Abstract
Fucosylated glycans on the surface of epithelial cells (ECs) regulate intestinal homeostasis by serving as attachment receptors and a nutrient source for some species of bacteria. We show here that epithelial fucosylation in the ileum is negatively regulated by IL-10-producing CD4+ T cells. The number of fucosylated ECs was increased in the ileum of mice lacking T cells, especially those expressing αβ T cell receptor (TCR), CD4, and IL-10. No such effect was observed in mice lacking B cells. Adoptive transfer of αβTCR+ CD4+ T cells from normal mice, but not IL-10-deficient mice, normalized fucosylation of ECs. These findings suggest that IL-10-producing CD4+ T cells contribute to the maintenance of the function of ECs by regulating their fucosylation.
The mammalian gastrointestinal tract is colonized by a community of bacteria1, and the host establishes physical, chemical, and immunological barriers as a shield to limit the exposure to these bacteria2,3. As the first barrier in the intestine, many different subsets of epithelial cells (ECs) reside in the intestinal epithelial monolayer. These subsets include absorptive enterocytes, goblet cells, Paneth cells, enteroendocrine cells, and antigen-sampling M cells2. Several lines of evidence point to the fact that both host-derived factors (e.g., cytokines and chemokines) and gut environmental factors (e.g., commensal bacteria, dietary products, and their metabolites) affect the intestinal barrier function2. For example, luminal bacteria induce the secretion of anti-microbial proteins (e.g., regenerating islet–derived protein 3γ) by ECs; this secretion limits bacterial load on the intestinal epithelium4,5.
The surfaces of ECs bear a coating (the glycocalyx) consisting of various glycoproteins and glycolipids, and ECs also secrete a large amount of glycosylated mucins, which act as a protective barrier in the intestine6. In addition to its protective function, the glycocalyx on the ECs also provides attachment sites for commensal bacteria, as exemplified by the attachment of Lactobacillus to glycolipids or glycoproteins7,8, and pathogens, as seen in Helicobacter pylori attachment to fucosylated or sialylated glycans9. Moreover, certain species of commensal and pathogenic bacteria have evolved to utilize the glycosylated molecules produced by the ECs10. Fucose is a residual sugar frequently present at the termini of glycoconjugates in the intestinal epithelium3,11. Some indigenous bacteria preferentially induce fucosylation of the intestinal epithelium12, and some reports, including ours, have proposed that fucosylation of intestinal ECs provides a niche for a stable microbial ecosystem13,14.
Mammals possess multiple sets of fucosyltransferases that mediate fucosylation through the transfer of guanosine-diphosphate fucose to acceptor molecules including oligosaccharides, glycoproteins, and glycolipids15,16. In the intestine, fucosylation is achieved by the addition of α(1,2)-fucose to terminal galactose residues by fucosyltransferase-1 (Fut1) and fucosyltransferase-2 (Fut2)15,16. It was reported that ECs in the small intestine selectively express the Fut1 and Fut2 genes: M cells of the Peyer’s patches express Fut1, whereas goblet cells and enterocytes express Fut217,18. The expression of Fut1 seems to be constitutive, whereas the expression of Fut2 can be induced by environmental stimuli and stresses, such as bacterial colonization18,19. Experiments with germ-free mice have demonstrated that Fut2-mediated α(1,2)-fucosylation was induced after weaning together with the appearance of commensal bacteria12. In addition, colonization by a single type of commensal bacteria, such as segmented filamentous bacteria Bacteroides thetaiotaomicron and Bacteroides fragilis, was sufficient to induce Fut2-mediated epithelial fucosylation12,14,20,21. Recently, mutations in Fut2 have been shown to be associated with inflammatory and autoimmune diseases such as Crohn’s disease and type 1 diabetes22,23, suggesting the involvement of host immune cells in the regulation of fucosylation. We have recently reported that IL-22 produced by type 3 innate lymphoid cells is critical for the induction and regulation of epithelial fucosylation. In the present study, we show that IL-10-producing CD4+ T cells play a pivotal role in the negative regulation of epithelial fucosylation in the intestine.
Results
T cell–deficient mice have increased numbers of fucosylated epithelial cells (F-ECs) with increased Fut2 expression in the intestine
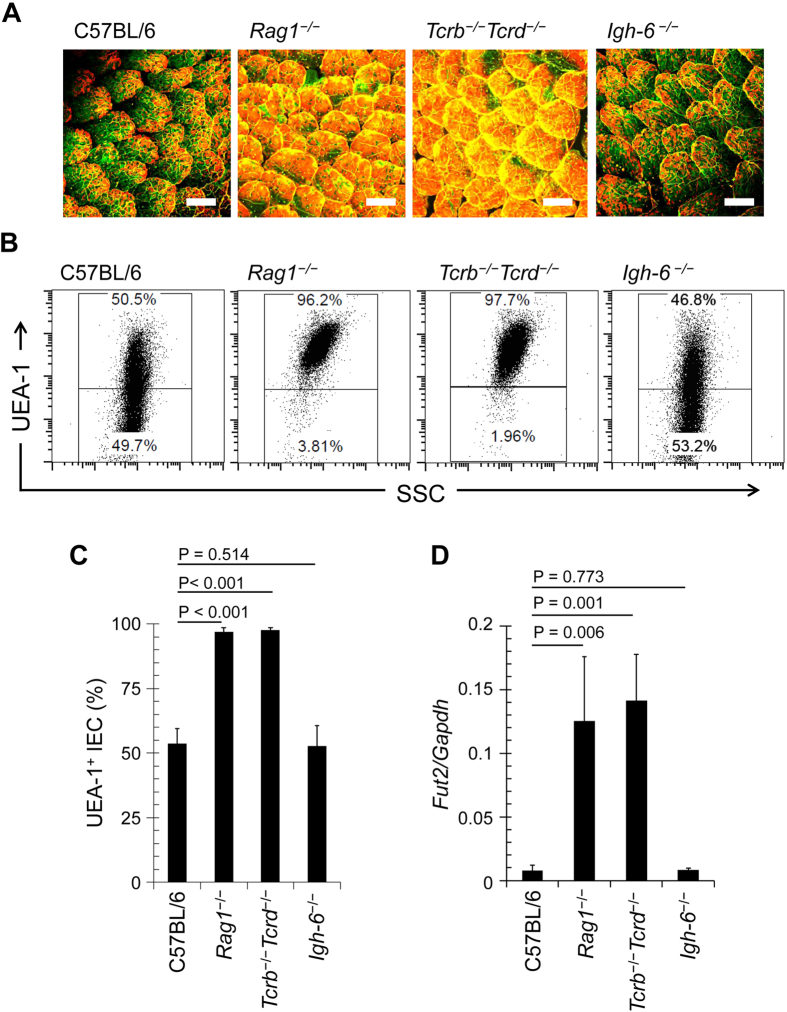
In this study, we focused on the ileum for the analysis of epithelial fucosylation because duodenal ECs have minimal fucosylation, whereas ileal ECs are highly fucosylated14. To assess whether epithelial fucosylation is affected by lymphocytes, we examined epithelial fucosylation in the ileum of recombinase-activating gene-1–deficient mice (Rag1−/−) and scid mice that lack mature T and B cells24,25. Higher numbers of fucosylated ECs (F-ECs) were found on the ileal epithelium of both Rag1−/− and scid mice than on that of control mice (Fig. 1A–C and Supplementary Figure 1A). In line with these findings, the increase of F-ECs in Rag1−/− and scid mice was accompanied by increased Fut2 mRNA expression in ileal ECs (Fig. 1D and Supplementary Figure 1B).
Figure 1. T cell deficiency enhances epithelial fucosylation in the ileum.
(A) Whole-mount staining of the ileum of C57BL/6, Rag1−/−, Tcrb−/−Tcrd−/−, and Igh-6−/− mice was performed with UEA-1 (red) for α-1,2 fucose staining and wheat germ agglutinin (green) for epithelial cell counterstaining. Scale bars: 100 μm. Data are representative of 4 independent experiments. (B,C) Ileal ECs were stained with CD45 antibody and UEA-1. CD45− cells were gated and analyzed for UEA-1 binding by flow cytometry. Representative dot plots are shown in (B). Mean percentages ± s.d. (n = 6) of UEA-1+ ECs are shown in (C). (D) Quantitative PCR analysis of Fut2 expression in ileal ECs. Data normalized against the expression of Gapdh are shown as mean ± s.d. (n = 6 from 2 independent experiments).
We then aimed to determine whether T or B cells are responsible for the regulation of epithelial fucosylation. To address this question, we examined βδ T cell receptor (TCR) knockout (Tcrb−/−Tcrd−/−) mice, which lack T cells, and Igh-6−/− mice, which lack B cells. The number of F-ECs and Fut2 mRNA expression were not significantly altered in ileal ECs of Igh-6−/− mice but were increased in Tcrb−/−Tcrd−/− mice (Fig. 1A–D), suggesting that T cells are pivotal in the epithelial fucosylation in the ileum.
CD4+ effector T cells expressing αβTCR are critical for the regulation of epithelial fucosylation in the intestine
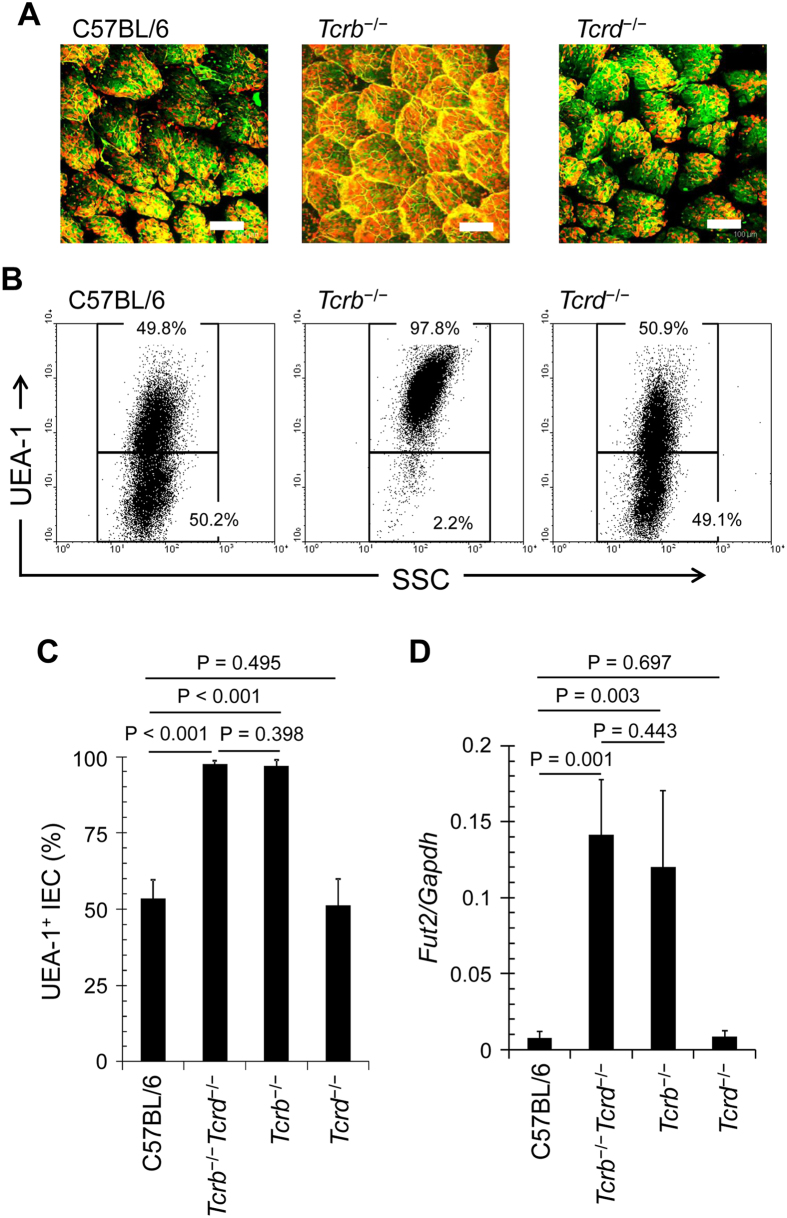
The intestine contains T cells expressing either αβTCR (αβT cells) or γδTCR (γδT cells)26. To determine whether epithelial fucosylation is regulated by αβT cells, γδT cells, or both, we analyzed epithelial fucosylation in the ileum of Tcrb−/− and Tcrd−/− mice. Like Rag1−/− and scid mice, Tcrb−/− mice showed increased numbers of F-ECs and Fut2 mRNA expression, whereas these parameters were similar in Tcrd−/− and control mice (Fig. 2A–D), suggesting that the regulation of epithelial fucosylation in the ileum is solely mediated by αβT cells. Athymic nu/nu mice also had a high number of F-ECs and increased Fut2 expression in ileal ECs in comparison with control mice (Supplementary Figure 2A,B). These data imply that thymus-derived αβT cells downregulate Fut2 expression and associated epithelial fucosylation in the ileum.
Figure 2. αβT cells regulate fucosylation of ileal ECs.
(A) Whole-mount staining of the ileum of C57BL/6, Tcrb−/−, and Tcrd−/− mice was performed with UEA-1 (red) and wheat germ agglutinin (green). Scale bars: 100 μm. Data are representative of 4 independent experiments. (B,C) Ileal ECs were stained with CD45 antibody and UEA-1. CD45− cells were gated and analyzed for UEA-1 binding by flow cytometry. Representative dot plots are shown in (B). Mean percentages of UEA-1+ ECs are shown ± s.d. (n = 6) in (C). (D) Quantitative PCR analysis of Fut2 expression in ileal ECs. Data normalized against the expression of Gapdh are shown as mean ± s.d. (n = 6 from 2 independent experiments).
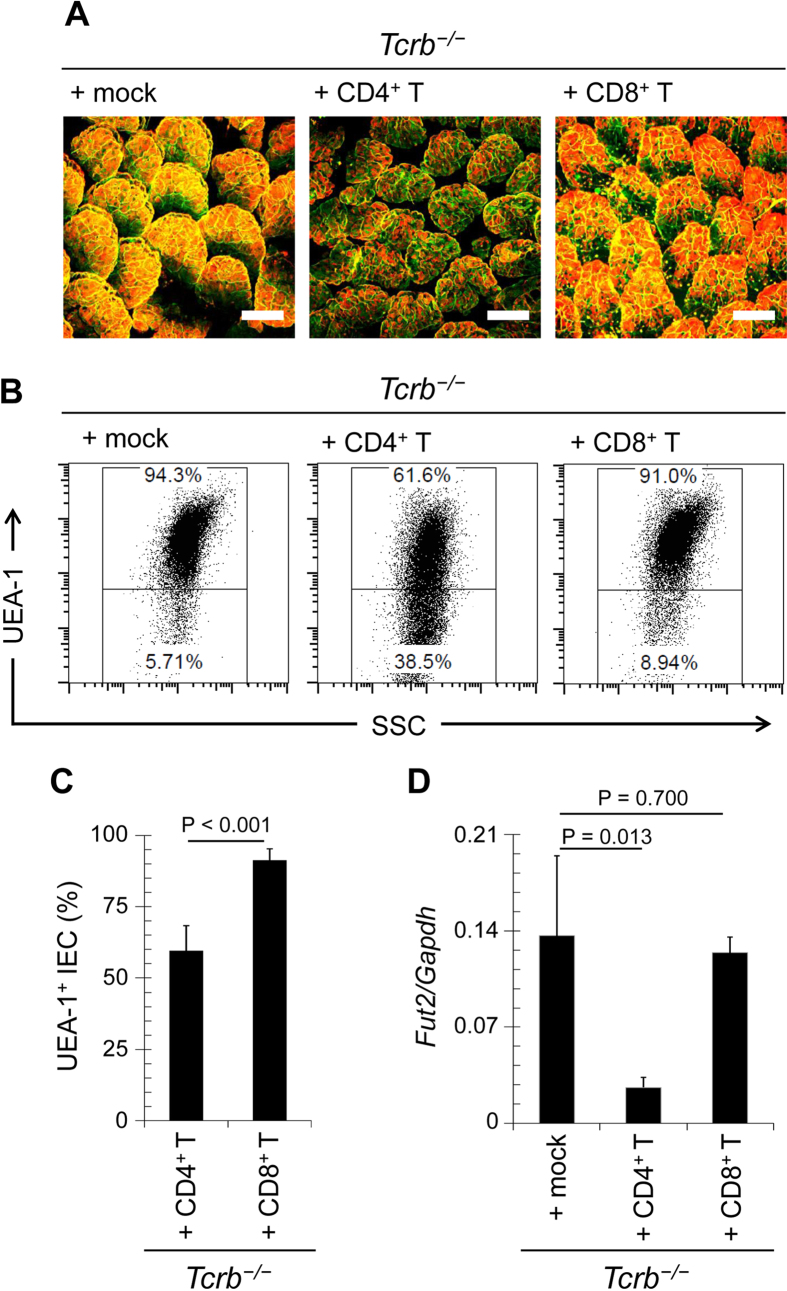
The αβT cells express either CD4 or CD8. To identify the sub-population of T cells responsible for the regulation of epithelial fucosylation, we adoptively transferred purified CD4+ or CD8+ T cells into Tcrb−/− mice. The number of F-ECs was reduced in mice that received CD4+ T cells, but not CD8+ T cells, in comparison with the number in mock-treated Tcrb−/− mice (Fig. 3A–C). Fut2 mRNA expression in ECs was also decreased by adoptive transfer of CD4+ T cells (Fig. 3D). These results indicate that epithelial fucosylation is negatively regulated by CD4+ αβT cells.
Figure 3. Epithelial fucosylation of ECs is downregulated by CD4+ T cells.
CD4+ or CD8+ T cells were purified from the spleens of C57BL/6 mice and adoptively transferred into Tcrb−/− mice. After 4 weeks, ileal ECs were analyzed for fucosylation by whole-mount staining (A) and flow cytometry (B,C), and for Fut2 expression by quantitative PCR (D). Scale bars in (A): 100 μm. Data in (A,B) are representative of 6 independent experiments. Data in (C,D) are shown as mean ± s.d. (n = 6 from 2 independent experiments).
IL-10-producing CD4+ T cells negatively regulate F-ECs
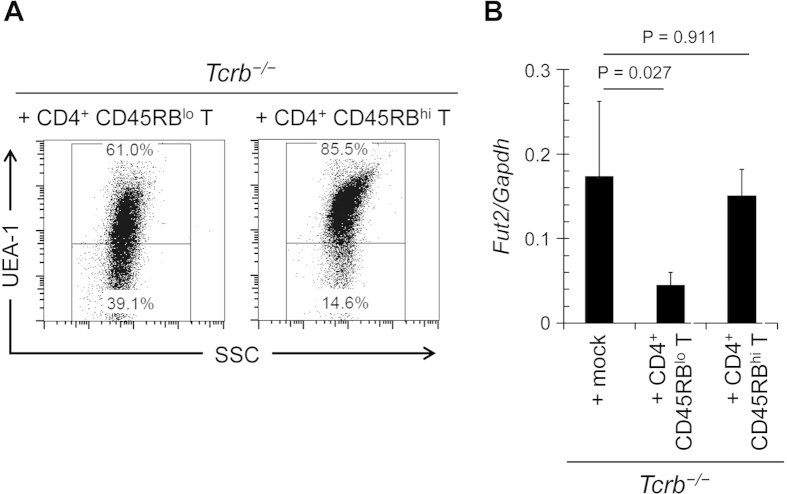
We then examined whether naïve CD4+ T cells or effector CD4+ T cells downregulate epithelial fucosylation. To address this question, we isolated CD4+CD45RBlo effector T cells and CD4+CD45RBhi naïve T cells from normal mice. Adoptive transfer of CD4+CD45RBlo effector T cells, but not CD4+CD45RBhi naïve T cells, into Tcrb−/− mice down-regulated epithelial fucosylation and decreased the expression of Fut2 mRNA in ileal ECs (Fig. 4A,B), suggesting that effector CD4+ T cells are responsible for the reduction of epithelial fucosylation.
Figure 4. Effector T cells are involved in the downregulation of epithelial fucosylation in the ileum.
CD4+ CD45RBhi or CD4+ CD45RBlo T cells were purified from the spleens of C57BL/6 mice and adoptively transferred into Tcrb−/− mice. After 4 weeks, ileal ECs were analyzed for fucosylation by flow cytometry (A) and for Fut2 expression by quantitative PCR (B). Data in (A) are representative of 6 independent experiments. Data in (B) are shown as mean ± s.d. (n = 6 from 2 independent experiments).
The intestinal effector CD4+ T cells produce various cytokines such as interferon γ (IFN-γ), IL-17, and IL-1027. In Ifng−/− mice, epithelial fucosylation was not affected (Fig. 5A). Additionally, adoptive transfer of CD4+ T cells from Rorcgfp/gfp mice (which fail to develop the IL-17-producing Th17 population28) or from wild-type (WT) mice had the same effect on epithelial fucosylation in Tcrb−/− mice; the F-EC number was still reduced upon adoptive transfer of the Th17-deficient CD4+ T cell population (Fig. 5B). Thus, it is likely that neither IFN-γ nor IL-17 is involved in the inhibition of epithelial fucosylation in the ileum.
Figure 5. IFN-γ and IL-17 are not involved in the regulation of ileal EC fucosylation.
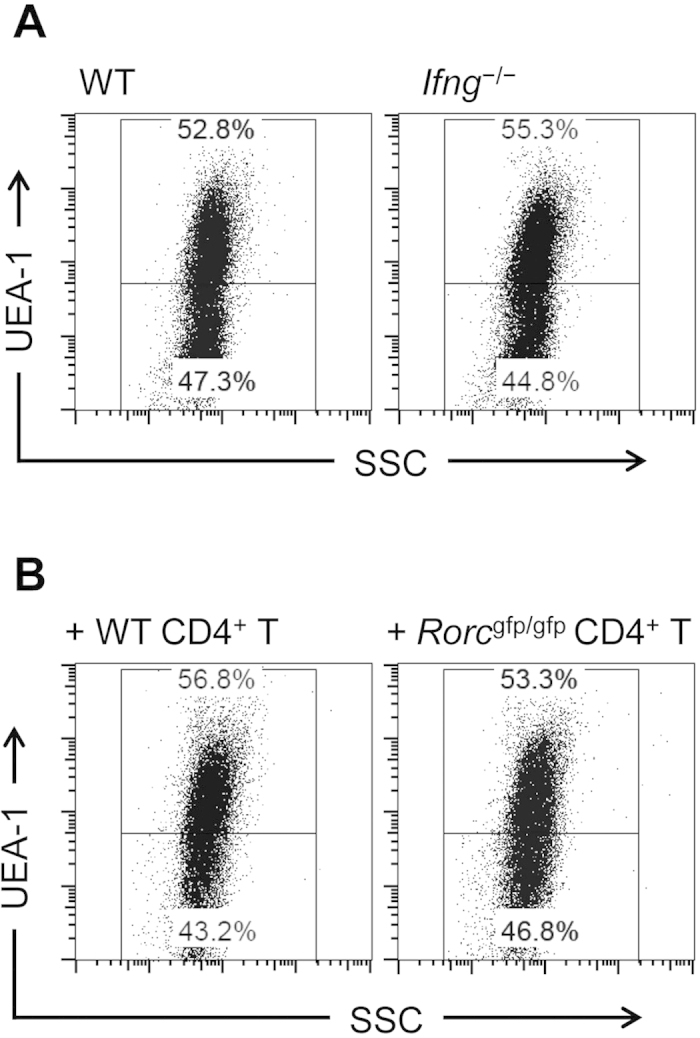
(A) Ileal ECs from wild-type (WT) and Ifng−/− C57BL/6 mice were analyzed for UEA-1 binding. The data are representative of 3 independent experiments. (B) CD4+ T cells were purified from spleens of WT and Rorcgfp/gfp C57BL/6 mice and adoptively transferred into Tcrb−/− mice. Ileal ECs were isolated 4 weeks after transfer and used for the analysis of UEA-1 binding. The data are representative of 4 individual experiments.
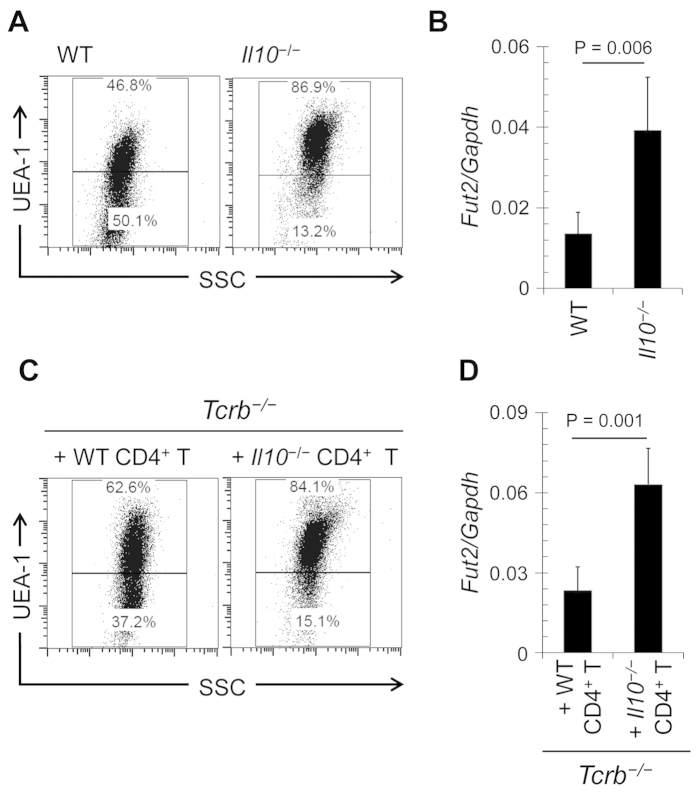
These results led us to focus on the possible involvement of IL-10-producing T cells29 in downregulation of epithelial fucosylation. The number of F-ECs and Fut2 mRNA expression were increased in the ileum of IL-10-deficient (Il10−/−) mice in comparison with WT mice (Fig. 6A,B). These parameters were also increased upon treatment of WT mice with anti-IL-10 receptor–neutralizing antibody (Supplementary Figure 3).
Figure 6. IL-10 derived from CD4+ T cells regulates epithelial fucosylation.
(A) Flow cytometric analysis of the ileum of wild-type (WT) and Il10−/− mice was performed with UEA-1. (B) Quantitative PCR analysis of Fut2 expression in ileal ECs of WT and Il10−/− mice. (C,D) CD4+ T cells were purified from the spleens of WT and Il10−/− mice and adoptively transferred into Tcrb−/− mice. After 4 weeks, ileal ECs were analyzed for fucosylation by flow cytometry (C) and for Fut2 expression by quantitative PCR (D). Data in (A,C) are representative of 6 independent experiments. Data in (B,D) are shown as mean ± s.d. (n = 6 from 2 independent experiments).
To determine whether CD4+ T cells were the major source of IL-10, we analyzed Il10 expression in different subsets of immune cells in the ileum of WT mice. We found that Il10 mRNA expression in CD4+ T cells was higher than that in other major immune cell populations (CD8+ T cells, IgA+ plasma cells, eosinophils, macrophages, and dendritic cells) (Supplementary Figure 4A). IL-10 production in ileal CD4+ T cells was confirmed in IL-10Venus reporter mice (Supplementary Figure 4B). These findings allowed us to determine whether IL-10 produced by CD4+ T cells was responsible for the downregulation of epithelial fucosylation. CD4+ T cells were purified from Il10−/− and WT mice and adoptively transferred into Tcrb−/− mice. The ability to downregulate epithelial fucosylation in Tcrb−/− mice was weaker in the case of Il10−/− CD4+ T cells than in the case of WT CD4+ T cells (Fig. 6C,D). These results suggest that IL-10-producing CD4+ T cells play an important role in the downregulation of epithelial fucosylation in the intestine.
Discussion
Several lines of evidence including ours suggested that commensal bacteria and type 3 innate lymphoid cells are a prerequisite for the induction of epithelial fucosylation12,13,14. Our current study extends our knowledge of the control of epithelial fucosylation by demonstrating its negative regulation by IL-10-producing CD4+ T cells.
In the intestine, several regulatory cell types maintain homeostasis by preventing excessive inflammatory responses; these cells recognize intestinal antigens including commensal bacteria30. IL-10 orchestrates intestinal immune homeostasis by controlling T cells31, macrophages32, and ECs33, including goblet cells34. Our current finding suggests an additional role for IL-10-producing CD4+ T cells in the regulation of Fut2 expression and consequently epithelial fucosylation. Among T cells, Foxp3+ Treg cells and CD4+ Foxp3− Tr1 cells are predominant in the gut and produce IL-1035,36,37. Under some conditions, IL-10 is also produced by other CD4+ T cells (e.g., Th1 cells)38. Although adoptive transfer of IL-10-producing CD4+ T cells decreased the frequency of F-ECs in T cell–deficient mice, intraperitoneal administration of recombinant IL-10 alone did not affect the frequency of F-ECs in the ileum (Supplementary Figure 5). Therefore, it is plausible that IL-10 is necessary but not sufficient for the downregulation of epithelial fucosylation and that some additional molecules on the surface of CD4+ T cells and/or produced by these cells are required to maintain appropriate levels of epithelial fucosylation. Our current efforts are aimed at revealing which types of CD4+ T cells and what additional molecules are pivotal for the IL-10-mediated maintenance of epithelial fucosylation.
Fucose is involved in maintaining microbiota homeostasis in the mucosal epithelia14,39,40. Fucose moieties expressed on ECs and secreted into the lumen are used by commensal bacteria as nutrients. For example, Bacteroides species produce enzymes to cleave fucose from host glycans and use it as a nutrient41,42. Genetic changes in the ability to synthesize fucose are associated with altered gut microbial composition40,43. In addition to commensal bacteria, inactivating mutations in the Fut2 gene reduce susceptibility to human Norwalk virus, which uses α1,2-fucosylated glycans as receptors44. Changes in susceptibility to other pathogens (e.g., Helicobacter pylori45, Candida albicans46, and Streptococcus pneumoniae47) have been reported.
The role of glycans in ECs as a physical barrier between the host and microbes is also highlighted by findings in mice lacking functional core 1–derived O-glycans48. These mice show rapid induction of severe spontaneous colitis with progressive severity, possibly initiated by defective barrier function of the mucosa and greater translocation of bacteria into the mucosal tissue48. Similarly, mice lacking core 2–derived O-glycans show a baseline defect in intestinal permeability and are more susceptible to intestinal challenge with dextran sodium sulfate49. Although the role of fucose on ECs in intestinal inflammation is not clearly understood, recent genome-wide association studies have implicated Fut2 in the pathogenesis of many intestinal inflammatory diseases including Crohn’s disease in the human population22. Of note, αβTCR mutant mice and Il10−/− mice develop spontaneous intestinal inflammation50,51,52. The fact that epithelial fucosylation is highly upregulated in Tcrb−/− and Il10−/− mice implies that fucosylation is at least partly related to intestinal inflammation. In fact, Il10−/− mice with colonic inflammation are reported to show increased expression of the H antigen, which is a product of the Fut2 activity, on the colonic epithelium53. Enhanced expression of Fut2 and subsequent increase in the proportion of F-ECs in the ileum may lead to the development of intestinal inflammation.
The mechanism by which IL-10 regulates FUT2 expression still needs to be determined. Our experiments show that both IL-10 receptor (IL-10R) subunits, α and β, are expressed only in a fraction of F-ECs (Supplementary Figure 6). Although the functional difference between IL-10R+ and IL-10R− F-ECs remains unclear, this finding suggests that IL-10 acts directly on IL-10R+ ECs. An interesting possibility is that additional molecules expressed on the surface and/or produced by CD4+ T cells convert IL-10R− ECs or ECs positive for only one IL-10R chain into ECs expressing both types of IL-10R. In ECs, FUT2 expression is induced by stimulation of ERK–ATF2 and JNK–c-Jun signaling13. IL-10 inhibits the p38/MAPK-activated protein kinase-2 pathway54, which activates ATF255. A similar mechanism may inhibit glycosylation and may be responsible for IL-10-mediated inhibition of Fut2 expression and associated fucosylation of ECs.
Methods
Mice
C57BL/6, Balb/c, C.B-17/lcr-scid/scid, and Balb/c-nu/nu mice were purchased from CLEA (Japan). Ifng−/−, Igh-6−/−, Il10−/−, Rag1−/−, Rorcgfp/gfp, Tcrb−/−, and Tcrd−/− mice (all in the C57BL/6 background) were purchased from The Jackson Laboratory. The Il10Venus mice were generated as previously reported56. All animals were maintained in the experimental animal facility at the University of Tokyo, and the experiments were approved by the Animal Care and Use Committee of the University of Tokyo and conducted in accordance with the committee’s guidelines.
Whole-mount immunofluorescence staining
Whole-mount immunofluorescence staining was performed as previously described14,18. Briefly, the mucus layer was removed by flushing the ileum with phosphate-buffered saline, and then tissues were fixed in 4% paraformaldehyde. The fixed tissues were stained with fluorescence-labeled Ulex europaeus agglutinin I (UEA-1; Vector Laboratories) and wheat germ agglutinin (Invitrogen). Images were analyzed by using a confocal laser-scanning microscope (Leica TCS SP2).
Cell preparations
ECs were prepared as previously described14,18. Briefly, after removal of Peyer’s patches, the intestine was opened longitudinally and washed extensively several times with ice-cold phosphate-buffered saline. The intestine was then cut into 1-cm pieces, which were incubated in 1 mM ethylenediaminetetraacetic acid in phosphate-buffered saline at 37 °C for 15 min with gentle shaking. The suspension was passed through a 70-μm cell strainer, and ECs obtained were washed with Dulbecco’s modified Eagle’s medium containing 20% fetal calf serum.
To isolate mononuclear cells, tissues remaining after EC isolation were finely minced, stirred 3 times in RPMI-1640 containing 2% fetal calf serum and 1 mg/ml collagenase (Wako) for 20 min and centrifuged on a discontinuous Percoll (GE Healthcare) gradient (40% and 75%)57. Cells at the interface between the 40% and 75% fractions were collected.
To prepare single-cell suspensions of splenocytes, excised spleen fragments were pressed through a 70-μm cell strainer using the plunger end of a syringe. Collected cells were treated with lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM ethylenediaminetetraacetic acid) to remove red blood cells and washed with 2% fetal calf serum in RPMI-1640.
Flow cytometry and cell sorting
Isolated ileal ECs were stained with anti-mouse CD45 antibody (clone 30-F11; BioLegend) to distinguish between hematopoietic and non-hematopoietic cells. To exclude non-viable cells, Via-Probe cell-viability solution (BD Biosciences) was used. Staining with anti-mouse EpCAM antibody (clone G8.8; eBioscience) was used to confirm that viable CD45− cells were ileal ECs. UEA-1 was used to assess fucosylation; non-stained samples served as controls.
Mononuclear cells were pre-incubated with anti-CD16/32 (Fcγ RII/III) antibody (BD Biosciences) and stained with fluorescence- or biotin-labeled antibodies. The following antibodies were used: anti-mouse CD45, anti-mouse CD4 (clone RM4–4; BD Biosciences), anti-mouse CD8α (clone 53–6.7; BD Biosciences), anti-mouse CD3ε (clone 145–2C11; BD Biosciences), anti-mouse CD45R (B220; clone: RA3–6B2, BD Biosciences), anti-mouse CD45RB (clone 16A; BD Biosciences), anti-mouse CD11b (clone M1/70; BD Biosciences), anti-mouse CD11c (clone HL3; BD Biosciences), anti-mouse IgA (clone mA-6E1; eBioscience), anti-mouse F4/80 (clone BM8; eBioscience), anti-mouse CD25 (clone PC61; BioLegend), anti-mouse TCRβ (clone H57–597; BD Biosciences), anti-mouse γδ TCR (clone GL3; BD Biosciences), anti-mouse FR4 (clone 12A5; BioLegend), anti-mouse IL-10Rα (clone 3F9; BD Biosciences), and anti-mouse IL-10Rβ (clone 547324; R&D Systems). Cells stained with biotinylated antibodies were further stained with fluorescence-labeled streptavidin (BD Biosciences). Staining with isotype antibodies was carried out. To isolate specific subsets of T cells from splenocytes, B220+ B cells were initially depleted by magnetic-activated cell sorting using B220 microbeads (Miltenyi). Flow cytometric analysis and cell sorting were performed by using FACS Canto and FACS Aria systems (BD Biosciences), respectively. The purity of isolated cells was consistently >95%.
Adoptive transfer experiments
The sorted cells (1 × 106) were administered intraperitoneally to 4–6-week-old Tcrb−/− mice. Recipient mice were analyzed by whole-mount staining and flow cytometry 4 weeks after adoptive transfer. Reconstitution of adoptively transferred cells to the spleen and small intestine was confirmed by flow cytometry at the time of analysis.
Anti-IL-10 receptor antibody and recombinant IL-10 treatment
Mice were intraperitoneally injected with 250 μg of purified anti-IL-10 receptor monoclonal antibody (clone 1B1.3A; eBioscience) or purified rat IgG1 as isotype control on every third day 4 times. Two days after the final injection, mice were used for the analysis of epithelial fucosylation.
Mice were intraperitoneally injected with 4 μg of recombinant IL-10 (BioLegend) in 100 μL of PBS or with vehicle alone57,58. Injections were performed on days 0, 2, 4, 6, and 8, and epithelial fucosylation was analyzed on day 10.
Quantitative PCR analysis
Cells were lysed in TRIzol (Invitrogen), and total RNA was extracted according to the manufacturer’s instructions. Purified RNA was reverse-transcribed by using a SuperScript VILO cDNA Synthesis Kit (Invitrogen). Quantitative PCR was carried out on a Lightcycler II system (Roche Diagnostics) with primers and probe combinations designed by using the Roche Universal Probe Library Assay Design Center (Roche), and the mRNA levels were presented as the ratios of target mRNAs to the internal control, glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA. The following primers and probes were used: Gapdh (sense 5′-tgtccgtcgtggatctgac, antisense 5′-cctgcttcaccaccttcttg; probe #80), Fut2 (sense 5′-gcggttcgtccattccta, antisense 5′-aaaggtacctgggcactcg, probe #76), and Il10 (sense 5′-cagagccacatgctcctaga, antisense 5′-gtccagctggtcctttgttt; probe #41).
Statistical analysis
Statistical analyses were conducted by using Microsoft Excel and GraphPad PRISM. Results were compared by using two-tailed Student’s t-tests. Data were plotted as means ± standard deviation (s.d.).
Additional Information
How to cite this article: Goto, Y. et al. IL-10-producing CD4+ T cells negatively regulate fucosylation of epithelial cells in the gut. Sci. Rep. 5, 15918; doi: 10.1038/srep15918 (2015).
Supplementary Material
Acknowledgments
This work was supported by grants from the following sources: the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Agency (to H.K.); a Grant-in-Aid for Scientific Research on Priority Areas, Scientific Research (S) (to H.K.); Scientific Research (B) (to J.K.); Grants-in-Aid for Scientific Research on Innovative Areas (J.K.); for Challenging Exploratory Research (J.K. and S.S.); Research Activity start-up (YG) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; the Global Center of Excellence (COE) Program “Center of Education and Research for Advanced Genome-based Medicine” (to H.K.); the Ministry of Health, Labour and Welfare of Japan (to H.K. and J.K.); the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (to J.K.); Kishimoto Foundation Research Grant (to J.K.). The Ph.D. studentship of A.L. was sponsored by Chugai Pharmaceutical Company, Inc.
Footnotes
Author Contributions A.L. and Y.G. planned and performed immunological experiments, analyzed the data, and wrote the paper; M.K. performed immunological experiments and analyzed the data; S.S. analyzed and discussed the data; K.H. provided critical reagents and discussed the data; and J.K. and H.K. performed the experiments and wrote the paper.
References
- Hooper L. V. & Macpherson A. J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10, 159–169 (2010). [DOI] [PubMed] [Google Scholar]
- Kunisawa J. & Kiyono H. Immune regulation and monitoring at the epithelial surface of the intestine. Drug Discov Today 18, 87–92 (2013). [DOI] [PubMed] [Google Scholar]
- Goto Y. & Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol Rev 245, 147–163 (2012). [DOI] [PubMed] [Google Scholar]
- Cash H. L., Whitham C. V., Behrendt C. L. & Hooper L. V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S. et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattrup C. L. & Gendler S. J. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol 70, 431–457 (2008). [DOI] [PubMed] [Google Scholar]
- Mukai T., Arihara K. & Itoh H. Lectin-like activity of Lactobacillus acidophilus strain JCM 1026. FEMS Microbiol Lett 77, 71–74 (1992). [DOI] [PubMed] [Google Scholar]
- Yamamoto K. et al. Binding specificity of Lactobacillus to glycolipids. Biochem Biophys Res Commun 228, 148–152 (1996). [DOI] [PubMed] [Google Scholar]
- Ilver D. et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279, 373–377 (1998). [DOI] [PubMed] [Google Scholar]
- Hooper L. V. & Gordon J. I. Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11, 1R–10R (2001). [DOI] [PubMed] [Google Scholar]
- Finne J. et al. Novel polyfucosylated N-linked glycopeptides with blood group A, H, X, and Y determinants from human small intestinal epithelial cells. J Biol Chem 264, 5720–5735 (1989). [PubMed] [Google Scholar]
- Bry L., Falk P. G., Midtvedt T. & Gordon J. I. A model of host-microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383 (1996). [DOI] [PubMed] [Google Scholar]
- Meng D. et al. Bacterial symbionts induce a FUT2-dependent fucosylated niche on colonic epithelium via ERK and JNK signaling. Am J Physiol Gastrointest Liver Physiol 293, G780–787 (2007). [DOI] [PubMed] [Google Scholar]
- Goto Y. et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345, 1254009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Simala-Grant J. L. & Taylor D. E. Fucosylation in prokaryotes and eukaryotes. Glycobiology 16, 158R–184R (2006). [DOI] [PubMed] [Google Scholar]
- Becker D. J. & Lowe J. B. Fucose: biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R (2003). [DOI] [PubMed] [Google Scholar]
- Lin B. et al. GDP-fucose: β-galactoside α1,2-fucosyltransferase, MFUT-II, and not MFUT-I or -III, is induced in a restricted region of the digestive tract of germ-free mice by host-microbe interactions and cycloheximide. Biochim Biophys Acta 1487, 275–285 (2000). [DOI] [PubMed] [Google Scholar]
- Terahara K. et al. Distinct fucosylation of M cells and epithelial cells by Fut1 and Fut2, respectively, in response to intestinal environmental stress. Biochem Biophys Res Commun 404, 822–828 (2011). [DOI] [PubMed] [Google Scholar]
- Biol M. C. et al. Role of insulin and nutritional factors in intestinal glycoprotein fucosylation during postnatal development. Am J Physiol 275, G936–942 (1998). [DOI] [PubMed] [Google Scholar]
- Umesaki Y., Setoyama H., Matsumoto S., Imaoka A. & Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun 67, 3504–3511 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanthakumar N. N., Meng D. & Newburg D. S. Glucocorticoids and microbiota regulate ontogeny of intestinal fucosyltransferase 2 requisite for gut homeostasis. Glycobiology 23, 1131–1141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern D. P. et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet 19, 3468–3476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. J. et al. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes 60, 3081–3084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992). [DOI] [PubMed] [Google Scholar]
- Bosma M. J. & Carroll A. M. The SCID mouse mutant: definition, characterization, and potential uses. Annu Rev Immunol 9, 323–350 (1991). [DOI] [PubMed] [Google Scholar]
- Kunisawa J., Takahashi I. & Kiyono H. Intraepithelial lymphocytes: their shared and divergent immunological behaviors in the small and large intestine. Immunol Rev 215, 136–153 (2007). [DOI] [PubMed] [Google Scholar]
- Littman D. R. & Rudensky A. Y. Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010). [DOI] [PubMed] [Google Scholar]
- Ivanov I. I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). [DOI] [PubMed] [Google Scholar]
- Barnes M. J. & Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity 31, 401–411 (2009). [DOI] [PubMed] [Google Scholar]
- Lathrop S. K. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenbergen S. & Samsom J. N. Maintenance of small intestinal and colonic tolerance by IL-10-producing regulatory T cell subsets. Curr Opin Immunol 24, 269–276 (2012). [DOI] [PubMed] [Google Scholar]
- Ding L., Linsley P. S., Huang L. Y., Germain R. N. & Shevach E. M. IL-10 inhibits macrophage costimulatory activity by selectively inhibiting the up-regulation of B7 expression. J Immunol 151, 1224–1234 (1993). [PubMed] [Google Scholar]
- Bharhani M. S. et al. IL-10 protects mouse intestinal epithelial cells from Fas-induced apoptosis via modulating Fas expression and altering caspase-8 and FLIP expression. Am J Physiol Gastrointest Liver Physiol 291, G820–829 (2006). [DOI] [PubMed] [Google Scholar]
- Hasnain S. Z. et al. IL-10 promotes production of intestinal mucus by suppressing protein misfolding and endoplasmic reticulum stress in goblet cells. Gastroenterology 144, 357–368 e359 (2013). [DOI] [PubMed] [Google Scholar]
- Pot C., Apetoh L. & Kuchroo V. K. Type 1 regulatory T cells (Tr1) in autoimmunity. Semin Immunol 23, 202–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A. et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med 200, 1289–1297 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard C. L. et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3– precursor cells in the absence of interleukin 10. Nat Immunol 8, 931–941 (2007). [DOI] [PubMed] [Google Scholar]
- Saraiva M. et al. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity 31, 209–219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco A. R. et al. Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap P. C. et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci USA 110, 17059–17064 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. J., Reinap B., Lee M. M. & Comstock L. E. Human symbionts use a host-like pathway for surface fucosylation. Science 307, 1778–1781 (2005). [DOI] [PubMed] [Google Scholar]
- Xu J. et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003). [DOI] [PubMed] [Google Scholar]
- Wacklin P. et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One 6, e20113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L. et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med 9, 548–553 (2003). [DOI] [PubMed] [Google Scholar]
- Ikehara Y. et al. Polymorphisms of two fucosyltransferase genes (Lewis and Secretor genes) involving type I Lewis antigens are associated with the presence of anti-Helicobacter pylori IgG antibody. Cancer Epidemiol Biomarkers Prev 10, 971–977 (2001). [PubMed] [Google Scholar]
- Thom S. M. et al. Non-secretion of blood group antigens and susceptibility to infection by Candida species. FEMS Microbiol Immunol 1, 401–405 (1989). [DOI] [PubMed] [Google Scholar]
- Blackwell C. C. et al. Non-secretion of ABO antigens predisposing to infection by Neisseria meningitidis and Streptococcus pneumoniae. Lancet 2, 284–285 (1986). [DOI] [PubMed] [Google Scholar]
- Fu J. et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest 121, 1657–1666 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. L. et al. Glycosyltransferase function in core 2-type protein O glycosylation. Mol Cell Biol 29, 3770–3782 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober W. & Ehrhardt R. O. Chronic intestinal inflammation: an unexpected outcome in cytokine or T cell receptor mutant mice. Cell 75, 203–205 (1993). [DOI] [PubMed] [Google Scholar]
- Kuhn R., Lohler J., Rennick D., Rajewsky K. & Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993). [DOI] [PubMed] [Google Scholar]
- Takahashi I., Kiyono H. & Hamada S. CD4+ T-cell population mediates development of inflammatory bowel disease in T-cell receptor α chain-deficient mice. Gastroenterology 112, 1876–1886 (1997). [DOI] [PubMed] [Google Scholar]
- Miyoshi J. et al. Ectopic expression of blood type antigens in inflamed mucosa with higher incidence of FUT2 secretor status in colonic Crohn’s disease. J Gastroenterol 46, 1056–1063 (2011). [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D. et al. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J 20, 3760–3770 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sadi R. et al. Mechanism of IL-1β modulation of intestinal epithelial barrier involves p38 kinase and activating transcription factor-2 activation. J Immunol 190, 6596–6606 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisawa J. et al. Microbe-dependent CD11b+ IgA+ plasma cells mediate robust early-phase intestinal IgA responses in mice. Nat Commun 4, 1772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannigan K. L. et al. Impaired hydrogen sulfide synthesis and IL-10 signaling underlie hyperhomocysteinemia-associated exacerbation of colitis. Proc Natl Acad Sci USA 111, 13559–13564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy Norton S. et al. IL-10 suppresses mast cell IgE receptor expression and signaling in vitro and in vivo. J Immunol 180, 2848–2854 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.