Abstract
The bZIP transcription factor NFIL3 (Nuclear factor Interleukin 3 regulated, also known as E4 binding protein 4, E4BP4) regulates diverse biological processes from circadian rhythm to cellular viability. Recently, a host of novel roles have been identified for NFIL3 in immunological signal transduction, cancer, aging and metabolism. Elucidating the signaling pathways that are impacted by NFIL3 and the regulatory mechanisms that it targets, inhibits or activates will be critical for developing a clearer picture of its physiological roles in disease and normal processes. This review will discuss the recent advances and emerging issues regarding NFIL3-mediated transcriptional regulation of CEBPb and FOXO1 activated genes and signal transduction.
Keywords: NFIL3, CEBP, CEBPbeta, Metabolism, Cancer, Immunology
SUMMARY
Introduction
NFIL3 is a b-ZIP Transcriptional Regulator
Nfil3 Regulates Circadian Rhythm
Light and Nutrient Availability Regulate NFIL3 Expression
-
NFIL3 in Development and Cell fate
5.1 Immunological Role of Nfil3
5.2 NFIL3 is Involved in Heart Development and Aging
NFIL3 Influences Cellular Survival
NFIL3 in Cancer
Nfil3 in Neuronal Regeneration
Nfil3 in Osteoblast Signal Transduction
NFIL3-CEBPb Antagonism: A Recurring Theme
Conclusions and Future Directions
1. Introduction
The NFIL3 transcriptional repressor impacts many cellular processes and is widely expressed in normal murine and human tissues1-19. Often, NFIL3 action opposes that of transcriptional activators by competing for access to target sites on DNA2,7,13,15,20-22. NFIL3 is also commonly found within the context of feedback regulatory circuits21. The list of physiological roles for NFIL3 has evolved tremendously over the last few years. In addition to roles in circadian rhythm and B cell/neuronal survival, NFIL3 is now implicated in having far reaching activities from immunological signal transduction to metabolism, aging and cancer1-4,6,7,9-11,14,18,23,24. Here, we give an overview of NFIL3 transcriptional regulatory circuits and discuss emerging areas of NFIL3 research regarding cancer and diabetes.
2. NFIL3 is a b-ZIP Transcriptional Regulator
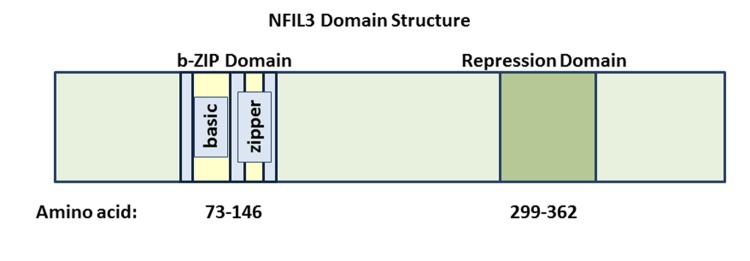
NFIL3 was initially identified by its ability to bind to and repress an E4 promoter sequence containing an ATF consensus site25,26. Several years later, NFIL3 was shown to bind to and activate the transcription of an IL3 (Interleukin 3) promoter sequence27. Although most of the published biological activities for NFIL3 involve its ability to repress transcription, a handful of mammalian cell culture studies as well as murine models indicate that it may also activate transcription by novel mechanisms27,28. The 462 amino acid sequence of NFIL3 includes a b-ZIP domain spanning amino acids 73-146 (Figure 1). The N-terminal portion of this domain (amino acids 79-95) contains the basic motif, which directly binds to DNA. The C-terminal portion of the b-ZIP domain (amino acids 99-106) contains an amphipathic leucine zipper region, which is responsible for homo-dimerization as well as hetero-dimerization interactions29. The NFIL3 protein also has a unique transcriptional repression domain that spans amino acids 299-363. This repressor domain function is transferable, since its fusion with the GAL4 DNA binding domain leads to transcriptional repression in reporter assays30. The molecular mechanisms employed by NFIL3 to repress transcription remain to be fully elucidated. In cancer cells, NFIL3 physically associates with Histone deacetylase 2 (HDAC2) to repress the TRAIL (Tumor Necrosis Factor Ligand Superfamily, Member 10), FAS (TNF receptor superfamily member 6) and GADD45a (Growth Arrest and DNA-damage-inducible, alpha) genes2. In hepatocytes Nfil3 represses Fgf21 (Fibroblast Growth Factor 21) expression in a histone methyltransferase G9a-dependent manner31. Nfil3 represses many additional genes including Period 2 (Per2), Arntl (Aryl Hydrocarbon Receptor Nuclear Translocator-Like), Usp2-45 (Ubiquitin-specific protease 2-45), BGL-GS (also known as BCL2-like 14), Cox2 (Cyclooxygenase 2), Runx2 (Runt-related transcription factor 2), Phex (Phosphate Regulating Endopeptidase Homolog), Ptgs2 (Prostaglandin-endoperoxide synthase 2, also known as Cox2), Pgr (Progesterone receptor), and Areg (amphiregulin), which are regulated in diverse tissues ranging from pancreatic beta cells to osteoblasts21,32-36.
Figure 1. NFIL3 is a bZIP Transcriptional Repressor.
This schematic depicts the overall domain structure of the NFIL3 transcriptional repressor. The bZIP and minimal repressor domains are indicated.
3. Nfil3 Regulates Circadian Rhythm
The Nfil3 gene is expressed in a circadian manner and encodes a regulator of circadian rhythm20,24,37-39. Organisms ranging from bacteria to humans have endogenous clocks or circadian rhythms that integrate intrinsic and extrinsic cues to optimize cellular growth and survival. These clocks are regulated by complex, interlocked feedback loops that are, in part, transcriptionally directed37. The three most common circadian transcription factor consensus sites are: 1) E/E’ box elements which are activated by the Clock (circadian locomotor output cycles kaput)/Bmal1 (brain and muscle ARNT-like protein 1) protein dimer 2) Rev-ErbA/ROR elements (RREs) which are repressed by NR1D1 (Nuclear Receptor subfamily 1, group D, member 1, also known as Rev/ErbA) and 3) D box elements which are activated by the bZIP transcriptional activator Dbp (D site albumin promoter binding protein) and repressed by the bZIP transcriptional repressor Nfil337. The mammalian circadian rhythm is initiated by the basic helix loop helix (bHLH)-PAS transcription factors Clock and Bmal1 which form heterodimers that induce the transcription of genes containing E box promoter elements including Period genes (Per1, Per2 and Per3), Cryptochrome genes (Cry1 and Cry2), Dbp, and Rev/ErbA40,41. Transcriptional programs that activate genes in circadian rhythm are eventually turned off by negative feedback mechanisms. Period and Cry proteins form heterodimers to ultimately hinder the induction of E box regulated genes, Rev/ErbA repress RRE regulated genes including the Bmal1 gene and NFIL3 represses D box regulated genes40,41. The Nfil3 promotercontains numerous RREs thereby facilitating its circadian expression37. Mammalian Nfil3 shows circadian expression in a large number of tissues including suprachiasmatic nuclei, liver, kidney, aorta, skeletal muscle, adrenal gland, and adipose tissue42.
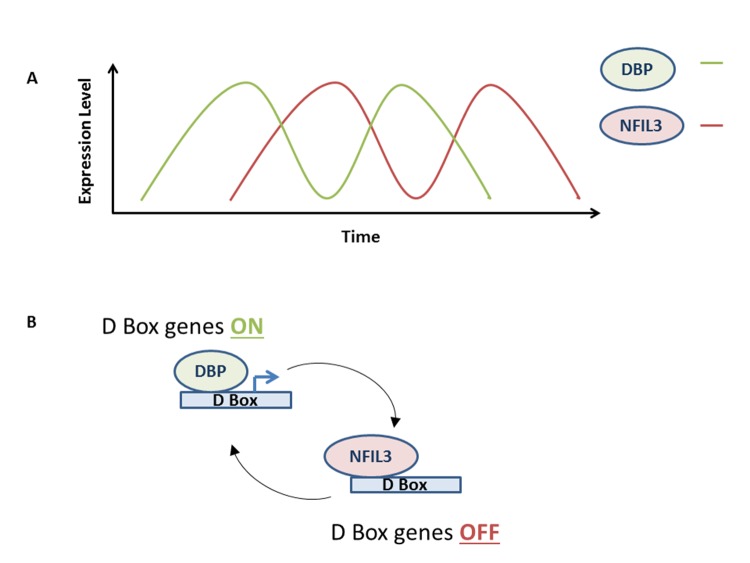
The mode of NFIL3 action on circadian rhythm may be prototypical of its action on other cellular processes. In circadian regulation, the NFIL3 repressor acts in an anti-phase manner with respect to the bZIP transcriptional activator DBP (Figure 2)20,37. NFIL3 and DBP compete for access to D box elements, exerting opposite effects on target genes20. NFIL3 expression peaks when DBP expression is at its lowest and vice versa (Figure 2 [A]). Therefore, D box regulated genes are repressed when levels of NFIL3 are high and are induced (by DBP) when NFIL3 levels are low (Figure 2 [B]). One of the most thought provoking roles of Nfil3 action in the circadian rhythm is that it has recently been shown to regulate period length (the time to complete a circadian cycle, which is normally 24 hours) in Rat1 fibroblast cells38. Specifically, the loss of Nfil3 lengthens period length whereas the overexpression of Nfil3 shortens period length. Levels of Dbp have opposite effects on period length38. Frequently, the circadian rhythm is coupled to cellular processes such as cell division and metabolism43-50. It would be informative to determine whether Rat1 cells grow more quickly with exogenous Nfil3 expression as one would predict with the altered circadian period length. In addition, with a shorter period, are all stages of the circadian rhythm and related cellular processes affected equally by exogenous Nfil3 or are certain aspects of this network differentially impacted? Finally, it will be important to determine whether NFIL3 affects period length in humans and mice.
Figure 2. NFIL3 regulates D-box genes anti-phase to DBP.
(A) Schematic shows how NFIL3 and DBP peak at different times during the circadian rhythm.
(B) DBP binds to and activates D-box genes, whereas NFIL3 binds to and represses D-box genes.
Another recent discovery links the circadian function of Nfil3 to the development of Interleukin 17 producing CD4+ helper T (TH17) cells in the immune system51. Th17 cells protect organisms from bacterial and fungal infections on mucosal membranes and are also associated with inflammatory disease52,53. Nfil3 was found to suppress TH17 cell development by binding to the promoter of the orphan nuclear receptor Rorgt geneand repressing transcription; Rorgt is required for the specification of TH17 cells51,54. Interestingly, Nfil3 was found to be highly expressed at night in mice whereas Rorgt was found highly expressed during the day51. Accordingly, TH17 cell frequencies were significantly higher during the day in with-type mice51. This diurnal difference in TH17 cells was abrogated in Nfil3 -/- mice51. This study is an exciting example that links the circadian and immunological functions of Nfil3. Overall, these emerging areas of Nfil3 regulated circadian processes highlight not only the basic mechanistic insights of its action, but also important connections within a broader cellular context.
4. Light and Nutrient Availability Regulate NFIL3 Expression
The circadian rhythm has been shown to be coupled to metabolic processes in a number of tissues to coordinate behaviors such as rest and locomotion with cues such as light and nutrient availability55-57. As a circadian regulator, Nfil3 impacts metabolism by being part of the cell intrinsic oscillator20-37. In addition to cell intrinsic regulation, Nfil3 stands out as a circadian component that is induced by environmental cues such as light and feeding, thereby enabling cells to integrate internal and external cues24,58. The chick pineal gland is used as a model system to study circadian oscillations in response to light due to the fact that chick pinealocytes possess a circadian oscillator and a photo-transduction pathway for light entrainment57. By exposing young chicks to varying light/dark cycles, it was found that NFIL3 had a role in light entrainment. Under normal conditions, NFIL3 expression in the chicken pineal gland peaks at early subjective night32. NFIL3 protein represses the expression of the PERIOD2 (PER2) gene, which is expressed in an anti-phase manner to NFIL3 and peaks in the morning32. Light exposure during early subjective night leads to an induction in NFIL3 gene expression and a delay in the expression of PER2 the next morning. Sterol Regulatory Element-Binding Protein (SREBP) is a transcription factor that induces NFIL3 expression in the chick pineal gland upon light exposure during early subjective night to ultimately cause a delay in PER2 expression the next morning58. In sum, studies in chicks have revealed a role for Nfil3 in shifting the expression of PER2 to a later time in response to light. It will be interesting to determine whether related mechanisms are employed in mammals.
Insulin and feeding are additional environmental cues that induce the expression of Nfil3. Feeding potently induces the expression of Nfil3 in the mouse liver, whereas fasting has the opposite effect24. Furthermore, insulin induces Nfil3 expression in Hepa1C1C-7 cells in a PI3K dependent manner. One of the Nfil3 repressed genes in the liver is Fgf21 encodes a potent anti-diabetic and triglyceride lowering hormone24. During fasting, Fgf21 is critical for lipolysis, gluconeogenesis, and ketogenesis. Mechanistically, Nfil3 physically associates with D box elements on the Fgf21 promoter in hepatocytes to repress transcription in a manner that depends on the histone methyltransferase G9a31. Thus, upon nutrient availability, Nfil3 action may lead to an epigenetic shift to maximize metabolism and biogenesis. Additionally, Ubiquitin-specific protease 2-45 (Usp2-45) was identified as another Nfil3 repressed gene in hepatocytes34. Usp2-45 encodes a deubiquitinase that regulates gluconeogenesis and glucose metabolism in the liver34. Under starvation conditions, Peroxisome proliferator-activated receptor-gamma coactivator alpha (Pgc1a) and Peroxisome proliferator-activated receptor-gamma coactivator beta (Pgc1b) along with Hepatocyte nuclear factor 4 (Hnf4) activate the transcription of Usp2-45, whereas Nfil3 strongly represses this gene under fed conditions. The ability of Pgc1a to induce Usp2-45 is strongly enhanced when cells are transduced with Nfil3 shRNA (to diminish Nfil3 expression)34. In summary, under fed conditions, Nfil3 expression is induced, leading to the repression of a number of genes (Fgf21 and Usp2-45) to shift metabolic processes. Taken as a whole, the ability of Nfil3 to regulate metabolism is becoming increasingly evident and suggests that it may have a role in diabetes.
5. NFIL3 in Development and Cell Fate
A number of breakthrough studies have revealed that NFIL3 not only regulates oscillatory mechanisms, but also has tremendous impacts on developmental processes. Some of the most significant advances relating to NFIL3 biology were recently made in the field of immunology. Nfil3 was recently found to be essentially required for Natural Killer (NK) cell development and function. Nfil3 has also been found to impact B-cell, T-cell, dendritic cell and macrophage responses. In addition to roles in immunological development, NFIL3 has also been recently found to significantly impact heart development and aging in numerous model organisms. These studies highlight completely novel roles for NFIL3 in the field of developmental biology.
5.1 Immunological Role of Nfil3
Recently it has become clear that Nfil3 has significant contributions to immunological development and function59. Initially shown to be critical for natural killer cell (NK) development, Nfil3 is also required for efficient Immunoglobulin E (IgE) class switching and for the attenuation of numerous inflammatory responses1,6,12,14,23. Arguably, the most striking immunological phenotype for mice that lack Nfil3 is the dramatic loss of mature NK cells14,23. Mice that were nfil3 -/- had normal B and T cell development, but showed defective development, maturation and function of NK cells. A 35 fold reduction of splenic NK cells was observed in nfil3 -/- mice. The few NK cells that did develop in nfil3 -/- mice were functionally defective in the ability to stimulate IFN-g (Interferon Gamma) upon IL2 (Interleukin 2) plus IL12 (Interleukin 12) stimulation. Of note, Nfil3 was recently found to be dispensable for the development of TRAIL+ NK cells60. Therefore, Nfil3 is not required for the development of all NK lineages. NK cells are best known for their cytotoxicity to stressed cells such as those that are virally infected or cancerous. NK cells also have important roles in immunological modulation. For example, NK cell action can dampen CD8+ T cell immune response to viruses, leading to chronic infections as seen with HIV and Hepatitis B61. A unique set of human decidual natural killer cells have recently been shown to express NFIL3 and have important roles in tissue remodeling, neoangiogenesis, and immune modulation to prevent fetal rejection62. These studies highlight the central role of Nfil3 in NK development and suggest that it has an impact in defending organisms against viral infections.
Aside from NK phenotype, Nfil3 also has roles in B cell, dendritic cell, T cell and macrophage-derived immunological responses59. In murine B cells, Nfil3 is required for IgE class switching and thus generation of IgE production, involved in allergic response11,12. Nfil3 binds to the Ie exon promoter, which is thought to promote the production of IgE12. Mice that are nfil3 -/- also lack mature CD8a+ dendritic cells that are crucial for antigen presentation and cross priming CD8+ T cells against cell presented antigens10. Other Nfil3 immunological events appear to involve potential feedback loops where Nfil3 may have roles in attenuating inflammation by either inhibiting the production of cytokines such as IL12 in macrophages or IL5 (Interleukin 5) and IL13 (Interleukin 13) in helper Th2 cells6,9,10,63. Nfil3 also augments the expression of the anti-inflammatory cytokine IL10 (Interleukin 10) in helper T cells (Th1 and Th2)28. Interestingly, NFIL3 is highly expressed in T cells that were isolated from patients with systemic lupus erythematosus (SLE); studies using human T cells showed that exogenous NFIL3 hindered T-cell activation and self-reactivity64. Overall, nfil3-/- mice are immunologically deficient with an almost complete absence of NK function, highly defective IgE production and defective antigen presentation from CD8+ T cells. These mice also lack important immunological regulatory mechanisms to hinder inflammation such as the production of IL10.
5.2 NFIL3 is Involved in Heart Development and Aging
The ability of NFIL3 to influence heart development and function has recently emerged. In Drosophila melanogaster, transcriptome analysis revealed that aged hearts express more of the NFIL3 homolog Vrille than younger hearts5. Aging in Drosophila heart tissue is characterized by an increase in heart period (HP; average time between successive end-diastolic positions). Vrille over-expression led to dilation of the heart. The loss of Vrille dramatically influenced HP: normal flies show a 55% increase in HP between ages 10 and 45 days, whereas Vrille loss of function flies only showed a 6% increase5. A significant number of the putative Vrille target genes, based on the presence of consensus sites and loss of expression in aged flies, were involved in mitochondrial function. Therefore, Vrille may hinder mitochondrial function to promote heart aging.
NFIL3 has also recently been investigated as having a role in heart development and disease in zebra fish (Danio rerio) and rats (Rattus norvegicus). Specifically, loss-of-function analyses of the NFIL3 homolog in zebra fish indicate that it is important for normal heart development65. RNAi targeting of the zebra fish NFIL3 homolog led to malformed looping of the embryonic heart tube which occluded blood flow and retarded cardiac growth65. This same study showed that the over-expression of Nfil3 in rat embryonic fibroblasts induced the expression of survival genes such as Igf1 (Insulin-like growth factor 1), Igf1r (Insulin-like growth factor 1 receptor) and Bcl2 (B-cell lymphoma 2) and hindered caspase 3 induction preventing apoptosis65. Yet another study showed that infarct volume and fibrosis were higher in mouse models subjected to ischemia/reperfusion during the time of day when Nfil3 was at its lowest, suggesting a survival role for Nfil3 in the heart66. Altogether, Nfil3 is emerging as an important cardiac signaling factor that on the one hand promotes aging and on the other hinders cell death.
6. NFIL3 Influences Cellular Survival
Programmed cell death is utilized throughout development and during homeostatic programs in tissues. For example, 50% of neurons undergo apoptosis during development67. NFIL3 has emerged as a survival factor that hinders the induction of apoptosis in numerous settings from B-cells to motor neurons2,19,68. Nfil3 was first shown to hinder apoptosis in murine pro-B lymphocytes which normally require the addition of IL3 (Interleukin 3) to the media for cellular survival19. IL3 binds to its cognate receptor to activate Ras as well as (B-cell lymphoma-extra large) Bcl-XL thereby mediating survival18. The removal of IL3 leads to cell death, which can be rescued by the exogenous expression of anti-apoptotic factors such as Bcl2, Bcl-XL and c-Myc18. Nfil3 is thought to have an important role in mediating the IL3 survival signal19. Nfil3 transcription is strongly induced by IL3 in a mechanism that involves the Ras pathway (activates GATA1 which binds to and activates NFIL3) and the Phosphatidylinositol 3 kinase (PI3K) pathway in pro-B cells18,19,69,70. Conversely, IL3 removal leads to a rapid loss in Nfil3 expression19. Exogenous Nfil3 expression strongly hinders IL3 deprivation induced cell death in FL5.12 murine pro-B lymphocytes19. In BAF-3 pro-B lymphocytes, Nfil3 was shown to rescue viability upon IL3 deprivation18,19, but not upon Interleukin 6 (IL6) deprivation71. Aside from the interleukin utilized in these rescue experiments (IL3 versus IL6), another key difference was that CD8 was exogenously expressed in the successful rescue experiments, which may have promoted the Nfil3 survival signal18,19,71. Loss-of-function experiments also indicate that Nfil3 is a survival factor in pro-B lymphocytes. Specifically, the expression of dominant negative NFIL3 (contains a mutated basic region that fails to bind DNA, but forms heterodimers with wild-type NFIL3) antagonized the ability of IL3 to promote cellular survival18. Higher amounts of IL3 were required to maintain Baf-3 pro-B cells in the presence of dominant-negative-NFIL3. Thus, numerous studies indicate that Nfil3 promotes IL3 mediated survival.
In addition to pro-B cells, Nfil3 also impacts the survival of motor neurons. During embryonic development more than half of the motor neurons produced undergo programmed cell death67. Nfil3 is highly expressed in vivo in embryonic rat and chicken motor neurons that survive developmental pruning68. Gain-of-function experiments showed that exogenous Nfil3 hindered motor neuron cell death upon trophic factor deprivation68. In addition to this, Fas ligand (FasL) induced motor neuron death was completely blocked by exogenous Nfil368. Gain-of-function experiments were also performed with chicken embryos. Specifically, E2.5 embryos were electroporated with NFIL3, which led to the survival of 45% more motor neurons68. These results highlight the importance of NFIL3 in neuronal programmed cell death. It will be exciting to determine whether NFIL3 impacts apoptosis during additional developmental processes, and particularly those that involve pruning.
Although many studies have shown an anti-apoptotic affect for Nfil3, this may not be the case for all physiologic settings. In glucocorticoid (GC) signal transduction, Nfil3 may induce cell death3,72,. The glucocorticoid receptor (GR) is normally regulated by a negative feedback mechanism upon activation; the GR mRNA and protein are down-regulated after GC addition3. However, in certain GC treated murine leukemic T cell lines, GR activation is not attenuated leading to glucocorticoid induced cell death3,72,73. The Nfil3 gene is induced under these conditions and is thought to be part of the mechanism that activates apoptosis. The knockdown of Nfil3 by RNA interference in CTLL-2 cytotoxic T lymphocyte cells partially rescued GC induced apoptosis3. Thus, Nfil3 regulates cell death in a context dependent manner.
7. NFIL3 in Cancer
In line with its ability to hinder cell death, NFIL3 has emerged as a novel survival factor in cancer. NFIL3 was found highly expressed in a number of poor prognosis cancers such as glioblastoma multiforme and basal like breast cancer. NFIL3 expression in breast cancer was found strikingly associated with poor prognosis by Kaplan Meier survival analysis2. Functionally, NFIL3 binds to and represses pro-apoptotic genes such as TRAIL to hinder the induction of apoptosis in cancer cells. The diminishment of NFIL3 by RNA interference led to cell death in the BT549, MDA-MB-468 and U87MG cancer cell lines2. Conversely, NFIL3 overexpression hindered H2O2 induced apoptosis in these same cancer cell lines2. On a mechanistic level, NFIL3 acted at least in part to physically block FOXO1 (Forkhead box O1) recruitment to apoptosis inducing genes by binding to promoter elements that have adjacent FOXO and NFIL3 consensus sites2. It is important to mention that NFIL3 did not block FOXO1 recruitment to all target genes, but just to a subset of genes involved in tumor suppression2. This data suggested that nuclear FOXO1 may still trans-activate non-NFIL3 target genes and that it may even promote cancer in this setting. Therefore, NFIL3 may physically block the ability of FOXO1 to induce cell death genes, so that FOXO1 can direct pro-oncogenic transcriptional programs without killing cells. At least two other recent studies have also found that nuclear FOXO may have pro-oncogenic roles. First, FOXO1 was found to be mutated to a presumably activated form in 8.6% of DLBCL (Diffuse large non-B-cell lymphoma); the FOXO1 mutations in this setting were strikingly associated with poor prognosis74. The FOXO1 mutations that were discovered in DLBCL clustered to two regions within the FOXO1 protein. One of the regions alters residues in the DNA binding domain, whereas the other affected region is proximal to Threonine 24 (T24), which is phosphorylated by AKT on the PI3K pathway74,75. T24 phosphorylation on FOXO1 promotes its interaction with the scaffolding protein 14-3-3, leading to cytoplasmic localization75. Interestingly, the M1L, R19Q, R21C, and T24 mutant forms of FOXO1 (found in DLBCL) failed to be phosphorylated on T24, failed to interact with 14-3-3 and were retained in the nucleus to presumably drive gene expression, suggesting a pro-oncogenic role for nuclear FOXO1 in this setting74. FOXO factors were also discovered to promote the development of AML in an MLL-AF9 murine model for leukemia76. In this model the loss of all three FOXO factors led to a significantly longer latency for the development of AML with a 16 fold reduction in leukemia initiating cells. It will be critical to determine whether NFIL3 is required in DLBCLs that contain activated FOXO1 and the MLL-AF9 cancers in order to block the activation of cell death by FOXO factors. The full spectrum of NFIL3 regulated transcriptional programs in cancer have yet to be elucidated. Most likely there will only be partial overlap with its output and the FOXO-induced cell death genes.
In addition to its ability to block FOXO recruitment to cell death genes, NFIL3 may also block the recruitment of Proline Acid Rich (PAR) transcription factors to pro-apoptotic genes in colon cancer77. NFIL3 was found to repress the pro-apoptotic, BH3-only gene BGL-GS (also known as BCL2-like 14) in colon cancer cells77. The BCL-GS protein binds to Bcl-XL to promote apoptosis77. In contrast, the PAR proteins: DBP, TEF (Thyrotroph Embryonic Factor) and HLF (Hepatic Leukemia Factor) can induce a BCL-GS promoter-driven reporter gene in cancer cell lines77. The BGL-GS gene was repressed by NFIL3 and activated by PAR bZIP factors by regulation through the same consensus site. This study also showed that TEF binding to the endogenous BCL-GS gene was induced by cisplatin and etoposide treatment77. Thus, NFIL3 may block the recruitment of bZIP transcriptional activators to pro-apoptotic genes to potentially hinder chemotherapeutic response. In addition to colon cancer, NFIL3 may have a role in esophageal cancer as irradiation of esophageal cancer cell lines leads to a loss in NFIL3 expression78. Delineating the full breadth of NFIL3 directed transcriptional programs such as those that counter FOXO and PAR transcription factors will be essential for understanding its contribution to cancer.
8. Nfil3 in Neuronal Regeneration
Neuronal regeneration in dorsal root ganglion cells is driven by transcriptional programs that include Nfil3-mediated repression of regeneration associated genes7,13. Nfil3 is overexpressed in regenerating DRG neurons in vivo, but has a negative impact on neurite outgrowth13. Knockdown of Nfil3 by siRNA and expression of dominant negative NFIL3 induced neurite outgrowth in primary adult rat DRG neurons13. The ability of NFIL3 to compete for access to consensus promoter sequences is becoming a recurring theme for its action in signaling circuits. In neurons, Nfil3 appears to compete for access to sites shared with cAMP-response element binding protein (CREB) and CCAAT/Enhancer Binding Protein (CEBPb) to regulate transcriptional programs during neuronal regeneration7,13. Nfil3 resides within a feedback loop that starts with cAMP-induced phosphorylation of CREB. Next, phosphorylated CREB activates Nfil3 expression, which represses CREB target genes as well as Cebpb targets by competing with transcriptional activators for access to target genes. At the same time, Nfil3 binds to its own promoter to repress its own expression7,13. This creates a highly regulated pulse of cAMP signaling in neurons. Of note, Nfil3 expression was specifically found in neurons that were able to regenerate, suggesting that it had a role in this process13. The experimental evidence points to a role for Nfil3 as a component that may promote a greater signaling sensitivity by being an attenuating factor. In sum, CREB activates transcriptional programs including the Nfil3 repressor which acts to attenuate the initial signal. In this manner, only a pulse of signal occurs instead of a sustained signal. It will be exciting to determine whether this type of feedback circuitry comes into play in additional Nfil3-mediated signaling pathways.
9. Nfil3 in Osteoblast Signal Transduction
Nfil3 was identified as a target of parathyroid hormone (PTH) in murine osteoblasts by representational display analysis15. The addition of PTH to primary mouse osteoblasts rapidly induced Nfil3 transcription in a manner that did not require new protein synthesis15. PTH treatment of osteoblasts induced Nfil3 binding to DNA probes that contained Nfil3 binding sites; this binding could be specifically super-shifted by NFIL3 antibody15. However, NFIL3 antibody was unable to super-shift the all PTH induced complexes that bound to the Nfil3 consensus site. Querying TRANSFAC (TRANScription FACtor database) revealed that the Nfil3 DNA binding element closely resembled the Cebpb binding site and it was known that PTH also induced Cebpb expression. Further experiments revealed that CEBPb antibody could super-shift the additional PTH-induced complexes15. A model was put forth in which Nfil3 and Cebpb compete for binding sites to repress and activate targets respectively in response to PTH. The signaling downstream of PTH includes adenylate cyclase and PKA (Protein kinase A), which were shown to be required for the ability of PTH to activate Nfil3 expression21. In addition, another activator of PKA, prostaglandin E2 (PGE2) was also able to activate Nfil3 expression. Forskolin, a direct activator of adenylate cyclase could also induce Nfil3 expression. Cyclooxygenase 2 (Cox2) is thought be one of the Nfil3 repressed genes downstream of PTH in mouse osteoblasts21. Additional Nfil3 targets in murine osteoblasts include Runx2, Osterix, and Phex21,35,36. These studies provide important clues about Nfil3 signal transduction in osteoblasts and place its action downstream of PKA.
10. NFIL3-CEBP b Antagonism: A Recurring Theme
The ability of Nfil3 to compete for DNA access with the transcriptional activator Cebpβ is becoming a commonly observed phenomenon. Nfil3 was found to repress Cebpβ target genes such as Ptgs2, Pgr and Areg in rat ovaries. Chromatin immunoprecipitation analysis with CEBPβ and NFIL3 specific antibodies revealed that hCG treatment led to Cebpβ promoter recruitment at 6 hours post-treatment, which was followed by increasing levels of Nfil3 promoter recruitment (between hours 6-12 post hCG treatment) accompanied by transcriptional repression of target genes and diminishing Cebpβ recruitment8. The recruitment of these factors to target sequences was further confirmed by EMSA8. In addition to a role in ovulation, NFIL3 was also found to hinder CEBPβ recruitment to Human Hepatitis B viral genes22. The overexpression of NFIL3 led to a loss in hepatitis B virion production22. In sum, the ability of NFIL3 to hinder CEBPβ promoter recruitment may have a tremendous impact on many cellular signaling circuits and introduces a new recurring paradigm for NFIL3 action.
11. Conclusions and Future Directions
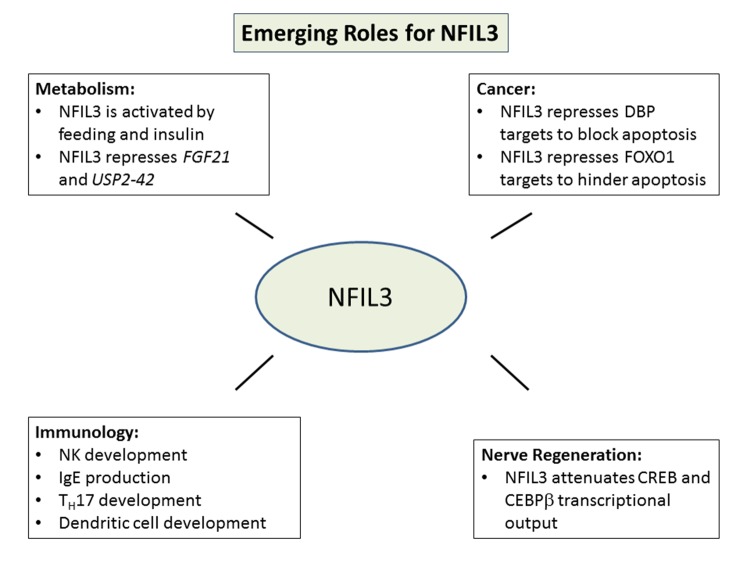
NFIL3 is emerging as a key signaling component in a myriad of cellular processes including metabolism, nerve regeneration, immune development and cancer (Figure 3). NFIL3 commonly antagonizes the recruitment of transcriptional activators to attenuate a signal. In addition to the well characterized ability of NFIL3 to act in an anti-phase manner to DBP with reference to the circadian rhythm, NFIL3 is recurrently found to act in an antagonistic manner to CEBPb7,13,15,20. Nfil3 hinders the recruitment of Cebpb to target genes in motor neurons and osteoblasts. It also appears to hinder CEBPb recruitment during virus production22. Very recently, NFIL3 has emerged as a novel factor that might promote human diseases such as diabetes and cancer. Strikingly, the expression of NFIL3 is significantly associated with poor outcome in cancer2. In cancer cells, NFIL3 was found to block FOXO1 access to cell death genes, potentially allowing FOXO1 to drive pro-oncogenic programs, a true paradigm shift for the PI3K pathway2. Many of the recent insights into NFIL3 biology have been gleaned from cell-based studies. New frontiers for NFIL3 studies will include more in depth in vivo models as well as trying to place NFIL3 into complex signaling cascades. Future research may reveal novel factors for NFIL3 to antagonize as well as mechanisms by which this factor may actually induce transcription as in the case of the IL3 gene. Finally, the regulation of NFIL3 will be an exciting new frontier for future research and will have broad implications for cellular processes discussed in this review as well as human diseases such as cancer and diabetes.
Figure 3. Emerging Cellular Roles for NFIL3.
Metabolism, nerve regeneration, cancer development and immunological development have recently been found to be impacted by NFIL3.
KEY POINT
NFIL3 transcription factor is emerging as a key signaling component in a myriad of cellular processes, such as: metabolism, nerve regeneration, immune development and cancer.
Acknowledgments
This work was funded in part by NCI R01 CA082783 (R.P.).
Footnotes
Conflict of interests: The authors declare no conflict of interest.
DISCOVERIES is a peer-reviewed, open access, online, multidisciplinary and integrative journal, publishing high impact and innovative manuscripts from all areas related to MEDICINE, BIOLOGY and CHEMISTRY
References
- 1.CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Seillet Cyril, Jackson Jacob T, Markey Kate A, Brady Hugh J M, Hill Geoffrey R, Macdonald Kelli P A, Nutt Stephen L, Belz Gabrielle T. Blood. 2013;121(9):1574–83. doi: 10.1182/blood-2012-07-445650. [DOI] [PubMed] [Google Scholar]
- 2.Survival factor NFIL3 restricts FOXO-induced gene expression in cancer. Keniry Megan, Pires Maira M, Mense Sarah, Lefebvre Celine, Gan Boyi, Justiano Karen, Lau Ying-Ka Ingar, Hopkins Ben, Hodakoski Cindy, Koujak Susan, Toole Joseph, Fenton Franklyn, Calahan Ashley, Califano Andrea, DePinho Ronald A, Maurer Matt, Parsons Ramon. Genes & development. 2013;27(8):916–27. doi: 10.1101/gad.214049.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nfil3 is a glucocorticoid-regulated gene required for glucocorticoid-induced apoptosis in male murine T cells. Carey Kirstyn T, Tan Kheng H, Ng Judy, Liddicoat Douglas R, Godfrey Dale I, Cole Timothy J. Endocrinology. 2013;154(4):1540–52. doi: 10.1210/en.2012-1820. [DOI] [PubMed] [Google Scholar]
- 4.The role of Nfil3 in zebrafish hematopoiesis. Progatzky Fränze, Taylor Harriet, Bugeon Laurence, Cassidy Sorcha, Radbruch Andreas, Dallman Margaret J, Lamb Jonathan R. Developmental and comparative immunology. 2012;38(1):187–92. doi: 10.1016/j.dci.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 5.dJun and Vri/dNFIL3 are major regulators of cardiac aging in Drosophila. Monnier Véronique, Iché-Torres Magali, Rera Michael, Contremoulins Vincent, Guichard Céline, Lalevée Nathalie, Tricoire Hervé, Perrin Laurent. PLoS genetics. 2012;8(11):e1003081. doi: 10.1371/journal.pgen.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A distal enhancer in Il12b is the target of transcriptional repression by the STAT3 pathway and requires the basic leucine zipper (B-ZIP) protein NFIL3. Smith Amber M, Qualls Joseph E, O'Brien Kevin, Balouzian Liza, Johnson Peter F, Schultz-Cherry Stacey, Smale Stephen T, Murray Peter J. The Journal of biological chemistry. 2011;286(26):23582–90. doi: 10.1074/jbc.M111.249235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genome-wide gene expression and promoter binding analysis identifies NFIL3 as a repressor of C/EBP target genes in neuronal outgrowth. MacGillavry Harold D, Cornelis Jochem, van der Kallen Loek R, Sassen Marion M, Verhaagen Joost, Smit August B, van Kesteren Ronald E. Molecular and cellular neurosciences. 2011;46(2):460–8. doi: 10.1016/j.mcn.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 8.A role for nuclear factor interleukin-3 (NFIL3), a critical transcriptional repressor, in down-regulation of periovulatory gene expression. Li Feixue, Liu Jing, Jo Misung, Curry Thomas E. Molecular endocrinology (Baltimore, Md.) 2011;25(3):445–59. doi: 10.1210/me.2010-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. Kobayashi Taku, Matsuoka Katsuyoshi, Sheikh Shehzad Z, Elloumi Houda Z, Kamada Nobuhiko, Hisamatsu Tadakazu, Hansen Jonathan J, Doty Kevin R, Pope Scott D, Smale Stephen T, Hibi Toshifumi, Rothman Paul B, Kashiwada Masaki, Plevy Scott E. Journal of immunology (Baltimore, Md. : 1950) 2011;186(8):4649–55. doi: 10.4049/jimmunol.1003888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NFIL3/E4BP4 is a key transcription factor for CD8α⁺ dendritic cell development. Kashiwada Masaki, Pham Nhat-Long L, Pewe Lecia L, Harty John T, Rothman Paul B. Blood. 2011;117(23):6193–7. doi: 10.1182/blood-2010-07-295873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The transcriptional regulator NFIL3 controls IgE production. Rothman Paul B. Transactions of the American Clinical and Climatological Association. 2010;121:156–71; discussion 171. [PMC free article] [PubMed] [Google Scholar]
- 12.IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Kashiwada Masaki, Levy Deborah M, McKeag Lisa, Murray Keri, Schröder Andreas J, Canfield Stephen M, Traver Geri, Rothman Paul B. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(2):821–6. doi: 10.1073/pnas.0909235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NFIL3 and cAMP response element-binding protein form a transcriptional feedforward loop that controls neuronal regeneration-associated gene expression. MacGillavry Harold D, Stam Floor J, Sassen Marion M, Kegel Linde, Hendriks William T J, Verhaagen Joost, Smit August B, van Kesteren Ronald E. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29(49):15542–50. doi: 10.1523/JNEUROSCI.3938-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. Kamizono Shintaro, Duncan Gordon S, Seidel Markus G, Morimoto Akira, Hamada Koichi, Grosveld Gerard, Akashi Koichi, Lind Evan F, Haight Jillian P, Ohashi Pamela S, Look A Thomas, Mak Tak W. The Journal of experimental medicine. 2009;206(13):2977–86. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parathyroid hormone-induced E4BP4/NFIL3 down-regulates transcription in osteoblasts. Ozkurt Ibrahim C, Tetradis Sotirios. The Journal of biological chemistry. 2003;278(29):26803–9. doi: 10.1074/jbc.M212652200. [DOI] [PubMed] [Google Scholar]
- 16.E4BP4/NFIL3, a PAR-related bZIP factor with many roles. Cowell Ian G. BioEssays. 2002;24(11):1023-1029. doi: 10.1002/bies.10176. [DOI] [PubMed] [Google Scholar]
- 17.Exclusion of NFIL3 as the gene causing hereditary sensory neuropathy type I by mutation analysis. Hulme D J, Blair I P, Dawkins J L, Nicholson G A. Human genetics. 2000;106(6):594–6. doi: 10.1007/s004390000306. [DOI] [PubMed] [Google Scholar]
- 18.Two distinct interleukin-3-mediated signal pathways, Ras-NFIL3 (E4BP4) and Bcl-xL, regulate the survival of murine pro-B lymphocytes. Kuribara R, Kinoshita T, Miyajima A, Shinjyo T, Yoshihara T, Inukai T, Ozawa K, Look A T, Inaba T. Molecular and cellular biology. 1999;19(4):2754–62. doi: 10.1128/mcb.19.4.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Ikushima S, Inukai T, Inaba T, Nimer S D, Cleveland J L, Look A T. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(6):2609–14. doi: 10.1073/pnas.94.6.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Genes & development. 2001;15(8):995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parathyroid hormone induces E4bp4 messenger ribonucleic acid expression primarily through cyclic adenosine 3',5'-monophosphate signaling in osteoblasts. Ozkurt Ibrahim C, Pirih Flavia Q, Tetradis Sotirios. Endocrinology. 2004;145(8):3696–703. doi: 10.1210/en.2003-1436. [DOI] [PubMed] [Google Scholar]
- 22.Transcriptional repression of human hepatitis B virus genes by a bZIP family member, E4BP4. Lai C K, Ting L P. Journal of virology. 1999;73(4):3197–209. doi: 10.1128/jvi.73.4.3197-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Gascoyne Duncan M, Long Elaine, Veiga-Fernandes Henrique, de Boer Jasper, Williams Owen, Seddon Benedict, Coles Mark, Kioussis Dimitris, Brady Hugh J M. Nature immunology. 2009;10(10):1118–24. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 24.Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding. Tong Xin, Muchnik Marina, Chen Zheng, Patel Manish, Wu Nan, Joshi Shree, Rui Liangyou, Lazar Mitchell A, Yin Lei. The Journal of biological chemistry. 2010;285(47):36401–9. doi: 10.1074/jbc.M110.172866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Transcriptional repression by the human bZIP factor E4BP4: definition of a minimal repression domain. Cowell I G, Hurst H C. Nucleic acids research. 1994;22(1):59–65. doi: 10.1093/nar/22.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Transcriptional repression by a novel member of the bZIP family of transcription factors. Cowell I G, Skinner A, Hurst H C. Molecular and cellular biology. 1992;12(7):3070–7. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Zhang W, Zhang J, Kornuc M, Kwan K, Frank R, Nimer S D. Molecular and cellular biology. 1995;15(11):6055–63. doi: 10.1128/mcb.15.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Motomura Yasutaka, Kitamura Hiroshi, Hijikata Atsushi, Matsunaga Yuko, Matsumoto Koichiro, Inoue Hiromasa, Atarashi Koji, Hori Shohei, Watarai Hiroshi, Zhu Jinfang, Taniguchi Masaru, Kubo Masato. Nature immunology. 2011;12(5):450–9. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Experimental identification of homodimerizing B-ZIP families in Homo sapiens. Acharya Asha, Rishi Vikas, Moll Jonathan, Vinson Charles. Journal of structural biology. 2006;155(2):130–9. doi: 10.1016/j.jsb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Recruitment of histone methyltransferase G9a mediates transcriptional repression of Fgf21 gene by E4BP4 protein. Tong Xin, Zhang Deqiang, Buelow Katie, Guha Anirvan, Arthurs Blake, Brady Hugh J M, Yin Lei. The Journal of biological chemistry. 2013;288(8):5417–25. doi: 10.1074/jbc.M112.433482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Protein-protein interaction between the transcriptional repressor E4BP4 and the TBP-binding protein Dr1. Cowell I G, Hurst H C. Nucleic acids research. 1996;24(18):3607–13. doi: 10.1093/nar/24.18.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negative control of circadian clock regulator E4BP4 by casein kinase Iepsilon-mediated phosphorylation. Doi Masao, Okano Toshiyuki, Yujnovsky Irene, Sassone-Corsi Paolo, Fukada Yoshitaka. Current biology : CB. 2004;14(11):975–80. doi: 10.1016/j.cub.2004.05.043. [DOI] [PubMed] [Google Scholar]
- 33.Clock-controlled output gene Dbp is a regulator of Arnt/Hif-1β gene expression in pancreatic islet β-cells. Nakabayashi Hiroko, Ohta Yasuharu, Yamamoto Masayoshi, Susuki Yosuke, Taguchi Akihiko, Tanabe Katsuya, Kondo Manabu, Hatanaka Masayuki, Nagao Yuko, Tanizawa Yukio. Biochemical and biophysical research communications. 2013;434(2):370–5. doi: 10.1016/j.bbrc.2013.03.084. [DOI] [PubMed] [Google Scholar]
- 34.Peroxisomal localization and circadian regulation of ubiquitin-specific protease 2. Molusky Matthew M, Ma Di, Buelow Katie, Yin Lei, Lin Jiandie D. PloS one. 2012;7(11):e47970. doi: 10.1371/journal.pone.0047970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Negative regulation of the osteoblast function in multiple myeloma through the repressor gene E4BP4 activated by malignant plasma cells. Silvestris Franco, Cafforio Paola, De Matteo Monica, Calvani Nicola, Frassanito Maria A, Dammacco Franco. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(19):6081–91. doi: 10.1158/1078-0432.CCR-08-0219. [DOI] [PubMed] [Google Scholar]
- 36.PTHrP(1-34)-mediated repression of the PHEX gene in osteoblastic cells involves the transcriptional repressor E4BP4. Pellicelli Martin, Taheri Maryam, St-Louis Mathieu, Bériault Véronique, Desgroseillers Luc, Boileau Guy, Moreau Alain. Journal of cellular physiology. 2012;227(6):2378–87. doi: 10.1002/jcp.22973. [DOI] [PubMed] [Google Scholar]
- 37.System-level identification of transcriptional circuits underlying mammalian circadian clocks. Ueda Hiroki R, Hayashi Satoko, Chen Wenbin, Sano Motoaki, Machida Masayuki, Shigeyoshi Yasufumi, Iino Masamitsu, Hashimoto Seiichi. Nature genetics. 2005;37(2):187–92. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 38.Cellular DBP and E4BP4 proteins are critical for determining the period length of the circadian oscillator. Yamajuku Daisuke, Shibata Yasutaka, Kitazawa Masashi, Katakura Toshie, Urata Hiromi, Kojima Tomoko, Takayasu Satoko, Nakata Osamu, Hashimoto Seiichi. FEBS letters. 2011;585(14):2217–22. doi: 10.1016/j.febslet.2011.05.038. [DOI] [PubMed] [Google Scholar]
- 39.Identification of negative transcriptional factor E4BP4-binding site in the mouse circadian-regulated gene Mdr2. Kotaka Maki, Onishi Yoshiaki, Ohno Tomoya, Akaike Toshihiro, Ishida Norio. Neuroscience research. 2008;60(3):307–13. doi: 10.1016/j.neures.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Mammalian circadian clock and metabolism - the epigenetic link. Bellet Marina Maria, Sassone-Corsi Paolo. Journal of cell science. 2010;123(Pt 22):3837–48. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coordination of circadian timing in mammals. Reppert Steven M, Weaver David R. Nature. 2002;418(6901):935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 42.Analysis of gene regulatory networks in the mammalian circadian rhythm. Yan Jun, Wang Haifang, Liu Yuting, Shao Chunxuan. PLoS computational biology. 2008;4(10):e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Circadian clocks and energy metabolism. Sancar Gencer, Brunner Michael. Cellular and Molecular Life Sciences. 2014;71(14):2667-2680. doi: 10.1007/s00018-014-1574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Peek Clara Bien, Affinati Alison H, Ramsey Kathryn Moynihan, Kuo Hsin-Yu, Yu Wei, Sena Laura A, Ilkayeva Olga, Marcheva Biliana, Kobayashi Yumiko, Omura Chiaki, Levine Daniel C, Bacsik David J, Gius David, Newgard Christopher B, Goetzman Eric, Chandel Navdeep S, Denu John M, Mrksich Milan, Bass Joseph. Science (New York, N.Y.) 2013;342(6158):1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Bookout Angie L, de Groot Marleen H M, Owen Bryn M, Lee Syann, Gautron Laurent, Lawrence Heather L, Ding Xunshan, Elmquist Joel K, Takahashi Joseph S, Mangelsdorf David J, Kliewer Steven A. Nature medicine. 2013;19(9):1147–52. doi: 10.1038/nm.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Circadian clock genes Per1 and Per2 regulate the response of metabolism-associated transcripts to sleep disruption. Husse Jana, Hintze Sophie Charlotte, Eichele Gregor, Lehnert Hendrik, Oster Henrik. PloS one. 2012;7(12):e52983. doi: 10.1371/journal.pone.0052983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The suprachiasmatic nucleus controls circadian energy metabolism and hepatic insulin sensitivity. Coomans Claudia P, van den Berg Sjoerd A A, Lucassen Eliane A, Houben Thijs, Pronk Amanda C M, van der Spek Rianne D, Kalsbeek Andries, Biermasz Nienke R, Willems van Dijk Ko, Romijn Johannes A, Meijer Johanna H. Diabetes. 2013;62(4):1102–8. doi: 10.2337/db12-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Circadian rhythms, metabolism, and insulin sensitivity: transcriptional networks in animal models. Kitazawa Masashi. Current diabetes reports. 2013;13(2):223–8. doi: 10.1007/s11892-012-0354-8. [DOI] [PubMed] [Google Scholar]
- 49.Cross-talks between circadian timing system and cell division cycle determine cancer biology and therapeutics. Lévi F, Filipski E, Iurisci I, Li X M, Innominato P. Cold Spring Harbor symposia on quantitative biology. 2007;72:465–75. doi: 10.1101/sqb.2007.72.030. [DOI] [PubMed] [Google Scholar]
- 50.Control mechanism of the circadian clock for timing of cell division in vivo. Matsuo Takuya, Yamaguchi Shun, Mitsui Shigeru, Emi Aki, Shimoda Fukuko, Okamura Hitoshi. Science (New York, N.Y.) 2003;302(5643):255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 51.TH17 cell differentiation is regulated by the circadian clock. Yu Xiaofei, Rollins Darcy, Ruhn Kelly A, Stubblefield Jeremy J, Green Carla B, Kashiwada Masaki, Rothman Paul B, Takahashi Joseph S, Hooper Lora V. Science (New York, N.Y.) 2013;342(6159):727–30. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.IL-17 and Th17 Cells. Korn Thomas, Bettelli Estelle, Oukka Mohamed, Kuchroo Vijay K. Annual review of immunology. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 53.IL-23 and Th17 cytokines in intestinal homeostasis. Maloy K J, Kullberg M C. Mucosal immunology. 2008;1(5):339–49. doi: 10.1038/mi.2008.28. [DOI] [PubMed] [Google Scholar]
- 54.A validated regulatory network for Th17 cell specification. Ciofani Maria, Madar Aviv, Galan Carolina, Sellars Maclean, Mace Kieran, Pauli Florencia, Agarwal Ashish, Huang Wendy, Parkhurst Christopher N, Muratet Michael, Newberry Kim M, Meadows Sarah, Greenfield Alex, Yang Yi, Jain Preti, Kirigin Francis K, Birchmeier Carmen, Wagner Erwin F, Murphy Kenneth M, Myers Richard M, Bonneau Richard, Littman Dan R. Cell. 2012;151(2):289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altered feeding differentially regulates circadian rhythms and energy metabolism in liver and muscle of rats. Reznick Jane, Preston Elaine, Wilks Donna L, Beale Susan M, Turner Nigel, Cooney Gregory J. Biochimica et biophysica acta. 2013;1832(1):228–38. doi: 10.1016/j.bbadis.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Expression profile of mRNAs encoding core circadian regulatory proteins in human subcutaneous adipose tissue: correlation with age and body mass index. Wu X, Xie H, Yu G, Hebert T, Goh B C, Smith S R, Gimble J M. International journal of obesity (2005) 2009;33(9):971–7. doi: 10.1038/ijo.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Light-induced phase-delay of the chicken pineal circadian clock is associated with the induction of cE4bp4, a potential transcriptional repressor of cPer2 gene. Doi M, Nakajima Y, Okano T, Fukada Y. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(14):8089–94. doi: 10.1073/pnas.141090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Light-dependent and circadian clock-regulated activation of sterol regulatory element-binding protein, X-box-binding protein 1, and heat shock factor pathways. Hatori Megumi, Hirota Tsuyoshi, Iitsuka Michiko, Kurabayashi Nobuhiro, Haraguchi Shogo, Kokame Koichi, Sato Ryuichiro, Nakai Akira, Miyata Toshiyuki, Tsutsui Kazuyoshi, Fukada Yoshitaka. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(12):4864–9. doi: 10.1073/pnas.1015959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.E4BP4: an unexpected player in the immune response. Male Victoria, Nisoli Ilaria, Gascoyne Duncan M, Brady Hugh J M. Trends in immunology. 2012;33(2):98–102. doi: 10.1016/j.it.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Differential requirement for Nfil3 during NK cell development. Seillet Cyril, Huntington Nicholas D, Gangatirkar Pradnya, Axelsson Elin, Minnich Martina, Brady Hugh J M, Busslinger Meinrad, Smyth Mark J, Belz Gabrielle T, Carotta Sebastian. Journal of immunology (Baltimore, Md. : 1950) 2014;192(6):2667–76. doi: 10.4049/jimmunol.1302605. [DOI] [PubMed] [Google Scholar]
- 61.Natural killer cell activation enhances immune pathology and promotes chronic infection by limiting CD8+ T-cell immunity. Lang Philipp A, Lang Karl S, Xu Haifeng C, Grusdat Melanie, Parish Ian A, Recher Mike, Elford Alisha R, Dhanji Salim, Shaabani Namir, Tran Charles W, Dissanayake Dilan, Rahbar Ramtin, Ghazarian Magar, Brüstle Anne, Fine Jason, Chen Peter, Weaver Casey T, Klose Christoph, Diefenbach Andreas, Häussinger Dieter, Carlyle James R, Kaech Susan M, Mak Tak W, Ohashi Pamela S. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(4):1210–5. doi: 10.1073/pnas.1118834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.CD34+ hematopoietic precursors are present in human decidua and differentiate into natural killer cells upon interaction with stromal cells. Vacca Paola, Vitale Chiara, Montaldo Elisa, Conte Romana, Cantoni Claudia, Fulcheri Ezio, Darretta Valeria, Moretta Lorenzo, Mingari Maria Cristina. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(6):2402–7. doi: 10.1073/pnas.1016257108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.IL-1 family members IL-18 and IL-33 upregulate the inflammatory potential of differentiated human Th1 and Th2 cultures. Blom Lars, Poulsen Lars K. Journal of immunology (Baltimore, Md. : 1950) 2012;189(9):4331–7. doi: 10.4049/jimmunol.1103685. [DOI] [PubMed] [Google Scholar]
- 64.E4BP4 overexpression: a protective mechanism in CD4+ T cells from SLE patients. Zhao Ming, Liu Qian, Liang Gongping, Wang Litao, Luo Shuangyan, Tang Qian, Zhao Hongjun, Su Yuwen, Yung Susan, Chan Tak Mao, Lu Qianjin. Journal of autoimmunity. 2013;41:152–60. doi: 10.1016/j.jaut.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 65.E4BP4 is a cardiac survival factor and essential for embryonic heart development. Weng Yi-Jiun, Hsieh Dennis Jine-Yuan, Kuo Wei-Wen, Lai Tung-Yuan, Hsu Hsi-Hsien, Tsai Chang-Hai, Tsai Fuu-Jen, Lin Ding-Yu, Lin James A, Huang Chih-Yang, Tung Kwong-Chung. Molecular and cellular biochemistry. 2010;340(1-2):187–94. doi: 10.1007/s11010-010-0417-6. [DOI] [PubMed] [Google Scholar]
- 66.Short communication: ischemia/reperfusion tolerance is time-of-day-dependent: mediation by the cardiomyocyte circadian clock. Durgan David J, Pulinilkunnil Thomas, Villegas-Montoya Carolina, Garvey Merissa E, Frangogiannis Nikolaos G, Michael Lloyd H, Chow Chi-Wing, Dyck Jason R B, Young Martin E. Circulation research. 2010;106(3):546–50. doi: 10.1161/CIRCRESAHA.109.209346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Programmed cell death and the control of cell survival: lessons from the nervous system. Raff M C, Barres B A, Burne J F, Coles H S, Ishizaki Y, Jacobson M D. Science (New York, N.Y.) 1993;262(5134):695–700. doi: 10.1126/science.8235590. [DOI] [PubMed] [Google Scholar]
- 68.The CES-2-related transcription factor E4BP4 is an intrinsic regulator of motoneuron growth and survival. Junghans Dirk, Chauvet Sophie, Buhler Emmanuelle, Dudley Keith, Sykes Toby, Henderson Christopher E. Development (Cambridge, England) 2004;131(18):4425–34. doi: 10.1242/dev.01313. [DOI] [PubMed] [Google Scholar]
- 69.GATA factors are essential for transcription of the survival gene E4bp4 and the viability response of interleukin-3 in Ba/F3 hematopoietic cells. Yu Yung-Luen, Chiang Yun-Jung, Yen Jeffrey J Y. The Journal of biological chemistry. 2002;277(30):27144–53. doi: 10.1074/jbc.M200924200. [DOI] [PubMed] [Google Scholar]
- 70.MAPK-mediated phosphorylation of GATA-1 promotes Bcl-XL expression and cell survival. Yu Yung-Luen, Chiang Yun-Jung, Chen Yu-Chun, Papetti Michael, Juo Chiun-Gung, Skoultchi Arthur I, Yen Jeffrey J Y. The Journal of biological chemistry. 2005;280(33):29533–42. doi: 10.1074/jbc.M506514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.The chimeric E2A-HLF transcription factor abrogates p53-induced apoptosis in myeloid leukemia cells. Altura R A, Inukai T, Ashmun R A, Zambetti G P, Roussel M F, Look A T. Blood. 1998;92(4):1397–405. [PubMed] [Google Scholar]
- 72.E4BP4 facilitates glucocorticoid-evoked apoptosis of human leukemic CEM cells via upregulation of Bim. Beach Jessica A, Nary Laura J, Hirakawa Yasuko, Holland Eli, Hovanessian Rebeka, Medh Rheem D. Journal of molecular signaling. 2011;6(1):13. doi: 10.1186/1750-2187-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calcium-dependent upregulation of E4BP4 expression correlates with glucocorticoid-evoked apoptosis of human leukemic CEM cells. Priceman Saul J., Kirzner Jonathan D., Nary Laura J., Morris Devin, Shankar Deepa B., Sakamoto Kathleen M., Medh Rheem D. Biochemical and Biophysical Research Communications. 2006;344(2):491-499. doi: 10.1016/j.bbrc.2006.03.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Analysis of FOXO1 mutations in diffuse large B-cell lymphoma. Trinh Diane L, Scott David W, Morin Ryan D, Mendez-Lago Maria, An Jianghong, Jones Steven J M, Mungall Andrew J, Zhao Yongjun, Schein Jacqueline, Steidl Christian, Connors Joseph M, Gascoyne Randy D, Marra Marco A. Blood. 2013;121(18):3666–74. doi: 10.1182/blood-2013-01-479865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.AKT/PKB signaling: navigating downstream. Manning Brendan D, Cantley Lewis C. Cell. 2007;129(7):1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Sykes Stephen M, Lane Steven W, Bullinger Lars, Kalaitzidis Demetrios, Yusuf Rushdia, Saez Borja, Ferraro Francesca, Mercier Francois, Singh Harshabad, Brumme Kristina M, Acharya Sanket S, Scholl Claudia, Schöll Claudia, Tothova Zuzana, Attar Eyal C, Fröhling Stefan, DePinho Ronald A, Armstrong Scott A, Gilliland D Gary, Scadden David T. Cell. 2011;146(5):697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.A novel role for proline- and acid-rich basic region leucine zipper (PAR bZIP) proteins in the transcriptional regulation of a BH3-only proapoptotic gene. Benito Adalberto, Gutierrez Olga, Pipaon Carlos, Real Pedro J, Gachon Frederic, Ritchie Alistair E, Fernandez-Luna Jose L. The Journal of biological chemistry. 2006;281(50):38351–7. doi: 10.1074/jbc.M607004200. [DOI] [PubMed] [Google Scholar]
- 78.Effect of ionizing irradiation on human esophageal cancer cell lines by cDNA microarray gene expression analysis. Bo Hideki, Ghazizadeh Mohammad, Shimizu Hajime, Kurihara Yuji, Egawa Seiko, Moriyama Yukichi, Tajiri Takashi, Kawanami Oichi. Journal of Nippon Medical School = Nippon Ika Daigaku zasshi. 2004;71(3):172–80. doi: 10.1272/jnms.71.172. [DOI] [PubMed] [Google Scholar]