo W. Handbook of Nuclear chemistry: Chemical applications of nuclear reactions and radiations. Springer; Dordrecht, Heidelberg, London, New York: 2011. [Google Scholar]
- [39].Allen AO HC, Ghormley JA, Davis TW. Decomposition of water and aqueous solutions under mixed fast neutron and gamma radiatio. Journal of Physical Chemistry. 1952;56:575–586. [Google Scholar]
- [40].CJ H. Effects of Cobalt gamma-Radiations on water and aqueous solutions. Journal of Physical Chemistry. 1952;56:587–594. [Google Scholar]
- [41].Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- [42].Brenerman BM, Illuzzi JL, Wilson DM., 3rd Base excision repair capacity in informing healthspan. Carcinogenesis. 2014;35:2643–2652. doi: 10.1093/carcin/bgu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Patrono C, Sterpone S, Testa A, Cozzi R. Polymorphisms in base excision repair genes: Breast cancer risk and individual radiosensitivity. World J Clin Oncol. 2014;5:874–882. doi: 10.5306/wjco.v5.i5.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Iyama T, Wilson DM., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst) 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Orimo H, Mei N, Boiteux S, Tokura Y, Kasai H. Analysis of 8-hydroxyguanine (8-OH-Gua) released from DNA by the formamidopyrimidine DNA glycosylase (Fpg) protein: a reliable method to estimate cellular oxidative stress. J Radiat Res. 2004;45:455–460. doi: 10.1269/jrr.45.455. [DOI] [PubMed] [Google Scholar]
- [46].Chen SK, Tsai MH, Hwang JJ, Chang WP. Determination of 8-oxoguanine in individual cell nucleus of gamma-irradiated mammalian cells. Radiat Res. 2001;155:832–836. doi: 10.1667/0033-7587(2001)155[0832:dooiic]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [47].Hazra TK, Izumi T, Maidt L, Floyd RA, Mitra S. The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- [49].Dhenaut A, Boiteux S, Radicella JP. Characterization of the hOGG1 promoter and its expression during the cell cycle. Mutat Res. 2000;461:109–118. doi: 10.1016/s0921-8777(00)00042-2. [DOI] [PubMed] [Google Scholar]
- [50].Singh B, Chatterjee A, Ronghe AM, Bhat NK, Bhat HK. Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC Cancer. 13 doi: 10.1186/1471-2407-13-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hyun JW, Cheon GJ, Kim HS, Lee YS, Choi EY, Yoon BH, Kim JS, Chung MH. Radiation sensitivity depends on OGG1 activity status in human leukemia cell lines. Free Radic Biol Med. 2002;32:212–220. doi: 10.1016/s0891-5849(01)00793-6. [DOI] [PubMed] [Google Scholar]
- [52].Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Lobrich M, Jeggo PA. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- [55].Liu T, Huang J. Quality control of homologous recombination. Cell Mol Life Sci. 2014;71:3779–3797. doi: 10.1007/s00018-014-1649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Daley JM, Kwon Y, Niu H, Sung P. Investigations of homologous recombination pathways and their regulation. Yale J Biol Med. 2013;86:453–461. [PMC free article] [PubMed] [Google Scholar]
- [57].Daley JM, Gaines WA, Kwon Y, Sung P. Regulation of DNA pairing in homologous recombination. Cold Spring Harb Perspect Biol. 2014;6:a017954. doi: 10.1101/cshperspect.a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol. 2014;34:1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R, Cornelius C, Wright WE, Pandita TK, Shay JW. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci U S A. 2012;109:E2949–2955. doi: 10.1073/pnas.1207718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Iwabuchi K, Hashimoto M, Matsui T, Kurihara T, Shimizu H, Adachi N, Ishiai M, Yamamoto K, Tauchi H, Takata M, Koyama H, Date T. 53BP1 contributes to survival of cells irradiated with X-ray during G1 without Ku70 or Artemis. Genes Cells. 2006;11:935–948. doi: 10.1111/j.1365-2443.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- [61].Nakamura K, Sakai W, Kawamoto T, Bree RT, Lowndes NF, Takeda S, Taniguchi Y. Genetic dissection of vertebrate 53BP1: a major role in non-homologous end joining of DNA double strand breaks. DNA Repair (Amst) 2006;5:741–749. doi: 10.1016/j.dnarep.2006.03.008. [DOI] [PubMed] [Google Scholar]
- [62].Stewart GS, Stankovic T, Byrd PJ, Wechsler T, Miller ES, Huissoon A, Drayson MT, West SC, Elledge SJ, Taylor AM. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci U S A. 2007;104:16910–16915. doi: 10.1073/pnas.0708408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan XY, Ried T, Deng CX. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- [64].Kang HJ, Hong YB, Kim HJ, Rodriguez OC, Nath RG, Tilli EM, Albanese C, Chung FL, Kwon SH, Bae I. Detoxification: a novel function of BRCA1 in tumor suppression? Toxicol Sci. 2011;122:26–37. doi: 10.1093/toxsci/kfr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gorrini C, Baniasadi PS, Harris IS, Silvester J, Inoue S, Snow B, Joshi PA, Wakeham A, Molyneux SD, Martin B, Bouwman P, Cescon DW, Elia AJ, Winterton-Perks Z, Cruickshank J, Brenner D, Tseng A, Musgrave M, Berman HK, Khokha R, Jonkers J, Mak TW, Gauthier ML. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med. 2013;210:1529–1544. doi: 10.1084/jem.20121337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Moynahan ME. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene. 2002;21:8994–9007. doi: 10.1038/sj.onc.1206177. [DOI] [PubMed] [Google Scholar]
- [67].Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- [68].Wu T, Wang XJ, Tian W, Jaramillo MC, Lau A, Zhang DD. Poly(ADP-ribose) polymerase-1 modulates Nrf2-dependent transcription. Free Radic Biol Med. 2014;67:69–80. doi: 10.1016/j.freeradbiomed.2013.10.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nowsheen S, Yang ES. The intersection between DNA damage response and cell death pathways. Exp Oncol. 2012;34:243–254. [PMC free article] [PubMed] [Google Scholar]
- [70].Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- [71].Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, Cimino F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem. 2006;281:39776–39784. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- [72].Gao Z, Sarsour EH, Kalen AL, Li L, Kumar MG, Goswami PC. Late ROS accumulation and radiosensitivity in SOD1-overexpressing human glioma cells. Free Radic Biol Med. 2008;45:1501–1509. doi: 10.1016/j.freeradbiomed.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Petkau A. Protection of bone marrow progenitor cells by superoxide dismutase. Mol Cell Biochem. 1988;84:133–140. doi: 10.1007/BF00421047. [DOI] [PubMed] [Google Scholar]
- [74].Chu K, Teele N, Dewey MW, Albright N, Dewey WC. Computerized video time lapse study of cell cycle delay and arrest, mitotic catastrophe, apoptosis and clonogenic survival in irradiated 14-3-3sigma and CDKN1A (p21) knockout cell lines. Radiat Res. 2004;162:270–286. doi: 10.1667/rr3221. [DOI] [PubMed] [Google Scholar]
- [75].Forrester HB, Albright N, Ling CC, Dewey WC. Computerized video time-lapse analysis of apoptosis of REC:Myc cells X-irradiated in different phases of the cell cycle. Radiat Res. 2000;154:625–639. doi: 10.1667/0033-7587(2000)154[0625:cvtlao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [76].Chen XL, Kunsch C. Induction of cytoprotective genes through Nrf2/antioxidant response element pathway: a new therapeutic approach for the treatment of inflammatory diseases. Curr Pharm Des. 2004;10:879–891. doi: 10.2174/1381612043452901. [DOI] [PubMed] [Google Scholar]
- [77].Stacy DR, Ely K, Massion PP, Yarbrough WG, Hallahan DE, Sekhar KR, Freeman ML. Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head Neck. 2006;28:813–818. doi: 10.1002/hed.20430. [DOI] [PubMed] [Google Scholar]
- [78].Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- [79].Kim JH, Thimmulappa RK, Kumar V, Cui W, Kumar S, Kombairaju P, Zhang H, Margolick J, Matsui W, Macvittie T, Malhotra SV, Biswal S. NRF2-mediated Notch pathway activation enhances hematopoietic reconstitution following myelosuppressive radiation. J Clin Invest. 2014;124:730–741. doi: 10.1172/JCI70812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chen Q, Xia X, Wu S, Wu A, Qi D, Liu W, Cui F, Jiao Y, Zhu W, Gu Y, Gao H, Zhang X, Cao J. Apoptosis, necrosis, and autophagy in mouse intestinal damage after 15-Gy whole body irradiation. Cell Biochem Funct. 2014;32:647–656. doi: 10.1002/cbf.3068. [DOI] [PubMed] [Google Scholar]
- [81].Travis EL, Rachakonda G, Zhou X, Korhonen K, Sekhar KR, Biswas S, Freeman ML. NRF2 deficiency reduces life span of mice administered thoracic irradiation. Free Radic Biol Med. 2011;51:1175–1183. doi: 10.1016/j.freeradbiomed.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Rube CE, Uthe D, Schmid KW, Richter KD, Wessel J, Schuck A, Willich N, Rube C. Dose-dependent induction of transforming growth factor beta (TGF-beta) in the lung tissue of fibrosis-prone mice after thoracic irradiation. Int J Radiat Oncol Biol Phys. 2000;47:1033–1042. doi: 10.1016/s0360-3016(00)00482-x. [DOI] [PubMed] [Google Scholar]
- [83].Epperly M, Bray J, Kraeger S, Zwacka R, Engelhardt J, Travis E, Greenberger J. Prevention of late effects of irradiation lung damage by manganese superoxide dismutase gene therapy. Gene Ther. 1998;5:196–208. doi: 10.1038/sj.gt.3300580. [DOI] [PubMed] [Google Scholar]
- [84].Oh CJ, Kim JY, Min AK, Park KG, Harris RA, Kim HJ, Lee IK. Sulforaphane attenuates hepatic fibrosis via NF-E2-related factor 2-mediated inhibition of transforming growth factor-beta/Smad signaling. Free Radic Biol Med. 2012;52:671–682. doi: 10.1016/j.freeradbiomed.2011.11.012. [DOI] [PubMed] [Google Scholar]
- [85].Ryoo IG, Ha H, Kwak MK. Inhibitory role of the KEAP1-NRF2 pathway in TGFbeta1-stimulated renal epithelial transition to fibroblastic cells: a modulatory effect on SMAD signaling. PLoS One. 2014;9:e93265. doi: 10.1371/journal.pone.0093265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rana T, Schultz MA, Freeman ML, Biswas S. Loss of Nrf2 accelerates ionizing radiation-induced bone loss by upregulating RANKL. Free Radic Biol Med. 2012;53:2298–2307. doi: 10.1016/j.freeradbiomed.2012.10.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Candeias SM, Testard I. The many interactions between the innate immune system and the response to radiation. Cancer Lett. 2015 doi: 10.1016/j.canlet.2015.02.007. [DOI] [PubMed] [Google Scholar]
- [88].Zhao C, Gillette DD, Li X, Zhang Z, Wen H. Nuclear factor E2-related factor-2 (Nrf2) is required for NLRP3 and AIM2 inflammasome activation. J Biol Chem. 2014;289:17020–17029. doi: 10.1074/jbc.M114.563114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Stone HB, Coleman CN, Anscher MS, McBride WH. Effects of radiation on normal tissue: consequences and mechanisms. Lancet Oncol. 2003;4:529–536. doi: 10.1016/s1470-2045(03)01191-4. [DOI] [PubMed] [Google Scholar]
- [90].Kong FM, Ten Haken R, Eisbruch A, Lawrence TS. Non-small cell lung cancer therapy-related pulmonary toxicity: an update on radiation pneumonitis and fibrosis. Semin Oncol. 2005;32:S42–54. doi: 10.1053/j.seminoncol.2005.03.009. [DOI] [PubMed] [Google Scholar]