Abstract
The objective of this study was to investigate the genetic basis of high level aminoglycoside resistance in Acinetobacter baumannii clinical isolates from Beijing, China. 173 A. baumannii clinical isolates from hospitals in Beijing from 2006 to 2009 were first subjected to high level aminoglycoside resistance (HLAR, MIC to gentamicin and amikacin>512 µg/mL) phenotype selection by broth microdilution method. The strains were then subjected to genetic basis analysis by PCR detection of the aminoglycoside modifying enzyme genes (aac(3)-I, aac(3)-IIc, aac(6′)-Ib, aac(6′)-II, aph(4)-Ia, aph(3′)-I, aph(3′)-IIb, aph(3′)-IIIa, aph(3′)-VIa, aph(2″)-Ib, aph(2″)-Ic, aph(2″)-Id, ant(2″)-Ia, ant(3″)-I and ant(4′)-Ia) and the 16S rRNA methylase genes (armA, rmtB and rmtC). Correlation analysis between the presence of aminoglycoside resistance gene and HLAR phenotype were performed by SPSS. Totally 102 (58.96%) HLAR isolates were selected. The HLAR rates for year 2006, 2007, 2008 and 2009 were 52.63%, 65.22%, 51.11% and 70.83%, respectively. Five modifying enzyme genes (aac(3)-I, detection rate of 65.69%; aac(6′)-Ib, detection rate of 45.10%; aph(3′)-I, detection rate of 47.06%; aph(3′)-IIb, detection rate of 0.98%; ant(3″)-I, detection rate of 95.10%) and one methylase gene (armA, detection rate of 98.04%) were detected in the 102 A. baumannii with aac(3)-I+aac(6′)-Ib+ant(3″)-I+armA (detection rate of 25.49%), aac(3)-I+aph(3′)-I+ant(3″)-I+armA (detection rate of 21.57%) and ant(3″)-I+armA (detection rate of 12.75%) being the most prevalent gene profiles. The values of chi-square tests showed correlation of armA, ant(3″)-I, aac(3)-I, aph(3′)-I and aac(6′)-Ib with HLAR. armA had significant correlation (contingency coefficient 0.685) and good contingency with HLAR (kappa 0.940). The high rates of HLAR may cause a serious problem for combination therapy of aminoglycoside with β-lactams against A. baumannii infections. As armA was reported to be able to cause high level aminoglycoside resistance to most of the clinical important aminoglycosides (gentamicin, amikacin, tobramycin, etc), the function of aminoglycoside modifying enzyme gene(s) in A. baumannii carrying armA deserves further investigation.
KEY WORDS: Acinetobacter baumannii, HLAR, Aminoglycoside modifying enzyme, 16S rRNA methylase, Correlation analysis
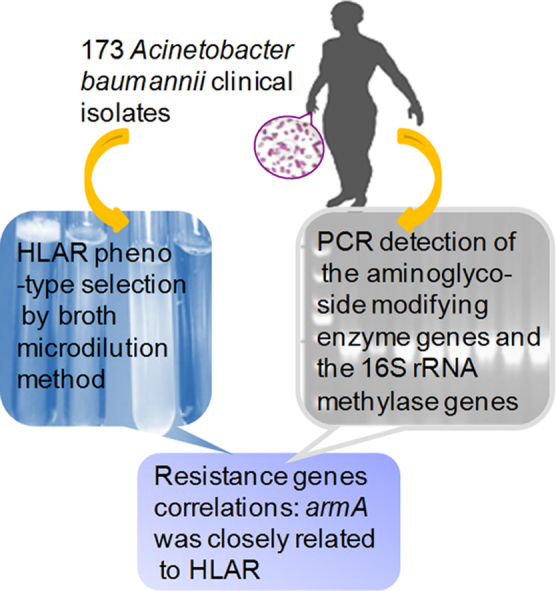
Graphical abstract
Using broth microdilution method and PCR detection analysis, genetic basis of high level aminoglycoside resistance was investigated in the 173 Acinetobacter baumannii clinical isolates from hospitals in Beijing from 2006 to 2009. The armA gene was closely related to HLAR.
1. Introduction
Acinetobacter baumannii is a notorious Gram-negative pathogen found in clinical settings due to its epidemic tendency and multidrug resistance (MDR)1, 2. It can cause serious infections like ventilator associated pneumonia (VAP), skin and soft tissue infection, wound infection, secondary meningitis, blood infection, etc2, 3. Since A. baumannii is commonly resistant to clinically available antimicrobial agents, including β-lactams, aminoglycosides and fluoroquinolones, the selection of appropriate antibiotics is increasingly limited3, 4, 5.
Aminoglycosides, which bind specifically to 16S rRNA in the 30S ribosomal subunits to inhibit protein synthesis, are often used in combination with broad spectrum β-lactams to treat Gram-negative bacterial infections6, 7, 8. Resistance to aminoglycosides is most commonly caused by aminoglycoside modifying enzymes, including acetyltransferases, phosphotransferases and nucleotidyltransferases9, 10. More recently, 16S rRNA methylases, ArmA, RmtA, RmtB, RmtC, RmtD, RmtE and NpmA have been reported among Enterobacteriaceae, Pseudomonas spp. and Acinetobacter spp.11, 12. Aminoglycoside modifying enzymes differ in aminoglycosides that they may modified, whereas 16S rRNA methylases confer high-level resistance to almost all aminoglycosides except streptomycin11, 13, 14.
The purpose of our study was to investigate the genetic basis of high level aminoglycoside resistance in A. baumannii clinical isolates from hospitals in Beijing, China.
2. Materials and methods
2.1. Bacterial Strains
173 A. baumannii clinical isolates collected from hospitals in Beijing, China between 2006 and 2009 were included in the current study, including 57 isolates in 2006, 23 isolates in 2007, 45 isolates in 2008 and 48 isolates in 2009. The strains were identified further in our laboratory by VITEK 2-compact bacteria identification system (Bio-Merieux Company) and by sequence analysis of the conserved region of 16S rRNA gene. Escherichia coli ATCC 25,922 and A. Baumannii ATCC 19606 were standard strains from American Type Culture Collection (ATCC).
2.2. Antimicrobial susceptibility to gentamicin and amikacin
The antimicrobial susceptibility of the isolates to gentamicin and amikacin were determined by microdilution method in CAMH broth (cation-adjusted Mueller–Hinton broth) according to CLSI recommendation. Three concentrations (1024, 512 and 256 µg/mL) were included in the experiment. The strains were recognized as high level aminoglycoside resistant (HLAR) if the MICs against gentamicin and amikacin were both higher than 512 µg/mL. E. coli ATCC 25922 and A. Baumannii ATCC 19606 were used as controls.
2.3. Polymerase chain reaction amplification of the aminoglycoside resistance genes
Polymerase chain reaction (PCR) was performed in a total volume of 25 µL containing one single colony, 0.6 µmol/L of each primer and 12.5 µL of 2×Go Taq Green Master Mix (Promega). The genes encoding the following aminoglycoside modifying enzymes were investigated: acetyltransferases AAC(3)-I, AAC(3)-IIc, AAC(6′)-Ib and AAC(6′)-II; phosphotransferases APH(4)-Ia, APH(3′)-I, APH(3′)-IIb, APH(3′)-IIIa, APH(3′)-VIa, APH(2″)-Ib, APH(2″)-Ic and APH(2″)-Id; nucleotidyltransferases ANT(2″)-Ia, ANT(3″)-I, ANT(4′)-Ia. The 16S rRNA methylase genes investigated included armA, rmtB and rmtC. The primer sequences, expected amplicon sizes and the annealing temperatures for PCR are shown in Table 1. The amplification reaction with a DNA thermal cycler (Perkin-Elmer Cetus, Foster City, CA) consisted of a predenaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 30 s, 55 or 58 °C for 30 s, extension at 72 °C for 1 min, and a final elongation at 72 °C for 5 min.
Table 1.
The primer sequences, amplicon sizes and annealing temperatures for PCR.
| Gene | Primer sequence | Amplicon size (bp) | Annealing temperature (°C) | Ref. |
|---|---|---|---|---|
| Aminoglycoside modifying enzyme gene | ||||
| ant(2″)-Ia | F: 5′-GCTCACGCAACTGGTCCA GA-3′ | 719 | 58 | 15 |
| R: 5′-GGCACGCAAGACCTCAACCT-3′ | ||||
| ant(3″)-I | F: 5′-TGATTTGCTGGTTACGGTGAC-3′ | 284 | 55 | 16 |
| R: 5′-CGCTATGTTCTCTTGCTTTTG-3′ | ||||
| ant(4′)-Ia | F: 5′-CTGCTAAATCGGTAGAAGC-3′ | 172 | 55 | 17 |
| R: 5′-CAGACCAATCAACATGGCACC-3′ | ||||
| aac(3)-I | F: 5′-TTACGCAGCAGCAACGATGT-3′ | 402 | 58 | 15 |
| R: 5′-GTTGGCCTCATGCTTGAGGA-3′ | ||||
| aac(3)-IIc | F: 5′-ACGCGGAAGGCAATAACGGA-3′ | 854 | 55 | 15 |
| R: 5′-TAACCTGAAGGCTCGCAAGA-3′ | ||||
| aac(6′)-Ib | F: 5′-CATGACCTTGCGATGCTCTA-3′ | 490 | 58 | 15 |
| R: 5′-GCTCGAATGCCTGGCGTCTT-3′ | ||||
| aac(6′)-II | F: 5′-TTCATGTCCGCGAGCACCCC-3′ | 178 | 55 | 18 |
| R: 5′-GACTCTTCCGCCATCGCTCT-3′ | ||||
| aph(2″)-Ib | F:5′-CTTGGACGCTGAGATATATGAGCAC-3′ | 867 | 55 | 19 |
| R:5′-GTTTGTAGCAATTCAGAAACACCCTT-3′ | ||||
| aph(2″)-Ic | F: 5′-CCACAATGATAATGACTCAGTTCCC-3′ | 444 | 55 | 19 |
| R: 5′-CCACAGCTTCCGATAGCAAGAG-3′ | ||||
| aph(2″)-Id | F: 5′-GTGGTTTTTACAGGAATGCCATC-3′ | 641 | 55 | 16 |
| R:5′-CCCTCTTCATACCAATCCATATAACC-3′ | ||||
| aph(3′)-I | F: 5′-ATGTGCCATATTCAACGGGAAACG-3′ | 816 | 55 | 16 |
| R:5′-TCAGAAAAACTCATCGAGCATCAA-3′ | ||||
| aph(3′)-IIb | F: 5′-ATGCATGATGCAGCCACCTCC-3′ | 804 | 55 | 17 |
| R: 5′-CTAGAAGAACTCGTCCAATAGCCT-3′ | ||||
| aph(3′)-IIIa | F: 5′-GGCTAAAATGAGAATATCACCGG-3′ | 278 | 55 | 17 |
| R: 5′-CTTTAAAAAATCATACAGCTCGCG-3′ | ||||
| aph(3′)-VIa | F: 5′-ATACAGAGACCACCATACAGT-3′ | 234 | 55 | 16 |
| R: 5′-GGACAATCAATAATAGCAAT-3′ | ||||
| aph(4)-Ia | F: 5′-CTGAACTCACCGCGACGTCT-3′ | 977 | 58 | 15 |
| R: 5′-TCCACTATCGGCGAGTACTT-3′ | ||||
| 16S rRNA methylase gene | ||||
| rmtB | F: 5′-GCTTTCTGCGGGCGATGTAA-3′ | 173 | 55 | 20 |
| R: 5′-ATGCAATGCCGCGCTCGTAT-3′ | ||||
| rmtC | F: 5′-CGAAGAAGTAACAGCCAAAG-3′ | 711 | 55 | 20 |
| R: 5′-ATCCCAACATCTCTCCCACT-3′ | ||||
| armA | F: 5′-ATTCTGCCTATCCTAATTGG-3′ | 315 | 55 | 20 |
| R: 5′-ACCTATACTTTATCGTCGTC-3′ | ||||
2.4. Correlation analysis between aminoglycoside resistance gene and HLAR phenotype
The correlations of aminoglycoside resistance gene with HLAR phenotype were statistically analyzed by chi-square test using SPSS 13.0. Based on the nature of the data, Pearson׳s chi-square test was used in correlation analysis of aac(3)-I, aac(6′)-Ib, aph(3′)-I or ant(3″)-I with HLAR phenotype, and Fisher׳s exact test was used in correlation analysis of aac(3)-IIc, aac(6′)-II, aph(3′)-IIb or armA with HLAR phenotype, respectively. Correlations were evaluated by P values. Contingency coefficient and kappa values obtained for each gene. The gene was considered correlated with HLAR if P value <0.05, and no correlation if P value ≥0.05. Contingency coefficient was used to measure the extent of the correlation, and higher value suggested stronger correlation. Kappa value was the scale of the correlation agreement (≥0.75, good agreement; 0.75>kappa value ≥0.4, general agreement;<0.4, poor agreement).
3. Results and discussion
3.1. Antimicrobial susceptibility to gentamicin and amikacin
Antimicrobial susceptibility of A. baumannii to gentamicin and amikacin was determined by broth microdilution method, and the results are summarized in Table 2. Totally 102 isolates showed high level aminoglycoside resistance (HLAR) with a HLAR rate of 58.96%. The HLAR rates for year 2006, 2007, 2008 and 2009 were 52.63%, 65.22%, 51.11% and 70.83%, respectively. The high rates of HLAR might cause a serious problem for combination therapy of aminoglycoside with β-lactams against A. baumannii infections.
Table 2.
High level aminoglycoside resistance among 173 Acinetobacter baumannii clinical isolates from Beijing, China.
| Year | Total strains | Strains of Sa |
Strains of Ib |
Strains of Rc |
Strains of non-HLARd | Strains of HLAR | HLAR rate (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Amke | Gmf | Amk | Gm | Amk | Gm | |||||
| 2006 | 57 | 27 | 22 | 0 | 0 | 30 | 35 | 27 | 30 | 52.63 |
| 2007 | 23 | 8 | 7 | 0 | 1 | 15 | 15 | 8 | 15 | 65.22 |
| 2008 | 45 | 22 | 14 | 0 | 0 | 23 | 31 | 22 | 23 | 51.11 |
| 2009 | 48 | 14 | 10 | 0 | 0 | 34 | 38 | 14 | 34 | 70.83 |
| Total | 173 | 71 | 53 | 0 | 1 | 102 | 119 | 71 | 102 | 58.96 |
S: susceptible
I: intermediate
R: resistance
HLAR: high level aminoglycoside resistance as demonstrated by MICs to amikacin and gentamicin higher than 512 µg/mL, non-HLAR: non-high level aminoglycoside resistance
Amk: Amikacin
Gm: Gentamicin
3.2. Polymerase chain reaction amplification of the aminoglycoside resistance genes
Totally 15 aminoglycoside modifying enzyme genes and three 16S rRNA methylase genes were investigated in all of the isolates. The results are shown in Table 3. Of the 15 aminoglycoside modifying enzyme genes investigated, seven were detected in the current A. baumannii isolates, with positive rates of 66.47%, 45.09%, 34.10%, 32.37%, 0.58%, 0.58% and 0.58% for ant(3″)-I, aac(3)-I, aph(3′)-I, aac(6′)-Ib, aac(3)-IIc, aac(6′)-II and aph(3′)-IIb, respectively. Among the positive aminoglycoside modifying enzyme genes, five were detected in the 102 HLAR isolates, with positive rates of 95.10%, 65.69%, 47.06%, 45.10% and 0.98% for ant(3″)-I, aac(3)-I, aph(3′)-I, aac(6′)-Ib and aph(3′)-IIb, respectively. The high detection rates of ant(3″)-I, aac(3)-I, aph(3′)-I and aac(6′)-Ib genes in the HLAR strains of our study were consistent with those reported by Cho et al.21. Among the three methylase genes, only armA was detected with a positive rate of 59.54% for all the strains and 98.04% in the 102 HLAR isolates (100 out of 102 strains showed positive results). rmtB or rmtC genes were not able to be detected in the current isolate group. These results were also in accordance with other reports which found armA to be the only 16S rRNA methylase gene detected in high level aminoglycoside resistant A. baumannii 13, 14, 21, 22.
Table 3.
Distribution of aminoglycoside resistance genes in 173 Acinetobacter baumannii isolates.
| Result | Aminoglycoside resistant genes |
|||||||
|---|---|---|---|---|---|---|---|---|
| armA | aac(3)-I | aac(3)-IIc | aac(6′)-Ib | aac(6′)-II | aph(3′)-I | aph(3′)-IIb | ant(3″)-I | |
| Positive isolates from HLAR | 100 | 67 | 0 | 46 | 0 | 48 | 1 | 97 |
| Positive rate from HLAR (%) | 98.04 | 65.69 | 0 | 45.10 | 0 | 47.06 | 0.98 | 95.10 |
| Positive isolates from non-HLAR | 3 | 11 | 1 | 10 | 1 | 11 | 0 | 18 |
| Positive rate from non-HLAR (%) | 4.23 | 15.49 | 1.41 | 14.08 | 1.41 | 15.49 | 0 | 25.35 |
| Total positive isolates | 103 | 78 | 1 | 56 | 1 | 59 | 1 | 115 |
| Positive rate (%) | 59.54 | 45.09 | 0.58 | 32.37 | 0.58 | 34.10 | 0.58 | 66.47 |
Aminoglycoside resistance genes were also detectable in non-HLAR strains, though at relatively low rates. The presence of the aminoglycoside resistance gene in non-HLAR strains suggested that complicated regulation mechanisms were involved in the onset of the HLAR phenotype.
3.3. Aminoglycoside resistance gene profile
The aminoglycoside resistance gene profiles of the 102 HLAR A. baumannii are shown in Table 4. As demonstrated, aac(3)-I+aac(6′)-Ib+ant(3″)-I+armA, aac(3)-I+aph(3′)-I+ant(3″)-I+armA and ant(3″)-I+armA were the most prevalent resistance gene profiles, with positive rates of 25.49%, 21.57% and 12.75%, respectively. Resistance gene profiles of secondary high detection rates were aac(3)-I+aac(6′)-Ib+aph(3′)-I+ant(3″)-I+armA, aac(3)-I+ant(3″)-I+armA and aph(3′)-I+ant(3″)-I+armA, and the corresponding positive rates were 8.82%, 7.84% and 7.84%. Other resistance gene profiles included aac(6′)-Ib+aph(3′)-I+ant(3″)-I+armA (detection rate of 4.90%), aph(3′)-I+armA (detection rate of 3.92%), aac(6′)-Ib+ant(3″)-I+armA (detection rate of 3.92%), aac(3)-I+aac(6′)-Ib+ant(3″)-I (detection rate of 0.98%) and aac(3)-I+aph(3′)-IIb+aac(6′)-Ib+ant(3″)-I+armA (detection rate of 0.98%).
Table 4.
Aminoglycoside resistance gene profiles of the 102 HLAR Acinetobacter baumannii.
| Aminoglycoside resistance gene profile | No. of isolate | Positive rate (%) |
|---|---|---|
| aac(3)-I+ aac(6′)-Ib+ant(3″)-I+armA | 26 | 25.49 |
| aac(3)-I+ aph(3′)-I+ant(3″)-I+armA | 22 | 21.57 |
| ant(3″)-I+armA | 13 | 12.75 |
| aac(3)-I+aac(6′)-Ib+aph(3′)-I+ant(3″)-I+armA | 9 | 8.82 |
| aac(3)-I+ant(3″)-I+armA | 8 | 7.84 |
| aph(3′)-I+ant(3″)-I+armA | 8 | 7.84 |
| aac(6′)-Ib+aph(3′)-I+ant(3″)-I+armA | 5 | 4.90 |
| aph(3′)-I+armA | 4 | 3.92 |
| aac(6′)-Ib+ant(3″)-I+armA | 4 | 3.92 |
| aac(3)-I+aac(6′)-Ib+ant(3″)-I | 1 | 0.98 |
| aac(3)-I+ aph(3′)-IIb+aac(6′)-Ib+ant(3″)-I+armA | 1 | 0.98 |
| None of 18 aminoglycoside resistance genes | 1 | 0.98 |
As shown in Table 4, the methylase gene armA was detected along with aminoglycoside modifying enzyme genes for most of the isolates investigated except 2 (one showed positive result for aac(3)-I+aac(6′)-Ib+ant(3″)-I gene, and the other showed no positive result for all the 18 aminoglycoside resistance genes). As armA was reported to be able to cause high level aminoglycoside resistance to most of the clinical important aminoglycosides (gentamicin, amikacin, tobramycin, etc.)1, the function of aminoglycoside modifying enzyme gene(s) in A. baumannii carrying armA deserves further investigation. The HLAR isolate with negative results for all of the 18 aminoglycoside resistance genes also needs our further study.
3.4. Correlation analysis between aminoglycoside resistance gene and HLAR phenotype
Data were statistically analyzed by chi-square test using SPSS 13.0, and the results are summarized in Table 5. The values of chi-square test showed armA, ant(3″)-I, aac(3)-I, aph(3′)-I and aac(6′)-Ib associated with HLAR. A contingency coefficient of 0.685 showed that armA was significantly correlated with HLAR. The contingency coefficients for ant(3″)-I, aac(3)-I, aph(3′)-I and aac(6′)-Ib were 0.588, 0.444, 0.311 and 0.310, respectively. Kappa values were further used to scale the correlation agreement. Among the 5 correlative genes, armA had good contingency (kappa value of 0.940), ant(3″)-I and aac(3)-I had general contingency (kappa values of 0.717 and 0.477), whereas aph(3′)-I and aac(6′)-Ib had poor consistency (kappa values of 0.289 and 0.282).
Table 5.
Correlation analysis between aminoglycoside resistance gene and HLAR (chi-square test).
| Aminoglycoside resistant genes | P value | Contingency coefficient | Kappa value |
|---|---|---|---|
| armA | 0.000 | 0.685 | 0.940 |
| aac(3)-I | 0.000 | 0.444 | 0.477 |
| aac(3)-IIc | 0.410 | 0.091 | −0.012 |
| aac(6′)-Ib | 0.000 | 0.310 | 0.282 |
| aac(6′)-II | 0.410 | 0.091 | −0.012 |
| aph(3′)-I | 0.000 | 0.311 | 0.289 |
| aph(3′)-IIb | 1.000 | 0.063 | 0.008 |
| ant(3″)-I | 0.000 | 0.588 | 0.717 |
4. Conclusions
A. baumannii clinical isolates collected between 2006 and 2009 from the hospitals in Beijing, China showed high levels of aminoglycoside resistance. Several resistance genes were detected in A. baumannii clinical isolates, and coexistence of resistance genes was found in most strains. Correlation analysis demonstrated that armA gene was closely related to HLAR. The high rates of HLAR in these clinical isolates may cause a serious problem for combination therapy of aminoglycoside with β-lactams against A. baumannii infections.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos. 81321004 and 81361138020) and the National Mega-project for Innovative Drugs (Nos. 2012ZX09301002-001, 2012ZX09301002-005 and 2014ZX09507009).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Contributor Information
Xiukun Wang, Email: xiukunwang@139.com.
Xuefu You, Email: xuefuyou@hotmail.com.
References
- 1.Thamlikitkul V., Tiengrim S. In vitro activity of colistin plus sulbactam against extensive-drug-resistant Acinetobacter baumannii by checkerboard method. J Med Assoc Thai. 2014;97 Suppl 3(Suppl 3):1–6. [PubMed] [Google Scholar]
- 2.Mortensen E., Trivedi K.K., Rosenberg J., Cody S.H., Long J., Jensen B.J. Multidrug-resistant Acinetobacter baumannii infection, colonization, and transmission related to a long-term care facility providing subacute care. Infect Control Hosp Epidemiol. 2014;35:406–411. doi: 10.1086/675612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gootz T.D. The global problem of antibiotic resistance. Crit Rev Immunol. 2010;30:79–93. doi: 10.1615/critrevimmunol.v30.i1.60. [DOI] [PubMed] [Google Scholar]
- 5.Cao J., Song W., Gu B., Mei Y.N., Tang J.P., Meng L. Correlation between carbapenem consumption and antimicrobial resistance rates of Acinetobacter baumannii in a university-affiliated hospital in China. J Clin Pharmacol. 2013;53:96–102. doi: 10.1177/0091270011435988. [DOI] [PubMed] [Google Scholar]
- 6.Lioy V.S., Goussard S., Guerineau V., Yoon E.J., Courvalin P., Galimand M. Aminoglycoside resistance 16S rRNA methyltransferases block endogenous methylation, affect translation efficiency and fitness of the host. RNA. 2014;20:382–391. doi: 10.1261/rna.042572.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naas T., Bentchouala C., Lima S., Lezzar A., Smati F., Scheftel J.M. Plasmid-mediated 16S rRNA methylases among extended spectrum beta-lactamase producing Salmonella enterica Senftenberg isolates from Algeria. J Antimicrob Chemother. 2009;64:866–868. doi: 10.1093/jac/dkp312. [DOI] [PubMed] [Google Scholar]
- 8.Doi Y., Adams J.M., Yamane K., Paterson D.L. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob Agents Chemother. 2007;51:4209–4210. doi: 10.1128/AAC.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He W., Zhang X., Zhang J., Jia X., Zhang J., Sun W. Riboswitch control of induction of aminoglycoside resistance acetyl and adenyl-transferases. RNA Biol. 2013;10:1266–1273. doi: 10.4161/rna.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labby K.J., Garneau-Tsodikova S. Strategies to overcome the action of aminoglycoside-modifying enzymes for treating resistant bacterial infections. Future Med Chem. 2013;5:1285–1309. doi: 10.4155/fmc.13.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y., Yu H., Guo Q., Xu X., Ye X., Wu S. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis. 2010;29:1349–1353. doi: 10.1007/s10096-010-1004-1. [DOI] [PubMed] [Google Scholar]
- 12.Livermore D.M. Current epidemiology and growing resistance of Gram-negative pathogens. Korean J Intern Med. 2012;27:128–142. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan M., Han H., Li C.R., Yang X.Y., Li G.Q., Cen S. Susceptibility of vertilmicin to modifications by three types of recombinant aminoglycoside-modifying enzymes. Antimicrob Agents Chemother. 2011;55:3950–3953. doi: 10.1128/AAC.00300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You X.F., Li C.R., Yang X.Y., Yuan M., Zhang W.X., Lou R.H. In vivo antibacterial activity of vertilmicin, a new aminoglycoside antibiotic. Antimicrob Agents Chemother. 2009;53:4525–4528. doi: 10.1128/AAC.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao L., Chen Q., Hu C., Jin S. Analysis of antibiotic resistance profile and gene patterns for aminoglycoside modifying enzymes in 64 strains of Escherichia coli. Chin J Microbiol Immunol. 2005;25:727–732. [Google Scholar]
- 16.Zhu J.M., Jiang R.J., Wu K.L., Wang J.M., Mo Y.S., Ma Z.L. Identification on a new subtype gene of aminoglycoside modifying enzyme in multi-drug resistant Acinetobacter baumannii. Chin J Nosocomiol. 2009;18:2371–2375. [Google Scholar]
- 17.Schmitz F.J., Fluit A.C., Gondolf M., Beyrau R., Lindenlauf E., Verhoef J. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J Antimicrob Chemother. 1999;43:253–259. [PubMed] [Google Scholar]
- 18.Tang Y., Liang Y., Dong S., Lu J. Detection of aminoglycosides resistance condition and the relative genes of Pseudomonas aeruginosa. Inter J Lab Med. 2006;27:677–690. [Google Scholar]
- 19.Vakulenko S.B., Donabedian S.M., Voskresenskiy A.M., Zervos M.J., Lerner S.A., Chow J.W. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob Agents Chemother. 2003;47:1423–1426. doi: 10.1128/AAC.47.4.1423-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doi Y., Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45:88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 21.Cho Y.J., Moon D.C., Jin J.S., Choi C.H., Lee Y.C., Lee J.C. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn Microbiol Infect Dis. 2009;64:185–190. doi: 10.1016/j.diagmicrobio.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Yamada Y., Suwabe A. Diverse carbapenem-resistance mechanisms in 16S rRNA methylase-producing Acinetobacter baumannii. J Med Microbiol. 2013;62:618–622. doi: 10.1099/jmm.0.048991-0. [DOI] [PubMed] [Google Scholar]



