Abstract
Translational regulation, exerted by the cytosolic ribosome, has been shown to participate in the establishment of abaxial-adaxial polarity in Arabidopsis thaliana: many hypomorphic and null alleles of genes encoding proteins of the cytosolic ribosome enhance the leaf polarity defects of asymmetric leaves1 (as1) and as2 mutants. Here, we report the identification of the SCABRA1 (SCA1) nuclear gene, whose loss-of-function mutations also enhance the polarity defects of the as2 mutants. In striking contrast to other previously known enhancers of the phenotypes caused by the as1 and as2 mutations, we found that SCA1 encodes a plastid-type ribosomal protein that functions as a structural component of the 70S plastid ribosome and, therefore, its role in abaxial-adaxial patterning was not expected.
The abaxial-adaxial patterning of plant lateral organs, such as the leaves, is known to depend on a complex regulatory network that involves microRNAs, trans-acting siRNAs and several families of transcription factors1. Translational regulation, exerted by the cytosolic ribosome, has also been shown to participate in the establishment of abaxial-adaxial polarity, although its role is much less well understood. In the model plant Arabidopsis thaliana (hereafter, Arabidopsis), loss-of-function mutations in many different subunits of the cytosolic ribosome specifically cause a dose-dependent syndrome, with phenotypes ranging from embryonic and gametophytic lethality to mild defects in organ growth and polarity. Many such mutations also enhance the leaf polarity defects of asymmetric leaves1 (as1) and as2 mutants2,3,4,5. One mechanism explaining how the cytosolic ribosome influences leaf polarity depends on the presence of uORFs (upstream open reading frames) in the transcripts of several ARF (auxin response factors) involved in abaxial-adaxial patterning6.
Considerable efforts have been devoted to the elucidation of the function of nucleus-encoded Plastid Ribosomal Proteins (PRPs) in Arabidopsis, most often using publicly available T-DNA insertion mutants. However, 16 out of 36 of the Arabidopsis nuclear-encoded PRPs are essential proteins, precluding ascertaining their role in post-embryonic tissues7.
Here we describe the identification of the Arabidopsis SCABRA1 (SCA1) gene, whose partial loss-of-function enhances the polarity defects caused by as2 alleles. However, contrary to other previously known as1 and as2 enhancers, we found that SCA1 encodes a plastid-type ribosomal protein that functions as a structural component of the 70S plastid ribosome and, therefore, its role in abaxial-adaxial patterning was unexpected. SCA1 was already annotated as EMBRYO DEFECTIVE 3113 (EMB3113), an embryonic lethal gene; the sca1-1 viable allele that we isolated has allowed us studying the role of SCA1 on leaf morphogenesis.
Results and Discussion
The scabra1-1 (sca1-1) mutant was isolated in a large-scale screen for ethyl methanesulfonate (EMS)-induced mutants with abnormal leaf morphology8. The sca1-1 mutant was assigned to the Scabra phenotypic class, which comprises six additional recessive mutants, all with pale green leaves, uneven leaf surface and prominent marginal teeth. The sca1-1 mutant exhibits a general reduction in size, which translates into rosettes with significantly reduced projected area when compared with its wild type, Landsberg erecta (Ler) (Fig. 1A,B; Supplementary Figure S1A).
Figure 1. Mutations in the SCA1 gene alter leaf morphology and pigmentation.

(A–G,I,J) Rosettes from the (A) Ler, (D) Col-0, and (G) No-0 wild types, the (B) sca1-1, (E) sca1-2, (F) sca1-1/sca1-2, (I) sca1-1/sca1-3, and (J) sca1-2/sca1-3 mutants, and (C) the sca1-1 35Spro:SCA1 transgenic line. (H) Dissected silique from a SCA1/sca1-3 plant showing a sca1-3/sca1-3 aborted seed. Unless otherwise stated, all plants are homozygous for the mutations shown. Pictures were taken (A–G,I,J) 16 and (H) 40 days after stratification (das). Scale bars indicate (A-G, I, J) 2 mm, and (H) 0.2 mm.
To identify the SCA1 gene, we followed a strategy combining map-based cloning and next-generation sequencing. We first mapped the sca1-1 mutation to a 760-kb candidate interval on chromosome 2 using 910 chromosomes (Fig. 2A). We next sequenced the sca1-1 genome using the Illumina HiSeq2000 platform. After discarding all the putative Ler/Col-0 polymorphisms, we identified five nucleotide substitutions of the type induced by EMS (three G→A and two C→T transition mutations) within the candidate interval (Supplementary Table 1; see Methods). The C→T mutation in the At2g33800 gene was predicted to cause a Leu→Phe amino acid substitution at residue 233 of the protein. This substitution was confirmed using conventional Sanger sequencing in sca1-1 and Ler plants. Additional evidence that At2g33800 is the same gene as SCA1 was obtained using a construct carrying the coding sequence of At2g33800 placed downstream of the 35S promoter (35Spro:SCA1). This construct fully complemented the mutant phenotype of sca1-1 plants, demonstrating that their phenotype is a consequence of reduced At2g33800 function (Fig. 1A–C; Supplementary Figure S1A).
Figure 2. Fine-mapping of the sca1-1 mutation, structure of the SCA1 gene, and molecular characterization of the sca1-2 allele.
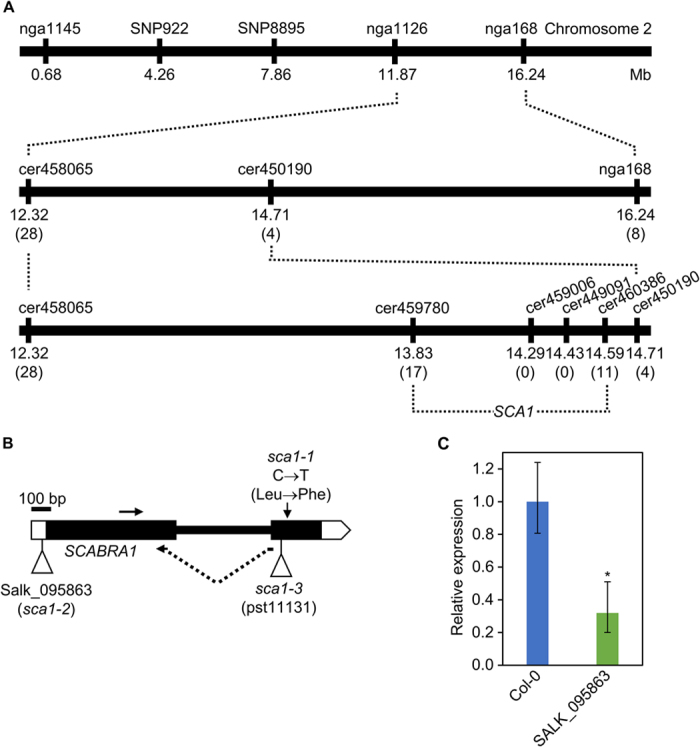
(A) Linkage analysis of the sca1-1 mutation. The names and physical map positions of the molecular markers used for linkage analysis are shown. All values not in parentheses indicate Mb. The number of recombinant chromosomes found are indicated in parentheses. (B) Structure of the SCA1 gene with indication of the position and nature of sca1 mutations. Horizontal arrows indicate the oligonucleotides (not drawn to scale) used as primers for determining the relative expression of SCA1. The vertical arrow marks the position of the point mutation in sca1-1. Triangles represent a T-DNA insertion in sca1-2 and a Ds transposon insertion in sca1-3. (C) qRT-PCR relative expression analysis of SCA1 in the SALK_095863 (sca1-2) line background. Bars indicate relative expression levels, determined by the comparative CT method, and normalized with the expression of the 18S rRNA housekeeping gene. Error bars indicate the interval delimited by 2–(ΔΔCT±SD). Asterisks indicate ΔCT values significantly different from those of Col-0 in a Mann–Whitney U-test (p < 0.01; n = 9).
At2g33800 encodes Plastid Ribosomal Protein S5 (PRPS5), a structural component of the plastid ribosome7. We studied two additional lines, which carry insertions in At2g33800: SALK_095863, with a T-DNA insertion in the 5′ untranslated region, and pst11131, with a Ds element inserted in the second exon (Fig. 2B). Plants homozygous for the SALK_095863 insertion showed a compact and small rosette with pale-green, roundish leaves and a general size reduction (Fig. 1D,E; Supplementary Figure S1B). The F1 progeny of sca1-1 x SALK_095863 crosses displayed a mutant phenotype, showing that both mutations are allelic (Fig. 1F). Using real-time quantitative RT-PCR (qRT-PCR), we showed that the levels of At2g33800 transcripts were reduced in SALK_095863 to ~40% of the wild-type levels (Fig. 2B,C), suggesting that the insertion behaves as a hypomorphic allele. No homozygotes were found for the pst11131 allele, but we observed 25% aborted seeds in the siliques of hemizygous plants, showing that this allele is embryo-lethal (Fig. 1G,H). Indeed, At2g33800 is also known as EMBRYO DEFECTIVE 3113 (EMB3113)9. The F1 progeny of crosses involving sca1-1 and hemizygous pst11131 plants also displayed defects more severe than those of sca1-1 (Fig. 1B,I). We named sca1-2 and sca1-3 the alleles present in the SALK_095863 and pst11131 lines, respectively. The sca1-2/sca1-3 heterozygotes were viable and exhibited stronger defects than the sca1-2 homozygotes (Fig. 1E,J).
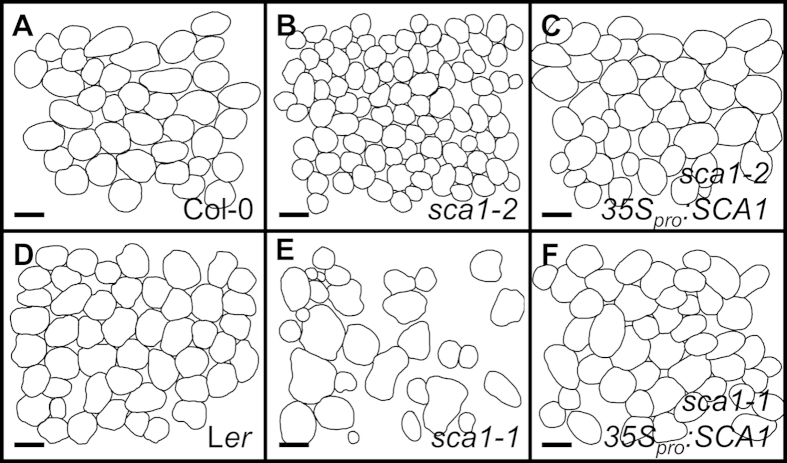
The viability of sca1-1 and sca1-2 mutants offers an opportunity to study the function of PRPS5 in post-embryonic tissues. Because sca1 leaves are pale green, we determined their levels of chlorophyll and carotenoids: both mutants had significantly lower levels of chlorophyll a and chlorophyll b. This reduction was more pronounced in sca1-2 than in sca1-1. The sca1-2 mutant also had reduced carotenoid levels (Supplementary Figure S2A). Consistent with these results, the maximum efficiencies of photosystem II, measured as Fv/Fm, were significantly reduced in sca1 mutants compared with their wild types (Supplementary Figure S2B). In addition to the defects in chloroplast function, mesophyll development was severely perturbed. In paradermal sections, sca1-2 exhibited significantly smaller palisade mesophyll cells (Fig. 3A,B; Supplementary Figure S3A). A more severe defect was observed in the mesophyll of sca1-1 (Fig. 3D,E), which had large intercellular spaces and a distribution of cell sizes wider than in the wild type (Supplementary Figure S3B). The mesophyll phenotype was fully complemented by the 35Spro:SCA1 transgene (Fig. 3C,F; Supplementary Figure S3A).
Figure 3. Representative diagrams of the sub-epidermal layer of palisade mesophyll cells from a first leaf of the (A) Col-0 and (D) Ler wild types, the (B) sca1-2, and (E) sca1-1 mutants, and the 35Spro:SCA1 transgenic line in the (C) sca1-2 and (F) sca1-1 genetic backgrounds, respectively.
Diagrams were drawn from differential interference contrast images taken from cleared leaves. Scale bars indicate 50 μm.
Previous authors have reported an enhancement of the phenotype of as1 mutants in some chloroplast-defective backgrounds, including the sca3 mutant found in our screen10. SCA3 encodes the plastid-targeted RpoTp RNA polymerase, which is required for the expression of plastid-encoded transcripts11. To investigate whether defects in plastid ribosomal proteins can also enhance the abaxial-adaxial polarity defects of as1 and as2 mutants (Fig. 4A–C), we isolated sca1 as1 and sca1 as2 double mutants. Both sca1-1 and sca1-2 enhanced the phenotype of as2-1 though to a different extent (Fig. 4E,H). A strong abaxialization was observed in sca1-1 as2-1 plants, which displayed as2-like cotyledons, radial leaves (Fig. 4E,F) and some trumpet-shaped leaves (Fig. 4G), an enhancement that occurred with full penetrance and similar expressivity in all double mutant plants. A milder enhancement was seen in sca1-2 as2-1 plants, which only occasionally had trumpet-shaped leaves (Fig. 4H,I), as expected if the sca1-2 allele is weaker than sca1-1, as suggested by our morphologic analysis. The same enhancement was not apparent in the sca1-1 as1-1 double mutant (Fig. 4J). To investigate the molecular basis of this interaction, we examined the expression of abaxial-adaxial polarity markers in sca1-1 as2-1 plants, including members of the KANADI (KAN), AUXIN RESPONSE FACTOR (ARF), YABBY (YAB) and HD-ZIPIII families of transcription factors, using qRT-PCR (Fig. 5A). Compared with Col-0, all the studied genes were upregulated in sca1-1 and as2-1, except for KAN1, which was upregulated only in sca1-1, specially the HD-ZIPIII gene REVOLUTA (REV) in sca1-1 and YAB5 in both mutants. On the contrary, in sca1-1 as2-1 plants, the KAN1, KAN2, ARF3 and REV genes exhibited transcript levels similar to those of the wild type, except for YAB5, which was upregulated.
Figure 4. Genetic interaction between the SCA1 and AS genes.

Rosettes from (A) Col-0, (B) as1-1, (C) as2-1, (D) sca1-1 introgressed in Col-0 mutants, and (E) sca1-1 as2-1, (H) sca1-2 as2-1 and (J) sca1-1 as1-1 double mutants. (G,I) Peltate leaves from the (G) sca1-1 as2-1 double mutant, showing the adaxial (ad) and abaxial (ab) sides of the leaf, and (I) sca1-2 as2-1. (F) Detail of the radialized leaves of the sca1-1 as2-1 double mutant. Pictures were taken (A–F,H,J) 16, (G) 30 and (I) 24 das. Scale bars indicate (A–E,G,J) 2 mm and (F) 0.5 mm.
Figure 5. Relative expression analysis of (A) the abaxial-adaxial polarity genes KANADI1 (KAN1), KAN2, AUXIN RESISTANT FACTOR3 (ARF3), REVOLUTA (REV) and YABBY5 (YAB5) in Col-0, sca1-1, as2-1 and sca1-1 as2-1 plants collected 10 das, and of (B) the plastid-encoded genes rbcL, psbA, rrn16, rrn23 and atpB, in Col-0 and sca1-1 plants collected 10 das.
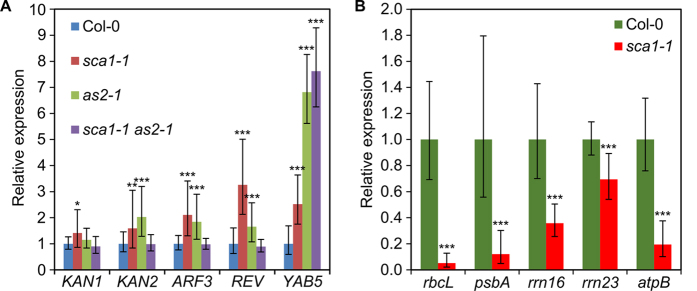
Bars indicate relative expression levels. Error bars indicate the interval delimited by 2–(ΔΔCT±SD). Asterisks indicate ΔCT values significantly different from those of Col-0 in a Mann–Whitney U-test (*p < 0.05, **p < 0.01, ***p < 0.001; n = 9).
We hypothesized that a dual role of SCA1 as a component of both the cytosolic and plastid ribosomes might explain the observed enhancement. However, this possibility is precluded by the predicted plastid localization signal in the SCA1 protein, the absence of SCA1 from the 80S ribosome in proteomic studies, and the very low similarity between the amino acid sequences of SCA1 and its counterpart in the cytosolic ribosome. In line with a role of SCA1 acting specifically in the plastids, we instead found that the sca1-1 mutation causes a general change in the expression of the plastid-encoded rbcL, psbA, rrn16, rrn23 and atpB genes, all of which were found to be downregulated in the mutant using qRT-PCR (Fig. 5B). Remarkably, mutations in components of the plastid transcriptional machinery (e.g. SCA3) and the plastid translational machinery [e.g. EMBRYO DEFECTIVE DEVELOPMENT1 (EDD1)] have been reported to cause similar defects in leaf abaxial-adaxial polarity, uncovering a role of plastids in the establishment of adaxial fate beyond other known roles in leaf morphogenesis10. The inhibition of plastid translation by lincomycin also altered abaxial-adaxial patterning, a defect that was dependent on the GENOME UNCOUPLED1 gene, which is involved in chloroplast retrograde signaling12.
The leaves are lateral organs highly specialized in light capture and photosynthesis, as reflected by their planar shape and the functional differentiation that is apparent along the abaxial-adaxial axis: the tight packing of the adaxial palisade mesophyll provides a solution to maximize light capture, while the abundant stomata and the spongy mesophyll on the abaxial side facilitate gas exchange. Considering that abaxial-adaxial polarity might have evolved as an innovation of land plants to optimize photosynthesis, a functional relationship between plastids and pattern formation is not fully unexpected. On the one hand, chloroplast biogenesis can be seen as a step towards the differentiation of the photosynthetically active palisade mesophyll. On the other, normal chloroplast activity might represent a checkpoint that controls the progression towards the acquisition of adaxial fate. Further study of mutants impairing chloroplast function, such as the ones described in this report, should help to understand how defective chloroplast development feeds back on the establishment of adaxial fate in plant leaves.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh. wild-type accessions No-0, Col-0 and Ler, and the T-DNA insertional line sca1-2 [SALK_095863 (N595863)] were obtained from the Nottingham Arabidopsis Stock Centre (NASC). The transposon tag line sca1-3 (pst11131) was obtained from the RIKEN collection. The sca1-1 mutant was isolated after EMS mutagenesis of Ler seeds as previously described by Berná et al.8. All plants were grown on half-strength Murashige and Skoog (MS) agar medium (2.15 g l−1 [Duchefa], pH 5.7, and 1% sucrose), at 20 ± 1 °C and 60–70% relative humidity under continuous fluorescent light (~75 μmol m−2 s−1) as previously described by Ponce et al.13. Crosses and allelism tests were performed as described by Berná et al.8. To analyze the genetic interactions of sca1-1 with as1-1 and as2-1, we standardized the genetic backgrounds, outcrossing sca1-1 three times to Col-0.
Cloning-by-sequencing
In order to clone the SCA1 gene, we used the approach described in Mateo-Bonmatí et al.14. In brief, we first defined a broad candidate interval of 760 kb (Fig. 2A) by linkage analysis as described in Ponce et al.15, using the primers listed on Supplementary Table 2, and then we re-sequenced the sca1-1 genome. After filtering all the putative Ler/Col-0 polymorphisms, a short list of EMS-type substitutions was obtained (Supplementary Table 1). A total of 3 G → A and 2 C → T substitutions were found within the candidate interval, of which only the C → T substitution in At2g33800 changed the protein sequence. DNA samples were sequenced by Fasteris (Geneva, Switzerland) using the Illumina HiSeq2000 platform. Paired-end reads were 100-bp long. A total of 89,797,616 reads were obtained, which correspond to a 50.38x sequencing depth. The raw data have been deposited at the Short Read Archive database (http://www.ncbi.nlm.nih.gov/sra) with accession number SRP050297.
Gene constructs and plant transformation
The 35Spro:SCA1 transgene was made amplifying the full-length coding sequence of At2g33800 from Col-0 cDNA using Phusion polymerase (Thermo Scientific) and primers containing attB1 and attB2 sites (Supplementary Table 2). The amplification product was cloned into the pGEM-T Easy221 donor vector (kindly provided by Prof. B. Scheres) using BP Clonase II (Life Technologies). The insert was sequenced using an ABI PRISM 3130xl Genetic Analyser (Applied Biosystems), and then transferred into the pMDC32 destination vector16 using LR Clonase II (Life technologies). The construct was then mobilized into Agrobacterium tumefaciens GV3101 (C58C1 RifR) electrocompetent cells, which were used to transform Arabidopsis plants by the floral dip method17. T1 transformants were selected on agar medium supplemented with 15 μg ml−1 hygromycin B (Invitrogen).
RNA isolation, cDNA synthesis and qRT-PCR
Total RNA was extracted using TRI Reagent (Sigma) from a single Col-0 seedling collected 21 days after stratification (das) to synthesize the cDNA used for preparing the 35Spro:SCA1 transgene and a pool of 10 seedlings (collected 10 das) for quantitative RT-PCR analysis, respectively. DNA was removed using the TURBO DNA-free Kit (Invitrogen). First-strand cDNA was synthesized using random hexamers and the Maxima Reverse Transcriptase system (Fermentas). The 18S rRNA18 and ACTIN210 genes were used as internal controls in the relative expression analyses of plastid-encoded and dorsoventrality genes, respectively. Three different biological replicates and triplicate reactions were used. PCR mixes were prepared in a volume of 20 μl by adding 7.5 μl of Maxima SYBR Green/ROX qPCR Master Mix (Fermentas), 5 μl of the corresponding primer pair (1.5 μM each), and 1 μl of cDNA template. Relative quantification of gene expression data was performed using the comparative CT method19 on a Step One Plus System (Applied Biosystems). The primer sets used are listed on Supplemental Table 2.
Morphometry
Rosette area measurements and the morphometric analysis of palisade cells of leaves were performed as described previously20,21. In brief, ten first-node leaves were manually excised and immediately kept in 70% ethanol. Samples were then incubated in a clearing solution (80 g chloral hydrate in 30 ml water) until photosynthetic tissues became transparent and veins were visible. Whole leaves were mounted on glass slides in solutions of 80 g chloral hydrate, 20 ml glycerol and 10 ml water. Pictures from palisade mesophyll were taken halfway along the primary vein and the leaf margin.
Pigment determination and photosynthesis analysis
For determination of chlorophylls and carotenoids, five independent samples of 100 mg each of fresh first-node and second-node leaves from rosettes collected 14 das were pooled, frozen in liquid N2, and homogenized with 4 ml of 80% acetone at 4 °C. The samples were centrifuged for 5 min at 2350 g and the pigment concentration in the supernatant was spectrophotometrically determined as previously described22. Photosynthetic maximum quantum yield was measured 20 das on plants dark-adapted for 30 min and after applying a 0.8-sec saturating light pulse (4000 μmol m−2 sec−1). Measurements were made with a DUAL-PAM/F fluorometer and a DUAL-BA leaf-positioning device (WALZ).
Additional Information
How to cite this article: Mateo-Bonmatí, E. et al. Plastid control of abaxial-adaxial patterning. Sci. Rep. 5, 15975; doi: 10.1038/srep15975 (2015).
Supplementary Material
Acknowledgments
We thank J.M. Serrano, F.M. Lozano, T. Trujillo, R. Sarmiento-Mañús, D. Navarro, L. Serna, J.M. Sánchez-Larrosa and A. Torregrosa for their excellent technical assistance. Research in the laboratory of J.L.M. was supported by grants from the Ministerio de Economía y Competitividad of Spain (BFU2011-22825 and BIO2014-53063-P) and the Generalitat Valenciana (PROMETEOII/2014/006). H.C. was a recipient of a Marie Curie International Reintegration Grant (PIRG03-GA-2008-231073). E.M.-B. held predoctoral fellowships from the Generalitat Valenciana (ACIF/2014/049) and the Ministerio de Educación of Spain (FPU13/00371).
Footnotes
Author Contributions J.L.M., V.Q. and H.C. conceived and designed the research. E.M.-B., R.C.-S., V.Q., A.H. and H.C. performed the research. E.M.-B., V.Q., H.C. and J.L.M. wrote the article.
References
- Byrne M. E. Making leaves. Curr. Opin. Plant Biol. 15, 24–30 (2012). [DOI] [PubMed] [Google Scholar]
- Casanova-Sáez R., Candela H. & Micol J. L. Combined haploinsufficiency and purifying selection drive retention of RPL36a paralogs in Arabidopsis. Sci. Rep. 4, 4122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V. et al. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135, 1315–1324 (2008). [DOI] [PubMed] [Google Scholar]
- Yao Y., Ling Q., Wang H. & Huang H. Ribosomal proteins promote leaf adaxial identity. Development 135, 1325–1334 (2008). [DOI] [PubMed] [Google Scholar]
- Horiguchi G. et al. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65, 724–736 (2011). [DOI] [PubMed] [Google Scholar]
- Rosado A., Li R., van de Ven W., Hsu E. & Raikhel N. V. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc. Natl. Acad. Sci. USA 109, 19537–19544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N. & Bock R. The translational apparatus of plastids and its role in plant development. Mol. Plant 7, 1105–1120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná G., Robles P. & Micol J. L. A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152, 729–742 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D., Muralla R., Sweeney C. & Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 13, 483–491 (2008). [DOI] [PubMed] [Google Scholar]
- Moschopoulos A., Derbyshire P. & Byrne M. E. The Arabidopsis organelle-localized glycyl-tRNA synthetase encoded by EMBRYO DEFECTIVE DEVELOPMENT1 is required for organ patterning. J. Exp. Bot. 63, 5233–5243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricová A., Quesada V. & Micol J. L. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol. 141, 942–956 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameshige T. et al. Pattern dynamics in adaxial-abaxial specific gene expression are modulated by a plastid retrograde signal during Arabidopsis thaliana leaf development. PLoS Genet. 9, e1003655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce M. R., Quesada V. & Micol J. L. Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J. 14, 497–501 (1998). [DOI] [PubMed] [Google Scholar]
- Mateo-Bonmatí E., Casanova-Sáez R., Candela H. & Micol J. L. Rapid identification of angulata leaf mutations using next-generation sequencing. Planta 240, 1113–1122 (2014). [DOI] [PubMed] [Google Scholar]
- Ponce M. R., Robles P., Lozano F. M., Brotons M. A. & Micol J. L. Low-resolution mapping of untagged mutations. Methods Mol. Biol. 323, 105–113 (2006). [DOI] [PubMed] [Google Scholar]
- Curtis M. D. & Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Yamauchi Y. et al. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16, 367–378 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D. & Livak K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- Ferrández-Ayela A. et al. Arabidopsis TRANSCURVATA1 encodes NUP58, a component of the nucleopore central channel. PLoS One 8, e67661 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez J. M. et al. Whole organ, venation and epidermal cell morphological variations are correlated in the leaves of Arabidopsis mutants. Plant Cell Environ. 34, 2200–2211 (2011). [DOI] [PubMed] [Google Scholar]
- Wellburn A. The spectral determination of chlorophyll a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–3013 (1994). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.