Abstract
Consumption of a protein-containing meal by a fasted animal promotes protein accretion in skeletal muscle, in part through leucine stimulation of protein synthesis and indirectly through repression of protein degradation mediated by its metabolite, α-ketoisocaproate. Mice lacking the mitochondrial branched-chain aminotransferase (BCATm/Bcat2), which interconverts leucine and α-ketoisocaproate, exhibit elevated protein turnover. Here, the transcriptomes of gastrocnemius muscle from BCATm knockout (KO) and wild-type mice were compared by next-generation RNA sequencing (RNA-Seq) to identify potential adaptations associated with their persistently altered nutrient signaling. Statistically significant changes in the abundance of 1,486/∼39,010 genes were identified. Bioinformatics analysis of the RNA-Seq data indicated that pathways involved in protein synthesis [eukaryotic initiation factor (eIF)-2, mammalian target of rapamycin, eIF4, and p70S6K pathways including 40S and 60S ribosomal proteins], protein breakdown (e.g., ubiquitin mediated), and muscle degeneration (apoptosis, atrophy, myopathy, and cell death) were upregulated. Also in agreement with our previous observations, the abundance of mRNAs associated with reduced body size, glycemia, plasma insulin, and lipid signaling pathways was altered in BCATm KO mice. Consistently, genes encoding anaerobic and/or oxidative metabolism of carbohydrate, fatty acids, and branched chain amino acids were modestly but systematically reduced. Although there was no indication that muscle fiber type was different between KO and wild-type mice, a difference in the abundance of mRNAs associated with a muscular dystrophy phenotype was observed, consistent with the published exercise intolerance of these mice. The results suggest transcriptional adaptations occur in BCATm KO mice that along with altered nutrient signaling may contribute to their previously reported protein turnover, metabolic and exercise phenotypes.
Keywords: branched-chain amino acids, leucine, α-ketoisocaproate, protein synthesis, protein degradation, glycolysis, mTOR, TCA cycle, eukaryotic initiation factor-2, integrin-linked kinase, signaling, myopathy, dystrophy
the mitochondrial branched-chain aminotransferase (BCATm, encoded by BCAT2 gene) catalyzes the rapid and reversible transamination of branched-chain amino acids (BCAAs, including Leu, Ile, and Val) in many peripheral tissues (36, 53). BCAAs are essential amino acids that frequently represent the most abundant component of dietary protein, and dietary supplementation with BCAA has been linked to positive health benefits such as satiety, lean body mass, muscle protein accretion, and glucose homeostasis (for review see Ref. 49). Paradoxically, increased plasma BCAAs have been shown to be predictive of an increased risk of diabetes mellitus and insulin resistance or have been associated with these disorders (1, 7, 10, 14, 21, 47, 58, 63, 73, 75, 84, 85, 87). While understanding the factors underlying this paradox is an active area of investigation, these positive and negative health associations have frequently been attributed, rightly or wrongly, to the ability of BCAAs to act as direct or indirect nutrient signals. For example, BCAAs appear to regulate the synthesis and/or release of hormones such as ghrelin, GLP-1, insulin, and leptin, which are able to affect eating behavior, glycemia, body composition, and energy balance. As direct nutrient signals they can, for example, activate protein synthesis and exert a brake on protein breakdown in muscle and other tissues (52).
Plasma BCAA concentrations represent a balance between their rate of appearance (Ra) and rate of disappearance (Rd). The factors effecting BCAA Ra include dietary intake (not a factor in fasting plasma studies) and protein breakdown in tissues arising from protein degradation (for review see Ref. 52). The Rd is affected by protein synthesis, BCAA catabolic metabolism (e.g., transamination, oxidation), and excretion. BCAA oxidative metabolism is a multistep process that begins with transamination. For example, Leu is transaminated to α-ketoisocaproate (KIC) by BCATm in many peripheral tissues including heart, skeletal muscle, kidney, and fat. This reaction is reversible, and thus, depending on substrate concentrations, BCATm can also convert KIC to Leu.
It has been unclear whether Leu, its metabolite, α-ketoisocaproate, or both participate in the activation of mammalian target of rapamycin (mTOR), increases in protein synthesis, and the inhibition of protein degradation that occurs after Leu treatment. Indeed, some of the negative associations between BCAAs and metabolic health have been recently hypothesized to be related to the effects of KIC, rather than Leu signaling (for review see Ref. 52). One approach that has been used to address this question was through inhibition of BCATm using transaminase inhibitors. While the specificity of those inhibitors is arguable, Tischler et al. (81) used them to determine whether Leu or its transaminated metabolite, KIC, were responsible for the effects of Leu on muscle protein accretion. They concluded that whereas Leu, but not KIC, stimulated protein synthesis, KIC, but not Leu, provided the brake to protein breakdown in muscle. Subsequent research identified phosphorylation of ribosomal proteins, such as ribosomal protein S6, and translation initiation factors in the actions of amino acids on protein synthesis along with mTOR and later its complex 1 (mTORC1) as the potential mediator of the activation of protein synthesis by nutrients, in particular, Leu (5, 6, 9, 11, 12, 18, 23, 31, 34, 39–43, 51, 54, 59, 67, 77, 86). However, how KIC might bring about a decrease in protein degradation is still not clear.
A second approach that has been used to assess the role of Leu and its metabolites in downstream signaling events involves genetic ablation of the gene encoding BCATm (e.g., 89). BCATm knockout (KO) mice fed standard rodent chow exhibit extraordinarily high plasma concentrations of BCAAs compared with wild-type mice. Remarkably, when given a choice of two diets (a normal diet and a diet deficient in BCAAs), the KO mice balanced their consumption such that plasma BCAA concentrations were reduced compared with KO mice fed the standard chow diet. However, even under this feeding regimen plasma BCAAs remained elevated in BCATm KO mice, and, as expected, KIC concentrations were far lower (74, 90). In contrast to the prevailing view that these mice might exhibit metabolic syndrome (62, 63), they instead exhibited decreased adiposity and body weight and resistance to diet-induced obesity, even though they ate more food than their wild-type counterparts. In addition, BCATm KO mice exhibited 33% reductions in their plasma glucose levels, a ∼50% reduction in glucose tolerance test (GTT) areas under the curve along with ∼50% reductions in plasma insulin during the GTT and improved insulin sensitivity, but not maximal insulin action (76, 79). However, these mice also had reduced exercise tolerance (76). The lower fasting glucose and exercise intolerance might be related in part to the important role of BCAA transamination in gluconeogenic amino acid production that is likely important during exercise.
She et al. (74) observed that an increase in energy expenditure in BCATm KO mice explained their ability to eat more food while maintaining lower body weight. However, factors usually associated with increased energy expenditure in other mouse models did not explain their increased energy expenditure and resistance to diet-induced obesity. For example, neither brown fat uncoupling protein 1 (Ucp1) nor muscle sarco/endoplasmic reticulum calcium ATPase 1 (SERCA1, encoded by Atp2a1 gene) was increased. Instead, increased energy expenditure in these mice was postulated to arise from an increased protein turnover (24, 74) arising from an increase in mTOR signaling brought about by elevated Leu along with loss of the putative KIC signal to protein degradation postulated originally by Tischler et al. (26, 81). Moreover, the increase in protein synthesis in these mice helped protect them from endotoxin-induced decrease in muscle protein synthesis and improve their survival during sepsis (48). While BCATm skeletal muscle exhibits increased protein synthesis and degradation, other tissues such as heart, kidney, and spleen appear to be protected as might occur in starvation (61). Finally, BCATm KO mice were exercise intolerant (76). This was largely postulated to be associated with their low circulating substrates and energy reserves, interruption of the muscle malate-aspartate cycle, along with the increased lactate-to-pyruvate ratio and ammonia in skeletal muscle observed after exercise. However, other changes in muscle could be having an influence on this phenotype.
In the present study, we have used mRNA sequencing to examine the adaptations in gene expression that might support and help explain the skeletal muscle phenotype of the BCATm KO mice. The data provide evidence of adaptations in protein synthetic and degradative pathways consistent with the previously observed protein turnover phenotype. Alterations in the expression of sarcomeric integrin-linked kinase (ILK) signaling and nutrient metabolism genes provide further insight into the metabolic improvements and exercise intolerance exhibited by these mice.
METHODS
Ethics statement.
All of the vertebrate animal procedures were approved by the Institutional Animal Care and Use Committee of Penn State University College of Medicine (Hershey, PA). The Animal Resource Program is accredited by American Association for Accreditation of Laboratory Animal Care International. All animal living conditions are consistent with standards laid forth in the Guide for the Care and Use of Laboratory Animals (2011), 8th edition, published by the National Research Council.
Isolation of gastrocnemius muscle from BCATm KO mice.
BCATm KO (−/−) mice and their homozygous wild-type (+/+) littermates on the C57BL/6 genetic background were genotyped at weaning and thereafter permitted free access to both normal rat chow and a purified amino acid diet lacking BCAAs. BCATm KO (−/−) mice and their homozygous wild-type (+/+) littermates on the C57BL/6 genetic background were genotyped at weaning and thereafter permitted free access to both normal rat chow and a purified amino acid diet lacking BCAAs. The breeding strategy is intercrossing of +/− heterozygotes. The heterozygotes are maintained by occasional backcrosses at one or two time(s) per year to new C57BL/6 representatives of the parental strain ordered from Jackson Laboratories.
Male mice (9–11 wk old; 4 wild-type and 4 KO genotypes) in the freely fed state were anesthetized under isoflurane anesthesia (carried with 100% O2) between 3:00 and 4:00 PM. With the mice under anesthesia, the gastrocnemius muscle was surgically removed and frozen between two aluminum blocks cooled to the temperature of liquid nitrogen and then stored at −80°C until RNA was isolated. While under continuous anesthesia, the animals were then euthanized by cutting the diaphragm and removing the heart.
Food deprivation and time of day are important considerations for sampling gene expression. It should be recognized that regulation of gene expression, especially for amino acid and other intermediary metabolism, is regulated in a circadian fashion and by nutrient sensors (28, 64, 78). For this reason, a narrow window of tissue sampling was used. We also used freely fed not food-deprived animals. BCATm KO mice have persistently elevated BCAAs. However, food deprivation in this model leads to even further BCAA elevations from the increased rate of appearance from protein breakdown, 1) which is part of the changes in this model and 2) which is associated with fasting, and the loss of the rate of disappearance owing to the BCATm gene KO. So in comparisons of this model to wild type, we avoid extended periods of food deprivation as much as possible and instead opt for either short periods of food deprivation in the afternoon when the animals are eating less or the freely fed state. Here we chose the freely fed state that the state the animals are normally in, and the 3:00–4:00 PM time period, a period when the amount of food intake is reduced compared with when the dark cycle begins.
RNA sequencing.
Total RNA was extracted as previously described with slight modification (22). Briefly, frozen muscle tissue was pulverized in liquid nitrogen with a mortar and pestle, followed by bead mill homogenization (Bullet Blender, Next Advance) using stainless steel beads (Next Advance, cat. #SSB14B) and mirVana RNA isolation kit (Life Technologies). Optical density values of extracted RNA were measured with NanoDrop (Thermo Scientific) to confirm an A260:A280 ratio >1.9. RNA integrity number was confirmed as >7 for each sample using Bioanalyzer RNA 6000 Nano Kit (Agilent Technologies). The cDNA libraries were prepared using the TruSeq RNA Sample Prep Kit v2 (Illumina) as per the manufacturer's instructions. The final product was assessed for its size distribution using Bioanalyzer DNA High Sensitivity Kit (Agilent Technologies) and for its concentration using Kapa library quantification kit (Kapa Biosystems). Eight libraries were pooled per HiSeq lane, followed by on-board cluster generation on a Rapid Run single-end flow cell and subsequent 50 cycles sequencing (v3 sequencing kit) according to the manufacturer's instructions (HiSeq 2500, Illumina). Demultiplexed and quality-filtered mRNA-Seq reads were then aligned to mouse reference assembly (mm10) using TopHat (v.2.0.9). The uniquely mapped reads were used to calculate the normalized expression level of genes, as fragments per kilobase of exon per million fragments mapped (FPKM), using Cufflinks (v.2.0.2).
Biostatistics and bioinformatic analysis.
Student's t-test was used to compare body weights. We used a Mann-Whitney test to determine whether statistical differences existed in the number count of genes detected by RNA-Seq (FPKM values ≥1) in the biological replicates in the control compared with the BCATm KO group. To determine significant differences in gene-specific FPKM values between wild-type control and BCATm KO groups, the DEGexp function of the DEGSeq 1.18.0 R package was used with the likelihood ratio test and default parameters. A P < 0.05 was used to determine significant differences. We manually inspected the list of the significantly different genes identified by DEGSeq compared with raw counts (Supplementary Tables S1 and S2).1 Based on this inspection and the flagging function provided with that program, it was decided to reduce the list used for bioinformatic analysis, and those genes are found in Supplementary Table S3. In scenario 1, a number of genes were removed for bioinformatic analysis because they were flagged by DEGSeq with the code “#VALUE!”, which indicates they lacked FPKM values in the wild-type or KO group. Many of these gene names had only one biological sample with an FPKM value (example - Cox20). Such genes were not added to Supplementary Table S3. An exception was made for a few genes that had multiple biological replicates with low biological variability in FPKM counts. In scenario 2, some genes designated as significantly different in Supplementary Table S2 were not further studied if the statistical significance appeared to be based on a single outlier FPKM value based on analysis of the values using ROUT routine in Graphpad Prism (example 5830428H23Rik). Finally a Cufflinks error in the analysis of one biological replicate for the gene, Mhy4, led to a reevaluation of the data set with an n = 3 for one group as indicated in the supplementary tables. In the line showing that data, one biological replicate was removed as indicated. Of the remaining 1,543 genes (Supplementary Table S3), 57 had “unmapped identifications” (unknown functions or poorly annotated genes) according to IPA and were eliminated from further analysis, leaving 1,486 statistically significant genes from DEGSeq. Those remaining with a normalized fold change of ±1.4 (838 genes) were subjected to bioinformatic analyses to narrow the number of pathways to focus upon.
Functional annotation clustering, pathway analysis, and gene ontology along with associated confidences indicating the strength of evidence for pathway effects were obtained using a series of tools including Ingenuity Pathway Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com), Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/), Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp), GeneCards (http://www.genecards.org), Panther Classification System (http://www.Pantherdb.org) and the Rat Genome Database (RGD, http://rgd.mcw.edu/). In contrast to IPA, neither Pantherdb nor the DAVID functional annotation tools takes into consideration the direction of gene expression change or its positive or negative influence on a pathway. Therefore we primarily used Pantherdb for its gene overrepresentation analyses at http://www.Pantherdb.org (release 20141219, PANTHER version 9.0; The Bonferroni correction was used for multiple testing for those analyses). In DAVID, we separated obtained insights from separately entering 1) the significantly different genes comparing BCATm KO to wild type, 2) the genes upregulated by BCATm KO, and 3) the downregulated genes. Recognizing the caveat that the products of some genes are inhibitory mediators, we integrated this information with the output from IPA analyses that does take into consideration the direction of change and function of proteins in pathways or cell functions. Some programs such as DAVID and Ingenuity provide a Z-score to assess the predicted activation or inhibited state of upstream regulators or downstream effects of the altered gene expression in known pathways. The magnitude of this number reflects the intensity/significance of the predicted change, whereas the − or + value indicates inhibition or stimulation respectively.
Data sharing.
The processed data (Supplementary Table S1), associated metadata, and the “raw” Illumina fastq file have been deposited with the National Center for Biotechnology Information Gene Expression Omnibus database (GEO accession number: GSE68915).
RESULTS
Four 9–11 wk old male BCATm KO (−/−) mice and four wild-type littermates were chosen for RNA-Seq. Their body weights are shown in Table 1. Consistent with previous studies (74, 76), there was a trend (P = 0.06) for BCATm KO to be ∼15–20% lighter than their wild-type (WT, +/+) littermates at this age (Table 1). The gastrocnemius muscle was removed from freely fed mice while under isoflurane anesthesia, and mRNA was isolated and was used to make cDNA libraries for Next Generation RNA sequencing. The mouse genome assembly, mm10, was used as reference for alignment of the sequencing data (Supplementary Table S1).
Table 1.
Characteristics of BCATm mice
| Genotype | Body Weight, g | n |
|---|---|---|
| Bcatm +/+, WT | 24 ± 0.70 | 4 |
| Bcatm −/−, KO | 19.3 ± 1.95* | 4 |
Bcatm, mitochondrial branched-chain aminotransferase; WT, wild type; KO, knockout.
P = 0.06 compared with control mice.
Initial analysis of mRNA sequencing.
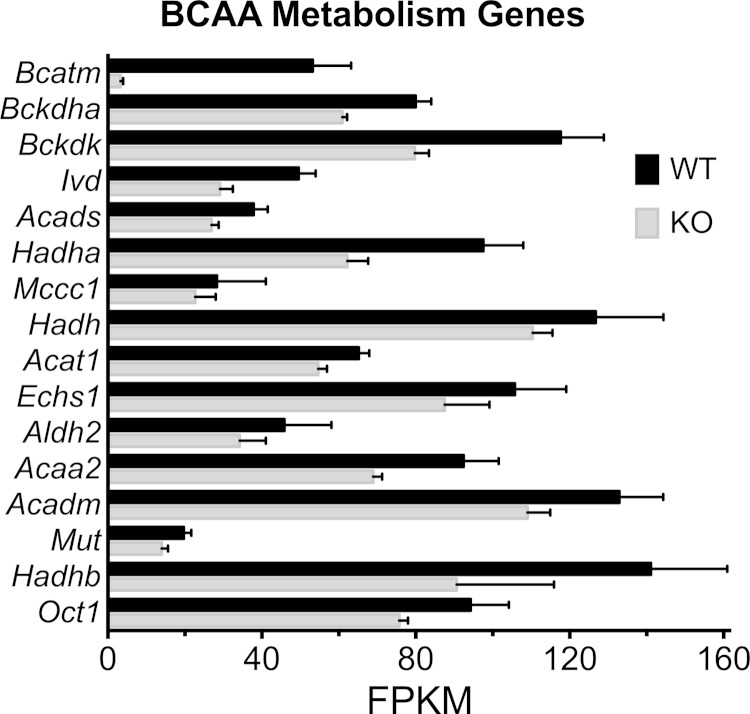
We examined the data for systematic issues such as whether a similar number of genes was detected in each tissue and whether the FPKM values were consistent with the tissue being from skeletal muscle. The number of genes detected with FPKM values ≥1 was 10,599 ± 187 for control and 10,485 ± 429 for the BCATm KO and was not significantly different (P = 0.8, Supplementary Table S1). Initial analysis of mRNA sequencing of gastrocnemius muscle from mice in the control group gave rise to FPKM values expected for striated skeletal muscle tissue. For example, of the 39,009 genes in the mouse genome build, the most highly abundant protein coding gene in gastrocnemius was Sepw1 (encoding selenoprotein W muscle 1), and the next 99 highest FPKM values were associated with genes coding proteins of the sarcomere, cation transporters, glycolysis, oxidative metabolism/phosphorylation, and protein synthetic machinery (Supplementary Table S1). DEGSeq was used to identify statistically differentially expressed muscle genes between WT and BCATm KO mice (Output in Supplementary Table S2). For example, that analysis shows that the BCATm KO mice expressed very little BCAT2 as expected (Fig. 1, Supplementary Tables S1–S3).
Fig. 1.
Statistically different branched-chain amino acid (BCAA) metabolism pathway genes expressed in mitochondrial branched-chain aminotransferase (BCATm) knockout (KO) and wild-type (WT) mice. Genes in the mouse BCAA metabolism KEGG pathway that were statistically different based on DEGSeq analysis (Supplementary Tables S2–S3) are plotted. The bars are mean fragments per kilobase of exon per million fragments mapped (FPKM) ± SE. Black bars, WT skeletal muscle; gray bars, BCATm KO.
A subset of the genes in Supplementary Table S2 identified as significantly differentially expressed between wild-type and KO mice was selected for bioinformatic analysis using the approaches described in the methods. These genes are listed in Supplementary Table S3. Analyzing this subset of genes using PANTHERdb overrepresentation analyses indicated that biological processes and functions associated with protein synthesis, metabolism, and the sarcomere were statistically overrepresented (Supplementary Table S4). The results from IPA analyses of the data from Supplementary Table S3 are summarized in Table 2.
Table 2.
IPA analysis of RNA-Seq comparison of BCATm KO and WT mice gastrocnemius
| Top Canonical Pathways | |||
|---|---|---|---|
| Name | Z-score | Molecules, n | P Value |
| eIF2 signaling | +3.5 | 26 | 5.8e-09 |
| ILK signaling | −1.5 | 25 | 2.8e-08 |
| mTOR signaling | +0.3 | 25 | 3.5e-08 |
| TCA cycle | NAN | 9 | 6.7e-08 |
| Glycolysis | NAN | 9 | 1.6e-07 |
| Regulation of eIF4 and p70S6K | +1.0 | 19 | 2.1e-06 |
| Top Diseases and Bio Functions | |||
|---|---|---|---|
| Category | Name | Molecules, n | P Value Range |
| Diseases and disorders | skeletal and muscle disorders | 185 | 1.2e–03-2.4e–22 |
| Molecular and cellular functions | cell death and survival | 288 | 1.4e–03-2.3e–17 |
| cellular growth and proliferation | 296 | 1.4e–03-9.4e–15 | |
| cellular assembly and proliferation | 188 | 1.4e–03-1.7e–14 | |
| cell morphology | 232 | 1.4e–03-2.2e–13 | |
| protein synthesis | 135 | 1.1e–03-5.9e–12 | |
| Physiological system development and function | skeletal and muscle system development and function | 167 | 1.4e–0.3-1.4e–15 |
| Diseases and Functions | |||
|---|---|---|---|
| Name | Z-score | Molecules, n | P Value |
| Cell death | +3.8 | 273 | 2.3e-17 |
| Glucose metabolism disorder | +3.4 | 102 | 8.8e-07 |
| Necrosis | +3.2 | 217 | 2.7e-15 |
| Apoptosis | +3.2 | 221 | 2.8e-08 |
| Insulin sensitivity | +2.7 | 15 | 3.64e-04 |
| Myopathy | +3.0 | 91 | 2.4e-22 |
| Atrophy of muscle | +2.5 | 18 | 2.5E-05 |
| Hypoglycemia | +2.1 | 14 | 2.0e-04 |
| Contractility of muscle | −2.8 | 27 | 3.4e-8 |
| Oxidation of fatty acid | −2.5 | 30 | 1.8e-11 |
| Size of body | −6.32 | 67 | 1.3e-04 |
The 838 genes from Supplementary Table S3 with a normalized fold change of ±1.4 were analyzed by IPA. The summarization related to general tissue functions and either muscle or myocytes function including heart muscle are shown (version 23814503). “Molecules, n” refers to the number of molecules with normalized fold changes of ±1.4 or greater; there may be more molecules with lower fold but statistically significant changes;
NAN, not a number.
All of the TCA cycle IDs had lower fragments per kilobase of exon per million fragments mapped (FPKM) values in BCATm KOs. Causal analysis approaches used to describe generation of P values have been described (46).
Evidence of elevated protein synthesis.
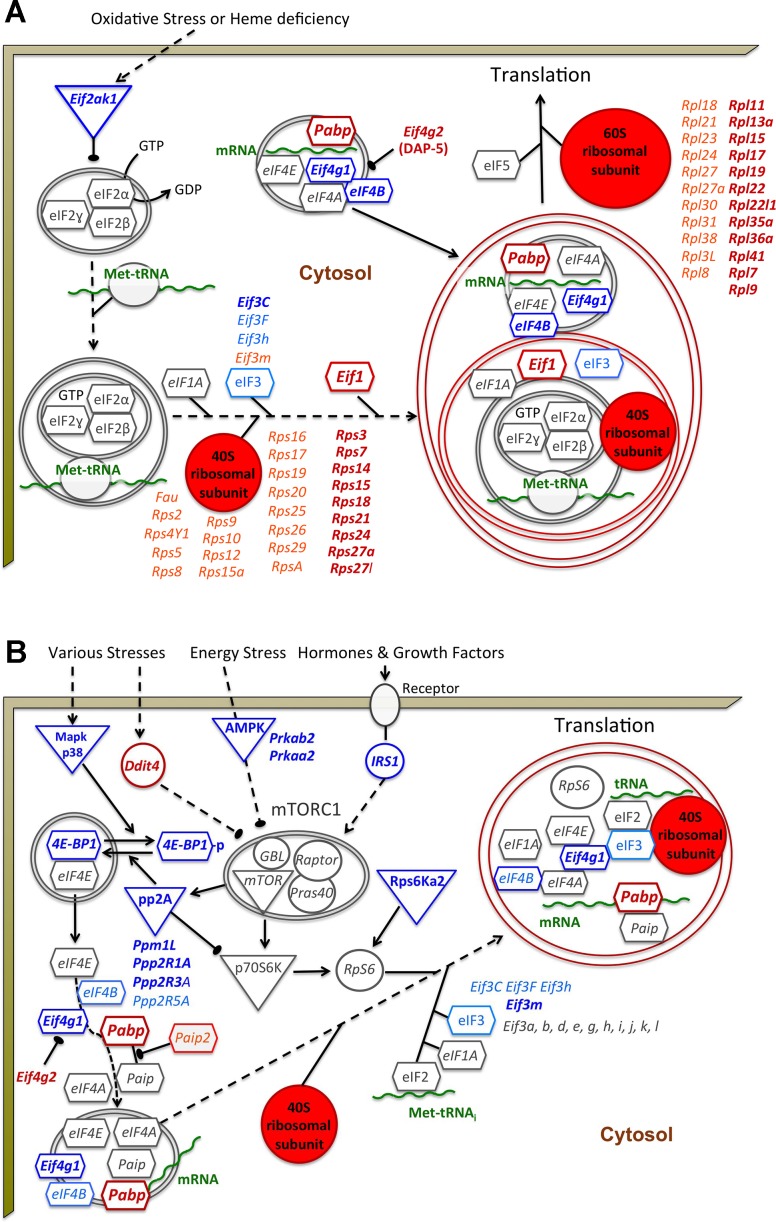
The functional annotation tool DAVID does not take into consideration the genes direction of change in gene expression. Therefore, we first used it to analyze significantly upregulated genes. This analysis indicated that genes upregulated by BCATm KO associated in the “very high” category (kappa = 1.0 and 0.85, respectively) with two functionally related terms: protein biosynthesis and ribosome. Canonical pathway and downstream pathway analysis results from IPA performed on the 838 up- and downregulated genes mentioned earlier were also consistent with increased protein synthesis. For example, two pathways associated with elevated protein synthesis had strong positive Z-scores. The first pathway was “EIF2 signaling” (P = 5.7e-9, Z-score 3.5, Fig. 2A). Contributing to that Z-score was the finding of decreased expression (blue color) of the negative regulator of EIF2, the heme-regulated eIF2α kinase (Eif2ak1, also called heme-regulated inhibitor, Hri), whose expression was decreased in BCATm KO mice. Also contributing to this conclusion was the general upregulation of 22 genes encoding protein components of the large and small ribosomal subunit proteins (Fig. 2A, red color or font). Notably, an additional 29 ribosomal protein genes were statistically altered but were below the 1.4-fold cut-off we used for IPA (Fig. 2A, pink font). A second pathway, “regulation of eIF4 and p70S6K signaling,” was also identified as being upregulated (P = 2.1e-6, Z-score 1), as was the related “mTOR Signaling” pathway (Fig. 2B). For example, there was reduced expression of 4E-BP1, a negative regulator of eIF4E, and a phosphatase that dephosphorylates it (PP2A), enhancing its inhibitory function. However, genes encoding upstream regulators of mTOR were either decreased or not affected, and members of the mTORC1 complex were not affected. Nevertheless, downstream targets of mTORC1 that are part of the translational machinery were frequently statistically elevated (Fig. 2B). Other gene expression changes that IPA indicated as a prediction of increased translation of protein were the increased expression of poly(A)-binding protein (Pabpc1) and eIF3g (88), decreased expression of eIF3F (88), and increased expression of Fxr1, Calr, Eprs, Cpeb1, and Ybx2 (Supplementary Tables S1–S3). Moreover, JunB was elevated about twofold in BCATm KO mice (Supplementary Tables S1–S3). JunB has been shown to increase skeletal muscle hypertrophy and mass by promoting FoxO3 binding to Fbxo32 and MuRF1 promoters and thereby reduces protein breakdown (68). The cyclin-dependent kinase inhibitor p21 (Cdkn1a), which plays an important role in skeletal muscle regeneration (20, 33), was also significantly elevated in BCATm KO mouse muscle (Supplementary Tables S1–S3). Interestingly, eIF4G2 (also known as death-associated protein 5, DAP-5) expression was almost twofold greater in muscle of KO compared with wild-type mice (Fig. 2, A and B; Supplementary Tables S1–S3). That isoform of eIF4G was recently shown to bind to eIF2S1 and eIF4A to promote cap-independent mRNA translation (50), and thus both cap-dependent and cap-independent mRNA translation may be affected in KO mice.
Fig. 2.
Effects of BCATm KO on protein synthesis pathways. Ingenuity Pathway Analysis (IPA) analysis results are shown for genes in the eukaryotic initiation factor (eIF)2 pathway (A) and eIF4 and p70S6K pathway (B). The beige border represents the sarcolemma. Arrows indicate a pathway direction, and a blunted arrow indicates an inhibitory factor or process. Blue color indicates that the expression of the gene was decreased. Red color indicates expression of a gene or a number of ribosomal protein (RP) genes composing 40S and 60S subunits were generally increased. Lighter shades of blue and red and lightfaced rather than boldfaced font indicates that the normalized fold change was significant but less than the ± 1.4 normalized fold change used as an arbitrary cut-off for IPA analysis (Supplementary Tables S1–S3). Gray font and equal sign (=) indicates no significant change.
Evidence of elevated protein degradation.
BCATm mice tend to have lower body weights compared with sibling controls attributed to a futile cycle consisting of increased rates of protein synthesis and degradation (Table 1 and Refs. 74, 76). Consistently, in the present study, gene ontology for size of the body also had a negative Z-score (Table 2). A number of gene expression changes also implied increased protein degradation in BCATm mice. Pathways associated with protein degradation, such as cell death, apoptosis, necrosis, and muscle myopathy had positive Z-scores with significant changes in hundreds of genes, suggesting that those pathways were activated (Table 2, Supplementary Table S5).
Gene ontology analyses also identified changes in the expression of a number of genes that normally contribute to protein turnover in skeletal muscle. For example, DNA damage-inducible transcript 4 (Ddit4, alias: Redd1) is a major inhibitor of mTOR during stress and a mediator of atrophy and target of glucocorticoids (13). Ddit4 gene expression was elevated almost threefold in the gastrocnemius of BCATm KO mice (Fig. 2B, Supplementary Tables S1–S3). Another glucocorticoid target, CCAAT/enhancer binding protein delta (Cebpd), the expression of which is associated with starvation and muscle atrophy (3), was elevated more than fourfold. The gene encoding atrogin-1 (Fbxo32, also called muscle atrophy F-box protein and MAFbx), which is activated in catabolic disorders, was elevated 1.7-fold in BCATm KO mouse muscles (Supplementary Tables S1–S3). BCATm KO was also associated with upregulation of the Hint family genes in skeletal muscle (Hint1, Hint2, Supplementary Tables S1–S3). Hint1 appears to regulate proteasomal degradation of proteins by the SKP2-CUL1-F-box protein-E3 ubiquitin-protein ligase complex, whereas the mitochondrial isoform, Hint2, has been described as an apoptotic sensitizer (16, 57). Increased expression of two genes that are linked to positive regulation of apoptosis, cytochrome C somatic (Cycs) and BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (Bbip3, the most highly expressed member of this family in muscle), were also increased (Supplementary Tables S1–S2, notably no caspase genes were affected though). The gene encoding the muscle-specific RING finger protein 1 (Trim63, alias: MuRF1), which regulates proteasomal degradation of sarcomeric proteins, was elevated 1.6-fold (Supplementary Tables S1–S3; Refs. 8, 29, 38, 69, 70). BCATm KO also led to increased expression of 20S proteasome core particle components (Pmsa7, Pmsa3, Psmb1, Psmb2, Psmb3, Psmb4, Psmb5, Psmb6; Supplementary Tables S1–S3). Cullins provide a scaffold for E3 ubiquitin ligases involved in proteasomal protein degradation; two of the most highly expressed cullins in muscle, Cul1 and Cul3, were significantly elevated in BCATm KO mice (Supplementary Tables S1–S3). Furthermore, expression of genes encoding two ubiquitin proteins, Rps27A (alias Uba80) and Uba52, was elevated, whereas a number of ubiquitin-specific protease genes that might be involved in reversing these effects (Usp2, Usp13, Usp15, Usp16, Usp19, Usp47) were decreased (Supplementary Tables S1–S3). Nedd4 the E3 ubiquitin ligase, which is elevated in atrophy associated with unloading and denervation, was also increased (45). There was an approximately twofold increase in degradation in endoplasmic reticulum protein 2 (Derlin2, Derl2), thought to be a component of the endoplasmic reticulum-associated channeling of unfolded luminal glycoproteins for proteasomal degradation (ERAD). However, the gene encoding the Ubiquitin COOH-terminal hydrolase-L3, Uchl3, which reduces stress in skeletal muscle and indirectly improves insulin signaling was increased (72).
Finally, one of the top canonical pathways, ILK signaling was predicted to be repressed in BCATm KO compared with wild-type mice (Table 2). Deletion of this pathway in skeletal muscle has been associated with a muscular dystrophy syndrome with multiple regenerating and degenerating fibers (25). Decreased activation of this pathway is also observed during mechanical unloading (Unloading models typically display muscle atrophy) as typified in reduced gene expression of Col1A1, Col1A2, Col3A1, Fn1, and Bgn (for review see Kjær Ref. 44). The expression of these genes was decreased about twofold or more in BCATm KO muscle (Supplementary Tables S1–S3), consistent with the exercise intolerance (76) and skeletal muscle atrophy pathways we observed to be activated in these mice (Table 2).
Metabolic genes.
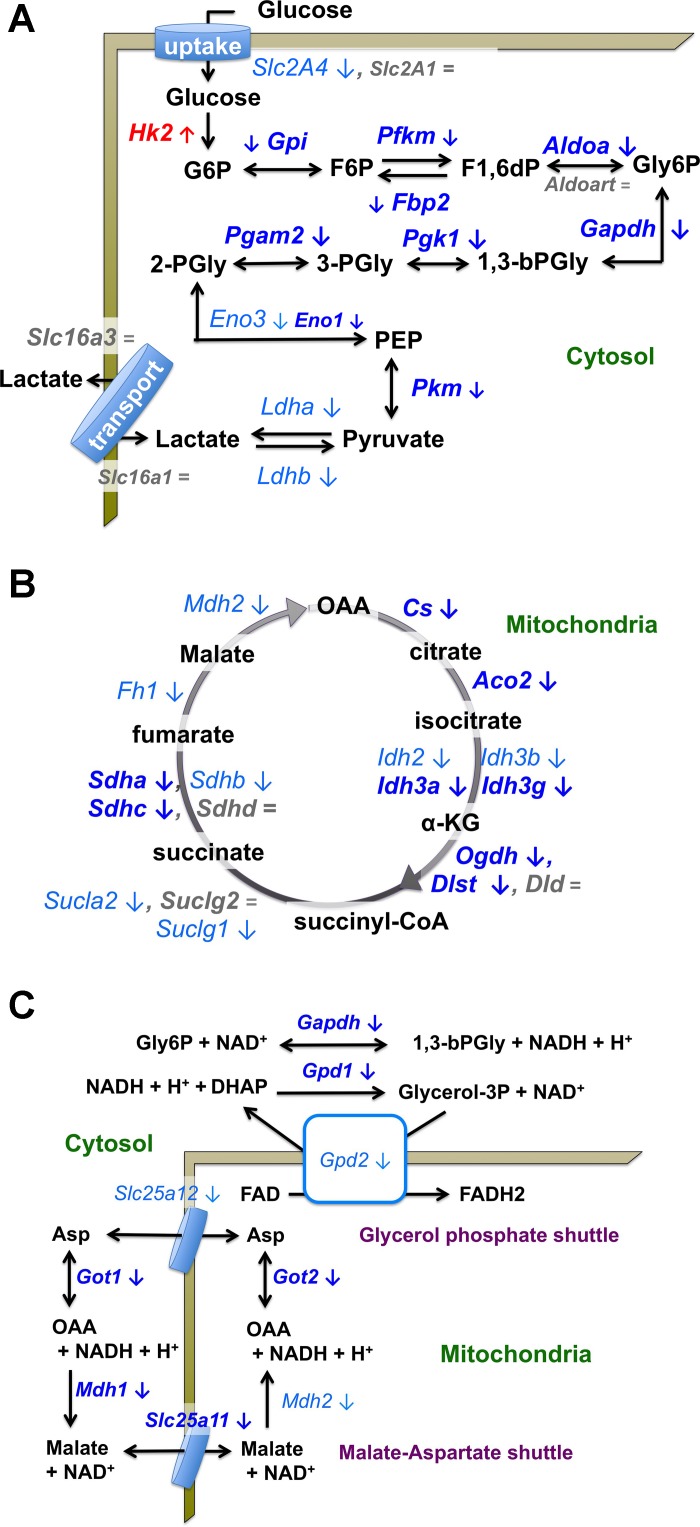
Pathway analyses implicated changes in several metabolic pathways in BCATm KO mice. This includes moderately reduced expression of genes in glucose and fatty oxidation pathways that become very significant when examined by pathway analysis (Table 2, Fig. 3, and Supplementary Table S5). Most genes in glycolysis were decreased as was the insulin responsive glucose transporter Glut4 (Slc2a4, Fig. 3A). An exception was hexokinase 2 (Hk2), whose expression was increased (Fig. 3A). A number of genes in the pentose phosphate pathway were also downregulated (Supplementary Tables S1–S3). Genes encoding proteins facilitating nonoxidative and oxidative pyruvate metabolism were also affected. Thus LDH gene isoforms and the Gpt gene encoding alanine amino transferase (Fig. 3A and Supplementary Tables S1–S3) were downregulated as were pyruvate dehydrogenase (PDH) component genes, Pdha, Pdhx, Dlat, and the inhibitory PDH kinase gene (Pdk2) in BCATm KO mice. Most genes coding for oxidative glucose metabolism in the TCA cycle were also modestly decreased (Fig. 3B), and there was a similar reduction in genes encoding the mitochondrial NADH shuttles (Fig. 3C). However, no consistent pattern of change was observed in the KEGG oxidative phosphorylation pathway for the electron transport chain. For example, genes encoding elements of complexes 1–4 were unaffected, upregulated, or downregulated (Supplementary Tables S1–S3).
Fig. 3.
Changes in genes encoding components of the glycolytic, TCA cycle, and mitochondrial NADH shuttle pathways in gastrocnemius muscle in response to BCATm KO. Effects of BCATm KO on muscle gene expression are shown for the glycolysis pathway (A), citric acid (TCA) cycle (B), and two mitochondrial NADH shuttles (C). Metabolites and glycolytic intermediates are shown in black font; gene names are italicized. The beige border represents the sarcolemma (A) or mitochondrial membrane (C). Black arrows indicate a pathway direction. Blue color indicates that the expression of the gene was decreased (emphasized with a down direction arrow). Red color indicates expression of a gene was increased (emphasized with an up direction arrow). Lighter shades of blue along with lightfaced rather than boldfaced font indicates that the normalized fold change was significant, but less than the −1.4 normalized fold change used as an arbitrary cut-off for IPA analysis (Supplementary Tables S1–S3). Gray font and equal sign (emphasized with = sign) indicates no significant change. Where the abundance of two isoforms is different, the font size is smaller for the isoform where the FPKM has a lower value (Supplementary Tables S1–S3).
Most genes involved in fatty acid oxidation were reduced by the KO of BCATm. Genes in the BCAA metabolic pathway downstream from BCATm (gene name: Bcat2) also displayed a generalized modest decrease in expression (Fig. 1, Table 3, Supplementary Tables S1–S3).
Table 3.
Significant changes in fatty acid oxidation gene expression in gastrocnemius of BCATm KO mice compared with WT sibling controls
| Gene Name | Control WT, FPKM | BCATm KO, FPKM | Normalized Fold Change | Molecular Identity |
|---|---|---|---|---|
| Acaa2 | 92 ± 9 | 69 ± 2 | −1.4 | acetyl-CoA acyltransferase 2 |
| Acad9 | 29 ± 2 | 19 ± 2 | −1.7 | acyl-CoA dehydrogenase family, member 9 |
| Acadl | 113 ± 14 | 76 ± 5 | −1.6 | acyl-CoA dehydrogenase, long chain |
| Acadm | 133 ± 11 | 109 ± 6 | −1.3 | acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| Acads | 38 ± 4 | 27 ± 2 | −1.5 | acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| Acat1 | 65 ± 3 | 55 ± 2 | −1.3 | acetyl-CoA acetyltransferase, mitochondrial also known as acetoacetyl-CoA thiolase |
| Acadvl | 147 ± 10 | 117 ± 2 | −1.3 | acyl-CoA dehydrogenase, very long chain; obesity/T2D susceptibiltiy |
| Acsl1 | 60 ± 10 | 45 ± 6 | −1.5 | acyl-CoA synthetase long-chain family member 1 |
| Cd36 | 115 ± 36 | 91 ± 18 | −1.4 | fatty acid translocase (transmembrane) |
| Cpt1b | 88 ± 8 | 73 ± 6 | −1.3 | carnitine palmitoyltransferase 1b, muscle |
| Cpt2 | 33 ± 7 | 24 ± 1 | −1.5 | carnitine palmitoyltransferase 2 |
| Eci1 | 107 ± 12 | 132 ± 9 | +1.2 | enoyl-CoA delta isomerase 1 |
| Fabp3 | 339 ± 62 | 246 ± 26 | −1.5 | fatty acid binding protein 3, muscle |
| Hadh | 127 ± 18 | 110 ± 5 | −1.2 | medium and short-chain L-3-hydroxyacyl-coenzyme A dehydrogenase, mitochondrial |
| Hadha | 96 ± 10 | 62 ± 5 | −1.7 | trifunctional protein, alpha subunit |
| Hadhb | 141 ± 20 | 91 ± 25 | −1.7 | trifunctional protein, beta subunit |
| Lpl | 197 ± 79 | 119 ± 22 | −1.8 | lipoprotein lipase |
| Slc25a20 | 32 ± 2 | 26 ± 1 | −1.3 | carnitine/acylcarnitine translocase (cytosol mitochondrial) |
| UCP3 | 23 ± 4 | 18 ± 1 | −1.4 | uncoupling protein 3, mitochondrial |
Data are means means ± SE of the FPKM. Normalized fold change values are shown, a positive value indicates a statistically significant increase, whereas a negative value indicates a decrease when BCATm KO mice are compared with WT control mice.
Muscle fiber type and function.
Muscle fiber phenotyping schemes can be devised with various levels of complexity and subdivisions of fiber type. A simple approach is to divide the fiber types into two (e.g., slow and fast) or three categories, slow (a.k.a., oxidative, type I fibers), fast oxidative-glycolytic (FOG; a.k.a. intermediate, type IIa and IIx), and fast-glycolytic (fastest in rodent, a.k.a., type IIb). This phenotyping may rely solely on evaluation of myosin heavy chain (Mhc family) isoform content. Alternatively more extensive muscle fiber phenotyping may consider other muscle fiber type-specific genes or proteins/isoforms coding for cation, oxygen handling, and sarcomeric components found in different muscle fiber types. We observed a number of significant changes in gene expression implying transitions occurring in the types of fibers present in gastrocnemius due to BCATm KO (Table 4). While we noted no change in the expression of the myosin heavy chain I gene (Myh7, found in slow oxidative/type I fibers), Myh1 (found in FOG fibers, IIA) and Myh2 (a fast glycolytic fiber, IIX) showed decreased expression. FPKM values for myosin heavy chain 4 (Mhy4), which is the most abundant myosin type in rodent gastrocnemius associated with the fastest most glycolytic fibers (IIb), was also decreased.2 Looking at other sarcomeric components, BCATm KOs expressed higher amounts of genes encoding slower fiber (I) types of troponin C and myosin binding protein C (Tnnc1, Mybpc1), whereas the faster twitch fiber types (Tnnc2, Mybpc2) were decreased (Table 4). The Mb gene encoding the O2-carrying protein, myoglobin, associated with more oxidative fiber types, was elevated (Table 4). Consistently, the gene encoding the myosin light chain associated with slower (Myl3) fiber types was elevated. However, those encoding the faster (Myl1, Mylpf) isoforms were also significantly increased.
Table 4.
Genes associated with skeletal muscle fiber type
| Fast Twitch or Type IIb Gene or Homologs | WT Control, FPKM | BCATm KO, FPKM | Normalized Fold Change | Slow Twitch or Non-IIb Type Genes or Homologs | WT Control, FPKM | BCATm KO, FPKM | Normalized Fold Change |
|---|---|---|---|---|---|---|---|
| Atp1b2* | 80 ± 12 | 48 ± 5 | −1.8 | Atp1b1 | 42 ± 5 | 33 ± 4 | −1.3 |
| Atp2a1 | 4,918 ± 657 | 4,344 ± 979 | −1.2 | Atp2a2* | 38 ± 12 | 32 ± 5 | N.S. |
| Casq1 | 1,226 ± 128 | 830 ± 54 | −1.6 | Casq2 | 13 ± 1 | 18 ± 3 | N.S. |
| Fhl3 | 89 ± 9 | 69 ± 2 | −1.4 | Fhl1 | 300 ± 26 | 274 ± 44 | −1.2 |
| Mb | 3,608 ± 444 | 4,657 ± 444 | +1.2 | ||||
| Mybpc2 | 845 ± 112 | 590 ± 54 | −1.5 | Mybpc1 | 169 ± 27 | 257 ± 56 | +1.4 |
| “324 ± 32, n = 4” | “300 ± 32, n = 4” | N.S. | |||||
| Myh4 | 3,792 ± 504 | 2,616 ± 201 | −2.9 | Myh1 (IIx) | 160 ± 30 | 63 ± 23 | −2.7 |
| (type IIb) | Myh2 (IIa) | 110 ± 19 | 75 ± 10 | −1.6 | |||
| Myh7 (I) | 13 ± 6 | 12 ± 4 | N.S. | ||||
| Myl1 | 35,460 ± 4,007 | 43,023 ± 4,981 | +1.1 | Myl2 | 221 ± 73 | 223 ± 67 | N.S. |
| Mylpf | 23,556 ± 1,413 | 31,291 ± 6,180 | +1.2 | Myl4 | 13 ± 4 | 33 ± 5 | +2.3 |
| Myl12a | 108 ± 8 | 110 ± 14 | N.S. | ||||
| Myom2 | 170 ± 25 | 102 ± 9 | −1.8 | Myom3 | 8 ± 1 | 8 ± 2 | N.S. |
| Myom1 | 174 ± 24 | 138 ± 17 | −1.4 | ||||
| Myoz1 | 1,868 ± 181 | 1,702 ± 12 | −1.2 | Myoz2 | 87 ± 14 | 103 ± 19 | N.S. |
| Myoz3 | 44 ± 8 | 24 ± 5 | −2.0 | ||||
| Obscn | 163 ± 18 | 100 ± 12 | −1.8 | ||||
| Tnnc2 | 12,424 ± 1,136 | 10,490 ± 798 | −1.3 | Tnnc1 | 68 ± 34 | 91 ± 33 | N.S. |
| Tnnt3 | 13,812 ± 868 | 12,412 ± 901 | −1.2 | Tnnt1 | 47 ± 20 | 50 ± 14 | N.S. |
| Tpm1 | 14,480 ± 693 | 13,726 ± 1,110 | −1.1 | Tpm2 | 2,020 ± 215 | 1,268 ± 148 | −1.7 |
Gene expression of isoforms of fast and slow twitch muscle fiber-associated genes from WT and BCATm KO mice are shown. Data are means means ± SE of the FPKM. Normalized fold change values are shown, a positive value indicates a statistically significant increase, whereas a negative value indicates a decrease when BCATm KO mice are compared to WT control mice.
N.S., no significant difference.
Other changes in gene expression also imply changes in muscle performance/function may be occurring based on calcium handling gene expression (4, 82). Thus the most highly expressed sarco/endoplasmic reticulum calcium ATPase in mouse gastrocnemius, SERCA1, was decreased. Furthermore, two SERCA inhibitory peptides, myoregulin (2310015B20Rik, Mln, Mrln) and sarcolipin (Sln), were increased (Supplementary Tables S1–S3, Ref. 4).
Upstream pathway analysis.
Upstream pathway analysis suggested a number of pathways in skeletal muscle appear to be affected by BCATm KO including a number of transcription factors and signaling pathways (Supplementary Table S6). This analysis does not require that expression of the upstream regulatory element of the pathway change. For example, our analysis predicted that the Ste20-like mitogen-activated protein 4 kinase 4 (Map4k4) pathway was activated (Z-score +4.4, p8.8e-12), based on the altered expression of 23 genes. However, Map4k4 expression was not affected (Supplementary Tables S1, S2, and S6). In C2C12 cells this pathway is a negative regulator of hypertrophy (83).
Activation was also predicted for other upstream regulators, some sharing the same pathway genes (Supplementary Table S6). For example IPA analysis predicted activation of α-Catenin (Ctnna1, 19 target genes affected, Z-score +2.9, P = 3.3e-08), the Fas cell surface death receptor pathways (Fas, 31 target genes, Z-score +2.5, P = 5.2e-06), and the small chromatin-binding protein p8 (Nupr1, 31 target genes, Z-score +2.5, P = 5.2e-06) pathways. Ctnna1 may play a role in muscle cell differentiation, Fas is a physiological regulator of programmed cell death, whereas Nupr1 promotes myogenic regeneration (71). All of these observations are consistent with various elements of our findings and the phenotype of these mice.
IPA analysis predicted inhibition of other pathways (Supplementary Table S6). For example, most of the various peroxisome proliferator-activated receptors (PPARs) were predicted to be downregulated, including Ppara (38 target genes affected, Z-score −3.3, P = 5.7e-08), Ppard (24 target genes affected, Z-score −2.2, P = 1.3e-08), Pparg (52 target genes affected, Z-score −4.3, P = 7.1e-16), and Ppargc1a (26 target genes affected, Z-score −2.0, P = 1.7e-08). Decreases in Ppargc1a activity in muscle have been linked to a generalized decrease in BCAA metabolism genes, as we observed in BCATm KO mice (Fig. 1, and Ref. 32). Pathway analysis also predicted downregulation of a master regulator of antioxidant defense (2), Nfe2l2 (35 target genes affected, Z-score −3.9, P = 1.7e-07), and Klf15 (14 target genes affected, Z-score −3.6, P = 1.6e-10), a master regulator of metabolic gene expression (78).
The insulin receptor (Insr, 44 target genes affected, Z-score −3.9, P = 1.7e-07, Supplementary Table S6) pathway was predicted to be downregulated, along with two other physiologically redundant “conditions” hypoglycemia and glucose metabolism disorder (Table 2). These findings are consistent with substantially lower plasma glucose and insulin found in BCATm KO mice (74, 76).
A number of upstream regulators previously associated with muscle degeneration and regeneration were affected. For example, a negative regulator of muscle protein synthesis and mTOR in skeletal muscle, Smad3, was decreased (Supplementary Tables S1–S3). However, two genes that Smad3 is known to regulate, atrogin-1 (Fbxo32 which increased by Smad3) and irisin (Fndc5, which is negatively regulated by Smad3), did not change in a direction that might be expected based on previous studies of Smad3 expression (27, 80). Muscle atrophy is associated with increased TGFb3 signaling, affecting a network of muscle genes including Gnb2l1 (alias RACK1), Cdkn1, and JunB (15). Each of these genes was elevated in BCATm KO mice (Supplementary Tables S1–S3). Another link to these genes and TGF-beta is decreased expression of Sparc (osteonectin) gene expression in BCATm KO mice (Supplementary Tables S1–S3), which is associated with myofibrillar atrophy (60). A gene encoding another protein, osteopontin (Spp1), is induced following muscle injury and is thought to be an important factor in the degenerative and regenerative events associated with muscle injury (for review see Ref. 65). Spp1 was elevated more than threefold in BCATm KO gastrocnemius (Supplementary Tables S1–S3 and S6).
DISCUSSION
Genetic ablation of BCATm leads to an overabundance of circulating BCAAs and a decrease in their metabolites, including the branched chain keto acids that appear to provide independent nutrient signals in BCATm KO. In this study we used a systems biology approach, next-generation mRNA-Seq, to determine the effect of global BCATm deletion on skeletal muscle gene expression to provide 1) insights into the potential adaptations arising from the altered nutrient signaling and 2) new research avenues for understanding the mechanisms of the increased protein turnover, metabolic phenotype, and exercise intolerance of these mice. Gene ontology analysis of the observed changes in gene expression implies that altered gene expression may contribute to the phenotype of these mice, as genes associated with both protein synthesis and regeneration, along with protein degradation and muscle degeneration, were upregulated.
Consistent with the previously observed increase in protein synthesis in these mice, we observed changes in gene expression in pathways involved in protein synthesis such as eIF2, mTOR, eIF4, and p70S6K1. In the eIF4 and p70S6K1 pathway, components of the mTORC1 complex were not affected, and there was a modest decrease in the upstream regulator Irs1. Surprisingly, expression of the mTORC1 repressor, Ddit4, was also increased. Although increased Ddit4 expression is typically associated with muscle wasting conditions, we note that Ddit4 expression is also increased in other models where mTOR signaling is persistently elevated such as TSC2 KO, where its increased expression can be inhibited by rapamycin (66). In such scenarios, increased Ddit4 expression may serve as a feedback mechanism to limit the magnitude of mTORC1 activation. Contributing to the impression of increased protein synthetic machinery was a generalized increase in the expression of the genes encoding the ribosomal protein (RP) genes that help form the 40S and 60S ribosomal subunits. Eukaryotic RP genes and ribosome biogenesis are frequently coordinately regulated at the transcriptional and translational level from yeast to mammals (17, 30, 35, 56). However, the mechanism underlying the transcriptional regulation does not appear to be evolutionarily conserved. In yeast, transcriptional regulation of RP genes can involve mTOR and the transcription factor FHL-1, both of which are expressed in mammals. In contradistinction to yeast, human and murine RP genes are scattered throughout their genomes, and while there is evidence of coordinate regulation of these genes in response to various stimuli (e.g., 35, 55), how this transcriptional regulation is coordinated in mammals has yet to be determined (35) but is thought to be mTOR independent (37). For example, this set of genes is not affected in a microarray dataset when TSC−/− cells (which have a persistently activated mTOR complex 1 that BCAAs normally activate) are treated with rapamycin that would inhibit mTORC1 (19).
BCATm KO mice have smaller, leaner bodies than wild-type mice along with increased global muscle protein turnover (74, 76). Our results identified a number of genes and pathways where increased protein degradation is implied, with proteasomal and apoptotic as opposed to autophagy pathways were predicted to be affected. We previously postulated that a relatively stable muscle protein pool was undergoing an increased rate of turnover in BCATm KO mice. While the results of the gene ontology analysis are consistent with that conclusion, they were also consistent with a different scenario in which fibers may be undergoing an increase in both degenerative and regenerative processes. Such processes can be activated when muscle is injured or dystrophic. In agreement with this concept, many of the affected genes or pathways were associated with terms such as apoptosis, injury, degeneration, necrosis, myopathy, atrophy, and dystrophy. None of these findings contradict the hypothesis that these differences in BCATm KO mice arise from a mixed messaging from the excess of one nutrient signal (Leu) and the loss of another, its metabolite KIC, thought to be signaling separate processes of protein synthesis and protein degradation (24, 81). Further studies are needed to address the role of persistently lower KIC and persistently elevated Leu in these changes.
BCATm KO mice exhibit increased energy expenditure (74, 76). It was surprising, therefore, that modest but significant and systematic declines in gene expression associated with anaerobic and oxidative metabolism of carbohydrate, fat, and amino acids occurred in these mice. It is unclear whether this is due to the fact that, while their energy expenditure increases, the circulating concentrations of metabolic fuels are decreased in BCATm KOs. While BCAA levels are elevated in BCATm KOs, they cannot be metabolized, so a reduction in the expression of enzymes that mediate steps beyond BCATm is not unexpected. Thus all of the observed changes, along with the pathway analysis indicating loss of fatty acid (PPAR) signaling and decreased insulin receptor signaling, may be related therefore to lower circulating metabolic substrate and consequent reduction in plasma insulin in this model (74, 76). Further studies at the protein level are needed to determine whether changes in these enzymes and transporters are in fact occurring in muscle of these mice. Glucose and insulin challenge along with clamp studies demonstrated improved, not worsened, disposal and insulin sensitivity in the muscle of these mice, with no change in capacity for disposal at very high insulin concentrations during a hyperinsulinemic, euglycemic clamp (74, 76).
Many changes occurred in sarcomeric, ion handling, O2 handling, and metabolic genes (oxidative and glycolytic) in BCATm KO mice consistent with muscles having slow, FOG, or fast twitch types. Our findings support our previous hypothesis that BCATm disruption may be affecting the malate-aspartate shuttle (76); the glycerol phosphate shuttle genes were affected as well. While a clear picture of muscle fiber type transition can be gleaned from our previous study in which the same RNA-Seq approach was employed (55), a seemingly uncoordinated pattern of changed emerged from BCATm gene deletion. Such lack of a coordinated change in muscle fiber specific genes and their fiber-specific isoforms that are meant to work together, if translated into protein, might lead to a dystrophic muscle situation predicted by the pathway analysis. Further studies are needed to evaluate this possibility along with the mechanisms underlying these transcriptional adaptations including the involvement of the upstream pathways implicated by the gene ontology analyses.
GRANTS
The study was funded by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-084428.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: C.J.L., Y.X., and Y.I.K. conception and design of research; C.J.L., S.R.K., Y.X., A.C.S., and Y.I.K. analyzed data; C.J.L. and S.R.K. interpreted results of experiments; C.J.L. prepared figures; C.J.L. drafted manuscript; C.J.L., S.R.K., Y.X., A.C.S., and Y.I.K. edited and revised manuscript; C.J.L., S.R.K., Y.X., A.C.S., and Y.I.K. approved final version of manuscript; Y.X. and Y.I.K. performed experiments.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Charles Lang for helpful discussions.
Footnotes
The online version of this article contains supplemental material.
During this part of our bioinformatics evaluation, it was noticed that Myh4, normally a major gene and protein component in muscle, had 0 FPKM values for several animals. Notably, Myh4 has multiple Ensembl entries and a complicated exon structure for these entries (39 and 38 exons). Using a new version of the Cufflinks software (v2.2.1) provided very similar values for the other genes, but resolved this situation for Myh4; however, wild-type animal 2 was still flagged with an error. These new FPKM values for Myh4 are reported here, and DEGSeq was rerun using these values without the ones for wild-type animal 2 to provide data for this gene. The flagged value is shown in Supplementary Table S1, and the other values from the wild-type n = 3 analysis appear in Supplementary tables S2-S3 and are noted in those tables.
REFERENCES
- 1.Adibi SA. Influence of dietary deprivations on plasma concentration of free amino acids of man. J Appl Physiol 25: 52–57, 1968. [DOI] [PubMed] [Google Scholar]
- 2.Al-Sawaf O, Fragoulis A, Rosen C, Keimes N, Liehn EA, Hölzle F, Kan YW, Pufe T, Sönmez TT, Wruck CJ. Nrf2 augments skeletal muscle regeneration after ischaemia-reperfusion injury. J Pathol 234: 538–547, 2014. [DOI] [PubMed] [Google Scholar]
- 3.Allen DL, Cleary AS, Hanson AM, Lindsay SF, Reed JM. CCAAT/enhancer binding protein-delta expression is increased in fast skeletal muscle by food deprivation and regulates myostatin transcription in vitro. Am J Physiol Regul Integr Comp Physiol 299: R1592–R1601, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160: 595–606, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthony JC, Anthony TG, Kimball SR, Jefferson LS. Signaling pathways involved in translational control of protein synthesis in skeletal muscle by leucine. J Nutr 131: 856S–860S, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Anthony JC, Yoshizawa F, Anthony TG, Vary TC, Jefferson LS, Kimball SR. Leucine stimulates translation initiation in skeletal muscle of postabsorptive rats via a rapamycin-sensitive pathway. J Nutr 130: 2413–2419, 2000. [DOI] [PubMed] [Google Scholar]
- 7.Badoud F, Lam KP, DiBattista A, Perreault M, Zulyniak MA, Cattrysse B, Stephenson S, Britz-McKibbin P, Mutch DM. Serum and adipose tissue amino Acid homeostasis in the metabolically healthy obese. J Proteome Res 13: 3455–3466, 2014. [DOI] [PubMed] [Google Scholar]
- 8.Bailey AN, Hocker AD, Vermillion BR, Smolkowski K, Shah SN, Jewett BA, Dreyer HC. MAFbx, MuRF1, and the stress-activated protein kinases are upregulated in muscle cells during total knee arthroplasty. Am J Physiol Regul Integr Comp Physiol 303: R376–R386, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balage M, Sinaud S, Prod'homme M, Dardevet D, Vary TC, Kimball SR, Jefferson LS, Grizard J. Amino acids and insulin are both required to regulate assembly of the eIF4E. eIF4G complex in rat skeletal muscle. Am J Physiol Endocrinol Metab 281: E565–E574, 2001. [DOI] [PubMed] [Google Scholar]
- 10.Belfiore F, Iannello S, Rabuazzo AM. Insulin resistance in obesity: a critical analysis at enzyme level. A review. Int J Obes 3: 301–323, 1979. [PubMed] [Google Scholar]
- 11.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 270: 2320–2326, 1995. [DOI] [PubMed] [Google Scholar]
- 12.Bolster DR, Vary TC, Kimball SR, Jefferson LS. Leucine regulates translation initiation in rat skeletal muscle via enhanced eIF4G phosphorylation. J Nutr 134: 1704–1710, 2004. [DOI] [PubMed] [Google Scholar]
- 13.Britto FA, Begue G, Rossano B, Docquier A, Vernus B, Sar C, Ferry A, Bonnieu A, Ollendorff V, Favier FB. REDD1 deletion prevents dexamethasone-induced skeletal muscle atrophy. Am J Physiol Endocrinol Metab 307: E983–E993, 2014. [DOI] [PubMed] [Google Scholar]
- 14.Caballero B, Finer N, Wurtman RJ. Plasma amino acids and insulin levels in obesity: response to carbohydrate intake and tryptophan supplements. Metabolism 37: 672–676, 1988. [DOI] [PubMed] [Google Scholar]
- 15.Calura E, Cagnin S, Raffaello A, Laveder P, Lanfranchi G, Romualdi C. Meta-analysis of expression signatures of muscle atrophy: gene interaction networks in early and late stages. BMC Genomics 9: 630, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cen B, Li H, Weinstein IB. Histidine triad nucleotide-binding protein 1 up-regulates cellular levels of p27KIP1 by targeting ScfSKP2 ubiquitin ligase and Src. J Biol Chem 284: 5265–5276, 2009. [DOI] [PubMed] [Google Scholar]
- 17.Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, Pende M. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33: 474–483, 2014. [DOI] [PubMed] [Google Scholar]
- 18.Dardevet D, Sornet C, Balage M, Grizard J. Stimulation of in vitro rat muscle protein synthesis by leucine decreases with age. J Nutr 130: 2630–2635, 2000. [DOI] [PubMed] [Google Scholar]
- 19.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endesfelder S, Krahn A, Kreuzer KA, Lass U, Schmidt CA, Jahrmarkt C, von Moers A, Speer A. Elevated p21 mRNA level in skeletal muscle of DMD patients and mdx mice indicates either an exhausted satellite cell pool or a higher p21 expression in dystrophin-deficient cells per se. J Mol Med (Berl) 78: 569–574, 2000. [DOI] [PubMed] [Google Scholar]
- 21.Felig P, Marliss E, Cahill GF Jr. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med 281: 811–816, 1969. [DOI] [PubMed] [Google Scholar]
- 22.Fertuzinhos S, Li M, Kawasawa YI, Ivic V, Franjic D, Singh D, Crair M, Sestan N. Laminar and temporal expression dynamics of coding and noncoding RNAs in the mouse neocortex. Cell Rep 6: 938–950, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox HL, Kimball SR, Jefferson LS, Lynch CJ. Amino acids stimulate phosphorylation of p70S6k and organization of rat adipocytes into multicellular clusters. Am J Physiol Cell Physiol 274: C206–C213, 1998. [DOI] [PubMed] [Google Scholar]
- 24.Fried SK, Watford M. Leucing weight with a futile cycle. Cell Metab 6: 155–156, 2007. [DOI] [PubMed] [Google Scholar]
- 25.Gheyara AL, Vallejo-Illarramendi A, Zang K, Mei L, St-Arnaud R, Dedhar S, Reichardt LF. Deletion of integrin-linked kinase from skeletal muscles of mice resembles muscular dystrophy due to α7β1-integrin deficiency. Am J Pathol 171: 1966–1977, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg AL, Tischler M, Libby P. Regulation of protein degradation in skeletal muscle. Biochem Soc Trans 8: 497, 1980. [DOI] [PubMed] [Google Scholar]
- 27.Goodman CA, McNally RM, Hoffmann FM, Hornberger TA. Smad3 induces atrogin-1, inhibits mTOR and protein synthesis, and promotes muscle atrophy in vivo. Mol Endocrinol 27: 1946–1957, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, Cline GW, Kim JK, Peroni OD, Kahn BB, Jain MK. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab 5: 305–312, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haddad F, Adams GR, Bodell PW, Baldwin KM. Isometric resistance exercise fails to counteract skeletal muscle atrophy processes during the initial stages of unloading. J Appl Physiol (1985) 100: 433–441, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23: 8862–8877, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem 273: 14484–14494, 1998. [DOI] [PubMed] [Google Scholar]
- 32.Hatazawa Y, Tadaishi M, Nagaike Y, Morita A, Ogawa Y, Ezaki O, Takai-Igarashi T, Kitaura Y, Shimomura Y, Kamei Y, Miura S. PGC-1alpha-mediated branched-chain amino acid metabolism in the skeletal muscle. PLoS One 9: e91006, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawke TJ, Meeson AP, Jiang N, Graham S, Hutcheson K, DiMaio JM, Garry DJ. p21 is essential for normal myogenic progenitor cell function in regenerating skeletal muscle. Am J Physiol Cell Physiol 285: C1019–C1027, 2003. [DOI] [PubMed] [Google Scholar]
- 34.Hinault C, Mothe-Satney I, Gautier N, Lawrence JC Jr, Van Obberghen E. Amino acids and leucine allow insulin activation of the PKB/mTOR pathway in normal adipocytes treated with wortmannin and in adipocytes from db/db mice. FASEB J 18: 1894–1896, 2004. [DOI] [PubMed] [Google Scholar]
- 35.Hu H, Li X. Transcriptional regulation in eukaryotic ribosomal protein genes. Genomics 90: 421–423, 2007. [DOI] [PubMed] [Google Scholar]
- 36.Hutson SM, Wallin R, Hall TR. Identification of mitochondrial branched chain aminotransferase and its isoforms in rat tissues. J Biol Chem 267: 15681–15686, 1992. [PubMed] [Google Scholar]
- 37.Iadevaia V, Liu R, Proud CG. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin Cell Dev Biol 36: 113–120, 2014. [DOI] [PubMed] [Google Scholar]
- 38.Kaisari S, Rom O, Aizenbud D, Reznick AZ. Involvement of NF-kappaB and muscle specific E3 ubiquitin ligase MuRF1 in cigarette smoke-induced catabolism in C2 myotubes. Adv Exp Med Biol 788: 7–17, 2013. [DOI] [PubMed] [Google Scholar]
- 39.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175, 2002. [DOI] [PubMed] [Google Scholar]
- 40.Kimball SR. Regulation of translation initiation by amino acids in eukaryotic cells. Prog Mol Subcell Biol 26: 155–184, 2001. [DOI] [PubMed] [Google Scholar]
- 41.Kimball SR, Jefferson LS. Molecular mechanisms through which amino acids mediate signaling through the mammalian target of rapamycin. Curr Opin Clin Nutr Metab Care 7: 39–44, 2004. [DOI] [PubMed] [Google Scholar]
- 42.Kimball SR, Jefferson LS. Regulation of global and specific mRNA translation by oral administration of branched-chain amino acids. Biochem Biophys Res Commun 313: 423–427, 2004. [DOI] [PubMed] [Google Scholar]
- 43.Kimball SR, Jefferson LS. Regulation of protein synthesis by branched-chain amino acids. Curr Opin Clin Nutr Metab Care 4: 39–43, 2001. [DOI] [PubMed] [Google Scholar]
- 44.Kjær M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol Rev 84: 649–698, 2004. [DOI] [PubMed] [Google Scholar]
- 45.Koncarevic A, Jackman RW, Kandarian SC. The ubiquitin-protein ligase Nedd4 targets Notch1 in skeletal muscle and distinguishes the subset of atrophies caused by reduced muscle tension. FASEB J 21: 427–437, 2007. [DOI] [PubMed] [Google Scholar]
- 46.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–530, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lackey DE, Lynch CJ, Olson KC, Mostaedi R, Ali M, Smith WH, Karpe F, Humphreys S, Bedinger DH, Dunn TN, Thomas AP, Oort PJ, Kieffer DA, Amin R, Bettaieb A, Haj FG, Permana P, Anthony TG, Adams SH. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am J Physiol Endocrinol Metab 304: E1175–E1187, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lang CH, Lynch CJ, Vary TC. BCATm deficiency ameliorates endotoxin-induced decrease in muscle protein synthesis and improves survival in septic mice. Am J Physiol Regul Integr Comp Physiol 299: R935–R944, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Layman DK, Anthony TG, Rasmussen BB, Adams SH, Lynch CJ, Brinkworth GD, Davis TD. Defining meal requirements for protein to optimize metabolic roles of amino acids. Am J Clin Nutr 101: 1330S–1338S, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liberman N, Gandin V, Svitkin YV, David M, Virgili G, Jaramillo M, Holcik M, Nagar B, Kimchi A, Sonenberg N. DAP5 associates with eIF2beta and eIF4AI to promote Internal Ribosome Entry Site driven translation. Nucleic Acids Res 43: 3764–3775, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z, Jahn LA, Long W, Fryburg DA, Wei L, Barrett EJ. Branched chain amino acids activate messenger ribonucleic acid translation regulatory proteins in human skeletal muscle, and glucocorticoids blunt this action. J Clin Endocrinol Metab 86: 2136–2143, 2001. [DOI] [PubMed] [Google Scholar]
- 52.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 10: 723–736, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynch CJ, Hutson SM, Patson BJ, Vaval A, Vary TC. Tissue-specific effects of chronic dietary leucine and norleucine supplementation on protein synthesis in rats. Am J Physiol Endocrinol Metab 283: E824–E835, 2002. [DOI] [PubMed] [Google Scholar]
- 54.Lynch CJ, Patson BJ, Anthony J, Vaval A, Jefferson LS, Vary TC. Leucine is a direct-acting nutrient signal that regulates protein synthesis in adipose tissue. Am J Physiol Endocrinol Metab 283: E503–E513, 2002. [DOI] [PubMed] [Google Scholar]
- 55.Lynch CJ, Xu Y, Hajnal A, Salzberg AC, Kawasawa YI. RNA sequencing reveals a slow to fast muscle fiber type transition after olanzapine infusion in rats. PLoS One 10: e0123966, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979, 2004. [DOI] [PubMed] [Google Scholar]
- 57.Martin J, Magnino F, Schmidt K, Piguet AC, Lee JS, Semela D, St-Pierre MV, Ziemiecki A, Cassio D, Brenner C, Thorgeirsson SS, Dufour JF. Hint2, a mitochondrial apoptotic sensitizer down-regulated in hepatocellular carcinoma. Gastroenterology 130: 2179–2188, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCormack SE, Shaham O, McCarthy MA, Deik AA, Wang TJ, Gerszten RE, Clish CB, Mootha VK, Grinspoon SK, Fleischman A. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes 8: 52–61, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mordier S, Deval C, Bechet D, Tassa A, Ferrara M. Leucine limitation induces autophagy and activation of lysosome-dependent proteolysis in C2C12 myotubes through a mammalian target of rapamycin-independent signaling pathway. J Biol Chem 275: 29900–29906, 2000. [DOI] [PubMed] [Google Scholar]
- 60.Nakamura K, Nakano SI, Miyoshi T, Yamanouchi K, Nishihara M. Loss of SPARC in mouse skeletal muscle causes myofiber atrophy. Muscle Nerve 48: 791–799, 2013. [DOI] [PubMed] [Google Scholar]
- 61.Neishabouri SH, Hutson SM, Davoodi J. Chronic activation of mTOR complex 1 by branched chain amino acids and organ hypertrophy. Amino Acids 47: 1167–1182, 2015. [DOI] [PubMed] [Google Scholar]
- 62.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab 15: 606–614, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS Jr, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9: 311–326, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oishi K, Yamamoto S, Itoh N, Miyazaki K, Nemoto T, Nakakita Y, Kaneda H. Disruption of behavioral circadian rhythms induced by psychophysiological stress affects plasma free amino acid profiles without affecting peripheral clock gene expression in mice. Biochem Biophys Res Commun 450: 880–884, 2014. [DOI] [PubMed] [Google Scholar]
- 65.Pagel CN, Wasgewatte Wijesinghe DK, Taghavi Esfandouni N, Mackie EJ. Osteopontin, inflammation and myogenesis: influencing regeneration, fibrosis and size of skeletal muscle. J Cell Commun Signal 8: 95–103, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pena-Llopis S, Vega-Rubin-de-Celis S., Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR, Brugarolas J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J 30: 3242–3258, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peyrollier K, Hajduch E, Blair AS, Hyde R, Hundal HS. L-leucine availability regulates phosphatidylinositol 3-kinase, p70 S6 kinase and glycogen synthase kinase-3 activity in L6 muscle cells: evidence for the involvement of the mammalian target of rapamycin (mTOR) pathway in the L-leucine-induced up-regulation of system A amino acid transport. Biochem J 350: 361–368, 2000. [PMC free article] [PubMed] [Google Scholar]
- 68.Raffaello A, Milan G, Masiero E, Carnio S, Lee D, Lanfranchi G, Goldberg AL, Sandri M. JunB transcription factor maintains skeletal muscle mass and promotes hypertrophy. J Cell Biol 191: 101–113, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rom O, Kaisari S, Aizenbud D, Reznick AZ. Cigarette smoke and muscle catabolism in C2 myotubes. Mech Ageing Dev 134: 24–34, 2013. [DOI] [PubMed] [Google Scholar]
- 70.Rom O, Kaisari S, Reznick AZ, Aizenbud D. Peroxynitrite induces degradation of myosin heavy chain via p38 MAPK and muscle-specific E3 ubiquitin ligases in C2 skeletal myotubes. Adv Exp Med Biol 832: 1–8, 2015. [DOI] [PubMed] [Google Scholar]
- 71.Sambasivan R, Cheedipudi S, Pasupuleti N, Saleh A, Pavlath GK, Dhawan J. The small chromatin-binding protein p8 coordinates the association of anti-proliferative and pro-myogenic proteins at the myogenin promoter. J Cell Sci 122: 3481–3491, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Setsuie R, Suzuki M, Tsuchiya Y, Wada K. Skeletal muscles of Uchl3 knockout mice show polyubiquitinated protein accumulation and stress responses. Neurochem Int 56: 911–918, 2010. [DOI] [PubMed] [Google Scholar]
- 73.She P, Olson KC, Kadota Y, Inukai A, Shimomura Y, Hoppel CL, Adams SH, Kawamata Y, Matsumoto H, Sakai R, Lang CH, Lynch CJ. Leucine and protein metabolism in obese Zucker rats. PLoS One 8: e59443, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.She P, Reid TM, Bronson SK, Vary TC, Hajnal A, Lynch CJ, Hutson SM. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab 6: 181–194, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.She P, Van Horn C, Reid T, Hutson SM, Cooney RN, Lynch CJ. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am J Physiol Endocrinol Metab 293: E1552–E1563, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.She P, Zhou Y, Zhang Z, Griffin K, Gowda K, Lynch CJ. Disruption of BCAA metabolism in mice impairs exercise metabolism and endurance. J Appl Physiol (1985) 108: 941–949, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shigemitsu K, Tsujishita Y, Miyake H, Hidayat S, Tanaka N, Hara K, Yonezawa K. Structural requirement of leucine for activation of p70 S6 kinase. FEBS Lett 447: 303–306, 1999. [DOI] [PubMed] [Google Scholar]
- 78.Shimizu N, Yoshikawa N, Ito N, Maruyama T, Suzuki Y, Takeda S, Nakae J, Tagata Y, Nishitani S, Takehana K, Sano M, Fukuda K, Suematsu M, Morimoto C, Tanaka H. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab 13: 170–182, 2011. [DOI] [PubMed] [Google Scholar]
- 79.Shin AC, Fasshauer M, Filatova N, Grundell LA, Zielinski E, Zhou JY, Scherer T, Lindtner C, White PJ, Lapworth AL, Ilkayeva O, Knippschild U, Wolf AM, Scheja L, Grove KL, Smith RD, Qian WJ, Lynch CJ, Newgard CB, Buettner C. Brain insulin lowers circulating BCAA levels by inducing hepatic BCAA catabolism. Cell Metab 20: 898–909, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tiano JP, Springer DA, Rane SG. SMAD3 negatively regulates serum irisin and skeletal muscle FNDC5 and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1alpha) during exercise. J Biol Chem 290: 7671–7684, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tischler ME, Desautels M, Goldberg AL. Does leucine, leucyl-tRNA, or some metabolite of leucine regulate protein synthesis and degradation in skeletal and cardiac muscle? J Biol Chem 257: 1613–1621, 1982. [PubMed] [Google Scholar]
- 82.Tonkin J, Rosenthal N. One small step for muscle: a new micropeptide regulates performance. Cell Metab 21: 515–516, 2015. [DOI] [PubMed] [Google Scholar]
- 83.Wang M, Amano SU, Flach RJR, Chawla A, Aouadi M, Czech MP. Identification of Map4k4 as a novel suppressor of skeletal muscle differentiation. Mol Cell Biol 33: 678–687, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med 17: 448–453, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wurtz P, Makinen VP, Soininen P, Kangas AJ, Tukiainen T, Kettunen J, Savolainen MJ, Tammelin T, Viikari JS, Ronnemaa T, Kahonen M, Lehtimaki T, Ripatti S, Raitakari OT, Jarvelin MR, Ala-Korpela M. Metabolic signatures of insulin resistance in 7,098 young adults. Diabetes 61: 1372–1380, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu G, Kwon G, Marshall CA, Lin TA, Lawrence JC Jr, McDaniel ML. Branched-chain amino acids are essential in the regulation of PHAS-I and p70 S6 kinase by pancreatic beta-cells A possible role in protein translation and mitogenic signaling. J Biol Chem 273: 28178–28184, 1998. [DOI] [PubMed] [Google Scholar]
- 87.Zeng YJ, Zeng FQ, Dai L, Yang C, Lin BZ, Zheng DH, Liu D, Yan L, Ren M, Cheng H. Characteristics and risk factors for hyperglycemia in Chinese female patients with systemic lupus erythematosus. Lupus 19: 1344–1350, 2010. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y, Guo K, LeBlanc RE, Loh D, Schwartz GJ, Yu YH. Increasing dietary leucine intake reduces diet-induced obesity and improves glucose and cholesterol metabolism in mice via multimechanisms. Diabetes 56: 1647–1654, 2007. [DOI] [PubMed] [Google Scholar]
- 89.Zhou Y, Jetton TL, Goshorn S, Lynch CJ, She P. Transamination is required for alpha-ketoisocaproate but not leucine to stimulate insulin secretion. J Biol Chem 285: 33718–33726, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zimmerman HA, Olson KC, Chen G, Lynch CJ. Adipose transplant for inborn errors of branched chain amino acid metabolism in mice. Mol Genet Metab 109: 345–353, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.