Abstract
Nucleotide pools in mammalian cells change due to the influence of antitumor drugs, which may help in evaluating the drug effect and understanding the mechanism of drug action. In this study, an ion-pair RP-HPLC method was used for a simple, sensitive and simultaneous determination of the levels of 12 nucleotides in mammalian cells treated with antibiotic antitumor drugs (daunorubicin, epirubicin and dactinomycin D). Through the use of this targeted metabolomics approach to find potential biomarkers, UTP and ATP were verified to be the most appropriate biomarkers. Moreover, a holistic statistical approach was put forward to develop a model which could distinguish 4 categories of drugs with different mechanisms of action. This model can be further validated by evaluating drugs with different mechanisms of action. This targeted metabolomics study may provide a novel approach to predict the mechanism of action of antitumor drugs.
Abbreviations: ADP, adenosine diphosphate; AMP, adenosine monophosphate; ANOVA, analysis of variance; ATP, adenosine triphosphate; AUC, area under the curve; CDP, cytidine diphosphate; CTP, cytidine triphosphate; dATP, deoxyadenosine triphosphate; dCDP, deoxycytidine diphosphate; dCTP, deoxycytidine triphosphate; dGMP, deoxyribonucleic monophosphate; dGTP, deoxyguanosine triphosphate; DMEM, Dulbecco׳s modified eagle׳s cell culture media; DMSO, dimethyl sulfoxide; DNA, deoxyribonucleic acid; dUDP, deoxyuridine diphpsphate; dUTP, deoxyuridine triphosphate; EC, energy charge; EDTA, ethylene diamine tetra-acetic acid; FCS, fetal calf serum; GDP, guanosine diphosphate; GMP, guanosine monophosphate; GTP, guanosine triphosphate; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PBS, phosphate buffered saline; PCA, principal component analysis; RNA, ribonucleic acid; ROC, receiver operating characteristic; RPMI-1640, Roswell Park Memorial Institute-1640; TBAHS, tetrabutylammonium hydrogen sulfate; TCA, trichloroacetic acid; UDP, uridine diphosphate; UTP, uridine triphosphate
KEY WORDS: Nucleotides, Targeted metabolomics analysis, Tumor cells, Potential biomarkers, Mechanisms of antitumor drug action, Antibiotic anticancer drugs, Principal component analysis, Ion-pair HPLC
Graphical abstract
A RP-HPLC method was used for determination 12 nucleotides in cells treated with antibiotic antitumor drugs. Four categories of drugs with different mechanisms of action were investigated by principal component analysis to develop a recognition model to predict a possible mechanism of an antitumor-compound action.
1. Introduction
Nucleotides are the foundation of all physiological functions. Cancer is a kind of genetic disease and genetic changes will bring about disturbances of nucleotide pools. Therefore, the fluctuation of nucleotide pools in cells can be considered as biomarkers to help reveal the occurrence and development of malignancies, and may be used to evaluate therapeutic effects in many different kind of cancers.
The relationship between nucleotide levels and malignancies has been verified by many studies. Growth stage changes in tumor cells are usually accompanied by alterations of nucleotide pools; for example, a higher rate of utilization of purine precursors has been shown during the proliferation phase than plateau phase1. Some studies have indicated that tumors are associated with abnormal concentrations of normal or modified nucleosides2, 3, 4. Moreover, the levels of nucleotide pools in tumor cells are different from normal cells. For example, the levels of GMP and GDP both were higher in colon cancer tissues than in normal colon tissues, while only GMP was increased in gastric carcinoma5. The activity of de novo synthesis of pyrimidine in MCF7 cells was 4.4 times higher than in MCF10A cells6.
Metabolomics, representing holistic thinking derived from genomics and proteomics, is widely used in the areas of disease diagnosis, drug development, plant metabolomics, nutritional science and microbial metabolomics7, 8, 9, 10, 11. It aims at quantitative analysis of all low molecular mass metabolites and searches for the relationship between metabolites and changes in physiology and pathophysiology by combining different analytical technologies with calculation methods. The levels of nucleotides in cells will be altered significantly by perturbations provided by pharmaceuticals or environmental factors12, 13. Clayton et al.14 first proposed the concept of “pharmaco-metabonomics” in 2006: analyzing metabolic profiles of biological samples could be a way to predict variations of metabolic phenotypes after treatment with drugs. Over the last several decades, many scholars have investigated the dynamic changes in cell metabolism, cell physiological status, and the activity of key enzymes after drug treatment. Zhang et al.15 did research on the effect of DNA synthetase inhibitor–aphidicolin on deoxyribonucleotide pools in human pancreatic carcinoma PANC1 cells and found that all components were increased except dUTP. Sokoloski et al.16 found that the level of ATP decreased and UTP increased in HL60 cell lines after inhibiting the activity of glycinamide nucleotidyltransferase. Studies by Iwasaki et al.17 showed that ribonucleotide reductase inhibitors could prevent de novo dCTP biosynthesis which benefited are-CTP mixing into DNA, suggesting that the antitumor activity of this category could be reinforced by combination with nucleotide reductase inhibitors. Studies on the effects of gemcitabine on ribonucleotides pools in leukemia cell lines and some solid tumors showed that the level of dNTP in cells was related to drug susceptibility, and wide differences among NTP levels in different tumor cells reflected different interactions between cells and drugs18, 19. Monitoring these changes in nucleotide pools will help to predict the therapeutic effects of antitumor drugs and reveal the targets of drug action. Although basic analysis methods applied to cells have long been used to aid in research on effects of drugs on metabolites, the study of tumor cells for distinguishing different mechanisms of drug action has not been investigated.
In our previous research, Zhang et al.20 developed a reliable, simple and reproducible method to determine the levels of 12 nucleotides simultaneously by using an ion-pair HPLC and analyzed the differences in nucleotide levels between normal cells and tumor cells. The method above was further used to explore three categories of chemotherapeutic drugs with different mechanisms of action by Liu et al.21. Three classes of antitumor drugs, including antimetabolic agents, agents directly acting on DNA and antimitotic agents, were selected to explore their effects on nucleotides in cells in order to find potential biomarkers associated with drugs. Based on the results of several analytic approaches, GMP and ATP were chosen as the best potential biomarkers for DNA-damaging drugs, while ATP, UDP and GMP were identified for two other categories of drugs. However, the method we used was not sufficient to develop a model to distinguish antitumor drugs with different mechanisms of action individually.
In this pilot study, daunorubicin (DNR), epirubicin (EPI) and dactinomycin D (ACD), with completely different mechanisms of action were evaluated and the best potential biomarkers were found. Based on the earlier study, all 12 chemotherapeutic drugs were classified into four groups by mechanism of drug action, leading to a new and discriminatory model. Moreover, different cell sensitivities to drugs were found which could affect the ability to accurately recognize mechanisms of drug action.
2. Methods
2.1. Chemicals and reagents
All standards (UTP-Na, ADP-Na2, AMP-Na2, ATP-Na2, CTP-Na2, GDP-Na2, GMP-Na2, UDP-Na2, dUTP-Na3, dATP-Na3, dCTP-Na3, dGTP-Na4) were purchased from Sigma (St. Louis, USA). Tetrabutylammonium hydrogen sulfate (TBAHS) was obtained from Tianjin Kermel Chemical Reagent Co., Ltd. Methanol (HPLC grade) was obtained from Shandong Yuwang Industrial Co., Ltd. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and trypsin were provided by Amresco LLC (Solon, USA). All other chemical reagents used, analytical grade, were purchased from Tianjin Bodi Chemical Holding Co., Ltd. Dulbecco׳s modified eagle׳s cell culture media (DMEM), Roswell Park Memorial Institute (RPMI) 1640 media, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and fetal calf serum (FCS) were purchased from Gibco-BRL (Grand Island, USA).
2.2. Instrument and chromatographic conditions
Separation was performed on an Elite-L2130 HPLC system (Hitachi Technologies, Japan) consisting of a binary solvent delivery system and an Elite-L2400 UV detector. The UV detection wavelength was set at 254 nm. The 12 nucleotides were separated simultaneously by using a Synergi Hydro-C18 column (250 mm×4.6 mm, 4.0 μm) at 25 °C. The mobile phases (A and B) consist of phosphate, methanol and TBAHS with a constant flow of 0.8 mL/min. A gradient program was used as follows: 0–50 min, 48%–60% A; 50–68 min, 60%–100% A; 68–77 min, 100% A.
2.3. Cell lines and culture conditions
All cell lines were kept in our laboratory. Human rhabdomyosarcoma cells (A204), hepatoma cell line (HepG2), human pulmonary carcinoma cells (A549), human ovarian carcinoma cells (SKOV3), human breast adenocarcinoma cells (MCF7) and human breast carcinoma cells (T47D) were cultured in DMEM medium containing 3.7 g/L NaHCO3, 10% FCS, penicillin (100 u/mL) and streptomycin (100 μg/mL). Human fibrosarcoma cells (HT1080), human prostate carcinoma cells (PC3), human prostatic cancer cells (DU145), human mouth epidermal carcinoma cells (KB), Henrietta Lacks strain of cancer cells (HeLa) and human gastric carcinoma cells (SGC7901) were maintained in RPMI-1640 medium containing 2.4 g/L HEPES, 2.0 g/L NaHCO3, 10% FCS, penicillin (100 u/mL) and streptomycin (100 μg/mL). The 12 tumor cell lines were incubated at 37 °C with 5% CO2 in a humidified air atmosphere. Cells were harvested in their exponential growth phase.
2.4. Determination of antitumor activity
The MTT assay was used to determine the antitumor activity of three drugs. 200 µL of cells (5×104 cells per well) were added into 96-well plates and cultivated for 24 h. The medium was replaced and the cells were treated with different concentrations of antitumor drugs for 48 h. 20 µL of MTT (5 mg/mL) was added after removing the supernatant and incubation continued for another 4 h. To each well was added 150 µL of dimethyl sulfoxide (DMSO) before measuring the absorbance (OD) at 492 nm using a Thermo MK3 Reader. The half maximal inhibitory concentration (IC50) values were determined.
2.5. Cell treatment and harvest
About 1×106 cells were added into each flask. After cultivation for 24 h they were divided into a control group and a drug-treated group. The drug-treated group was treated with antitumor drugs according to IC50 as determined by the MTT method and the control group was added into the same amount of medium. Cells were harvested after 48 h.
2.6. Nucleotides extraction for cells
Sample preparation was the same as the method reported previously20. Trichloroacetic acid (TCA, 6%) was used to extract the intracellular nucleotides on ice.
2.7. Data analysis and statistics
An external standard method was used to measure the concentrations of nucleotides in the tumor cells. One-way analysis of variance (ANOVA) was calculated by SPSS 16.0 software to show the difference of components between the control group and the drug-treated group. The P values were recalculated by Bonferroni correction. Principal component analysis (PCA) was performed by SIMCA-P 11.5 demo software to understand the link between significant components, search for potential biomarkers, and develop a discriminatory model. Moreover, the potential biomarkers were validated by receiver operating characteristic (ROC) curve.
3. Results and discussion
3.1. Effect of antibiotic antitumor drugs on nucleotides
DNR, EPI and ACD are antibiotics. They express their antitumor properties by interfering with the transcription process and inhibiting RNA synthesis. The content of nucleotides in different drug-treated groups varied in degree. The change trends of 12 nucleotides in cells compared with the control group were list in Table 1 and Supporting information. The level of AMP in all cells was increased after administration of drugs; ADP and UDP up-regulated were observed in most cells; the main trends of UTP and ATP were down. Based on past reports, the three drugs we chose could disorder the function of mitochondria22, 23, 24 and induce cell apoptosis25, 26, 27. The damage of mitochondria and apoptosis might be the reason why the ATP level was depleted. The lower ATP level promotes hydrolysis of ADP to supply energy for cellular metabolism, which leads to the decreased amount of GMP. Furthermore, the lower level of ATP also results in energy deficiency when GMP is generated and induces GMP down-regulation. Shepelevtseva et al.28 have shown that anthracycline ring antibiotics can cause glycogen storage in cardiac muscle tissue and decrease the content of RNA and DNA. Because glycogen synthesis utilizes UTP to provide energy, the lower content of UTP may be associated with glycogen synthesis. On the other hand, the synthesis of UTP is based on CTP, so UTP down-regulation must be accompanied with the same trend of CTP, which is well confirmed by our study. In addition, the decrease in dGTP brings about a weaker reduction reaction from ADP to dATP and results in a lower level of dATP.
Table 1.
| Sample | DNR |
EPI |
ACD |
|||
| Main trend | P value | Main trend | P value | Main trend | P value | |
| ADP | ↑ (11) | ⁎⁎⁎P<0.0011 | ↑ (12) | ⁎⁎⁎P<0.0011 | ↑ (10) | ⁎⁎P<0.0055 |
| AMP | ↑ (12) | ⁎⁎⁎P<0.0011 | ↑ (12) | ⁎⁎⁎P<0.0011 | ↑ (12) | ⁎⁎⁎P<0.0011 |
| ATP | ↓ (12) | ⁎⁎⁎P<0.0011 | ↓ (10) | ⁎⁎⁎P<0.0011 | ↓ (12) | ⁎⁎⁎P<0.0011 |
| UTP | ↓ (11) | ⁎⁎⁎P<0.0011 | ↓ (12) | ⁎⁎⁎P<0.0011 | ↓ (12) | ⁎⁎⁎P<0.0011 |
| CTP | ↓ (12) | ⁎⁎P<0.0055 | ↓ (10) | – | ↓ (12) | ⁎⁎⁎P<0.0011 |
| dGTP | ↓ (10) | – | ↓ (12) | – | ↓ (11) | ⁎⁎⁎P<0.0011 |
| dATP | ↓ (9) | – | ↓ (11) | – | ↓ (10) | ⁎⁎P<0.0055 |
| GDP | ↓ (8) | – | ↑ (7) | – | ↓ (9) | – |
| GMP | ↓ (9) | – | ↓ (8) | – | ↓ (9) | – |
| UDP | ↑ (7) | – | ↑ (9) | – | ↑ (8) | – |
| dCTP | ↓ (10) | – | ↓ (10) | – | ↓ (8) | – |
| dUTP | ↓ (8) | – | ↓ (9) | – | ↓ (9) | – |
Changes of trend compared with the control group: ↑ up-regulated; ↓ down-regulated. The number in the bracket represents the amounts of cells of which change of trends are in accordance with the main trend for each drug-treated group.
P value was calculated using one-way ANOVA with Bonferroni correction (significance at ⁎⁎P<0.0055, ⁎⁎⁎P<0.0011); –: no significant difference.
DNR is an anthracycline antibiotic whose structure has four combined aromatic nucleus and the shape of one molecular amino sugar is flat. As shown in Table 1, in this group, the dGMP pools of all cells were shrunk except for two cells lines; the main trends of dATP and dUTP were down, which suggested that the synthesis of DNA was abnormal. Furthermore, the disorder of dNTP pools was accompanied with other abnormal nucleotides pools that might be connected with mechanisms of drug action. There are several possible explanations for the mechanism of anthracyclines, and some controversy still exists among them. Firstly, the anthracene ring of drugs implants into DNA and its plane is perpendicular to the axis of the helix. The amino sugar on the side chain interacts with the sugar phosphate group of DNA by hydrogen bonds, which makes the adduct more stable, changes the DNA structure, and disturbs DNA replication and RNA synthesis29. DNR inhibits the precursors mixing into DNA and RNA30, and also inhibits DNA polymerase and RNA polymerase31. Secondly, DNR forms a ternary complex with topoisomerase II and DNA that keeps the reversible topoisomerase-DNA complexes in a stable state. Thus, it leads DNA fragments to be separated32. Thirdly, under the catalytic action of cytochrome P450 reductase, NADPH as an electron donor regenerates DNR and produces semiquinone free radicals, as well as active free radicals generated under aerobic conditions, such as superoxide anion free radicals and hydroxyl free radicals, which will cause cell membrane lipid oxidation, cytotoxicity, damage to DNA and cardiotoxicity33. DNR implants into a single-stranded DNA at the acting locus of CpG first and then ApG; DNR first embeds into (CpG)2 base pairs when DNR interacts with a double stranded DNA and then into (CpA)(TpG) and (CpT)(ApG) base pairs33. As a consequence, the unbalance of dGTP, dCTP and dATP pools may be associated with the chelation between DNR and DNA, while the disorder of NTP pools is connected with the inhibition of RNA synthesis. Vidal et al.34 had studied the energy deprivation and nucleotide degradation induced by doxorubicin and DNR using isolated cardiomyocytes. They found that DNR at higher concentration (150 µmol/L) caused a fast depletion of DNA (above 40%) and an up-regulated ADP in cytoplasm (6-fold) accompanied with released adenosine and inosine concurrently, while a lower concentration (0.15 µmol/L) could reduce the levels of ADP and NAD in cells. Our study is in accordance with the result of higher concentrations of DNR. Furthermore, glutathione (GSH) played an important role as mediated by MRP1 in multidrug resistance (MDR) associated with overexpression of P-glycoprotein (P-gp) and/or multidrug resistance-associated protein (MRP1). Benderra et al.35 had investigated the regulation of glutathione modulated nuclear accumulation of DNR in human MCF7 cell resistance VP-16 (MRP1 overexpression) and doxorubicin (P-gp overexpression). The research indicated that DNR clearly accumulated in the MCF/VP nucleus under the action of GSH-biosynthesis inhibitors. A previous study by Cao et al.36 showed that a glucose uptake inhibitor could increase the sensitivity of cancer cells to DNR and overcome the drug resistance of tumor cells in hypoxia. Moreover, AKR1B10 protein participated in the metabolism of DNR and associated with drug resistance mediated by MRP137. Consequently, drug concentration, the differential expression of GSH, glucose transporter and AKR1B10 among different tumor cells could bring about different cell sensitivities to DNR and finally lead to different responses of tumor cells to DNR during metabolic processes.
EPI is an isomeride of doxorubicin which has lower cardiotoxicity. As shown in Table 1, the changes in nucleotides pools were similar to DNR: the level of dGTP in cells was down-regulated; the main trends of dATP and dUTP were also decreasing; other metabolic changes in nucleotides were observed which might be associated with the mechanism of EPI action. EPI and DNR are anthracycline antibiotics. Their anthracene rings implant into G and C base pairs to form a covalent bond with DNA38, which may destroy the function of DNA template, interfere with transcription and also prevent the synthesis of mRNA. The interaction between EPI and DNA could induce DNA to disintegrate and make topoisomerase II-DNA complexes more stable.
In addition, EPI inhibits DNA helicase directly and DNA and RNA polymerase indirectly39. In our study, a decreased amount of dNTP was observed, which might be associated with the inhibition of DNA synthesis, and other unbalanced nucleotides pools may reflect abnormal RNA synthesis. Autophagy is a process of decomposition depending on lysosomes in cells. Lysosomes digest proteins or organelles and release large molecules which can be further metabolized, while the energy generated is useful to help cells adjust to hunger and stress, such as hypoxia, Γ-ray and antitumor drugs. Research40 showed that EPI could induce MCF7 cell autophagy which protected MCF7 cells from EPI-induced apoptosis. Autophagy would increase in EPI-resistance of MCF7 cell, but when autophagy was inhibited, the sensitivity of MCF7 cells would return. Cantoni et al.41 studied the effects of doxorubicin and EPI on nucleic acid metabolism and cytotoxicity. They found that after exposure to drug for 1 h, the extent and rate of DNA strand breaks induced by EPI were greater than with doxorubicin; the EPI-induced damage disappeared after removing EPI, but was maintained in the doxorubicin group. Accordingly, the differential of cell autophagy and the capacity of DNA repair induced by EPI after damage might result in the interaction between cells and drug differently, which might be the reason that nucleotides levels in each cell line differed.
In the ACD-treated group dNTP pools were abnormal as shown in Table 1, the content of dGTP in almost all cells was decreased and the main trends of dATP, dCTP and dUTP were decreasing. These results might be associated with mechanism of drug action. ACD is composed of a three-membered ring, a phenoxazine-3-ketone chromophore and a two side chains of five-cyclic peptide, action mechanism of which is to prevent DNA synthesis depending on concentrations or RNA synthesis, and inhibit RNA synthesis. Adducts formed also break single-stranded RNA. ACD combines with guanine and implants phenoxazine rings into G/C base-pairs according to the result of X-ray diffraction studies42. A cyclopeptide side chain in the minor groove of DNA double helices reacts with bottom and side walls of minor groove by hydrogen bonding or hydrophobic interaction to fix adducts. The peptide chain in the minor groove of DNA inhibits RNA polymerase activity and mRNA synthesis, but can not stop the replication of DNA. So the down regulations of dGTP and dCTP may be the result that ACD implants into DNA, while disturbances of other kinds of nucleotides pools also reflect RNA synthesis disruption. ACD gets into cells by passive diffusion. The uptake by cells of ACD is determined by the structure and composition of the cytomembrane, while some substances that change the structure of the cell membrane, such as amphotericin B, and reinforce permeability could raise the rate of ACD uptake and increase the lethal effect in drug-resistant cells. The differential uptake of ACD may cause cells to react to drugs differently and ultimately result in alterations of nucleotides pools in each cell to varying degrees.
3.2. Statistical analysis of metabolites
Fig. 1 shows the energy charge (EC) values of tumor cells which were calculated as follows: ([ATP]+1/2[ADP])/ ([ATP]+[ADP]+[AMP])43. The EC value, which can be a useful measure to estimate the balance among adenine nucleotides, quantitatively represents the energy status of high-energy phosphate bonds. As shown in Fig. 1, the energy charge values of all tumor cells were lower than the control group except for SGC7901 cells. Significant differences were observed between the drug-treated group and the control group (P<0.01), which suggested that drugs could interfere with energy metabolism of tumor cells and damage mitochondria. It might be a common event by which the three drugs of this category lead to cell death.
Figure 1.
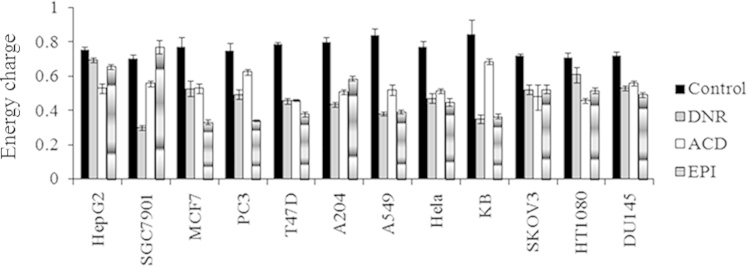
Changes in energy charge (EC) values of 12 tumor cells treated with three drugs for 48 h. Significant differences were observed between the drug-treated groups and the control group.
Significant differences were found between the control group and the drug-treated group by using ANOVA by SPSS 16.0. A Bonferroni correction was used to recalculate the P value. P<0.0055 is customarily considered as indicating statistically significant differences between two groups. The results (Table 1 and Supporting information) showed that levels of AMP, ADP, UTP, ATP and CTP (P<0.0011 for the first four and P<0.0055 for CTP) fluctuated observably in the DNR-treated group, AMP, ATP, ADP and UTP (all P<0.0011) in the EPI-treated group, AMP, ATP, dGTP, CTP, UTP, ADP and dATP (P<0.0011 for the first five and P<0.0055 for the last two) in the ACD-treated group. The contents of ATP, UTP and CTP mainly tended to decline; moreover, the levels of AMP and ADP mostly were elevated, which could indicate that significant differences in nucleotides of cells could be the potential biomarkers associated with these three antibiotic antitumor drugs.
To investigate the overall metabolic differences, Fig. 2 represents the PCA score plot of cells in the control group and the drug-treated groups using pattern recognition approaches by the SIMPCA-P 11.5 Demo software. Data points marked by squares represent the control group and the other shapes represent the drug-treated groups. A clear and significant separation could be observed between the control group and the drug-treated group which indicates that the nucleotide metabolite pattern was different. As shown in Fig. 2, the data points of the ACD-treated group were rather close and the other two groups were relatively scattered which might stem from the different structures among three drugs. DNR and EPI belong to anthracycline-based drugs, while ACD is a phenoxazine-based drug whose aromatic rings fit with DNA directly, which indicates that phenoxazine rings combine with DNA more tightly. This structural difference may lead to the differential efficacy as indicated by the much smaller IC50 values of ACD. The metabolites that contributed the most toward classification are shown in the loading plot (Fig. 3). Each point represented one nucleotide on the intracellular levels and majority of the nucleotides were clustered in the center. Outliers including ATP, AMP, ADP and UTP contributed more to the PCA model and might be potential biomarkers associated with the drug action of antibiotic agents.
Figure 2.
PCA score plot of cells in the control group and three drug-treated groups ( Control group;
Control group;  DNR-treated group;
DNR-treated group;  ACD-treated group;
ACD-treated group;  EPI-treated group). A significant separation could be observed between the control group and the drug-treated groups.
EPI-treated group). A significant separation could be observed between the control group and the drug-treated groups.
Figure 3.
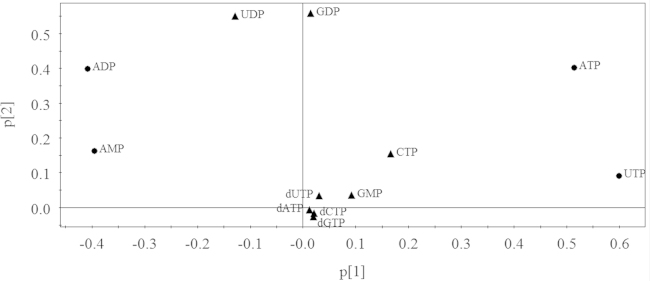
Loading plot of cells in the control group and three drug-treated groups. Each point represented one nucleotide on the intracellular levels. Tumor cells of the antibiotic antitumor drug-treated groups were correlated with ATP, AMP, UTP and ADP.
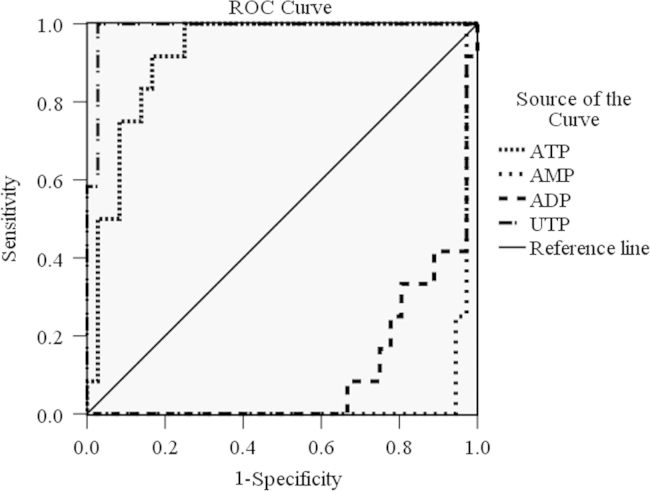
According to the results of ANOVA and PCA analysis, we chose ATP, AMP, UTP and ADP to further assess as potential biomarkers using ROC curve analysis by SPSS 16.0 software. ROC curve is considered a method for diagnosing and evaluating differences. The area under curve is used to assess the value of test. The closer a value is to 1.0, the better a test it will be. When the value is 0.5, it indicates that the test has no meaning. As shown in Fig. 4, the results of ROC curve are as follows: UTP (0.988)>ATP (0.921)>ADP (0.106)>AMP (0.035). AUCs above 0.9 mean a higher accuracy in diagnosis, therefore, UTP and ATP are most likely to be the biomarkers for diagnosing this mechanism of antitumor drugs.
Figure 4.
ROC curve of the ability of nucleotides to discriminate tumor cells in the control group and the drug-treated groups. UTP and ATP were the best biomarkers for antibiotic antitumor drugs.
Antibiotic anticancer drugs intercalate into G and C bases of DNA double helices to inhibit the function of RNA polymerase. As a result, they could interfere with the transcription process and inhibit RNA synthesis, especially for mRNA. Our results show that AMP, ATP, ADP and UTP might be the potential biomarkers for this agent. To meet the huge demand for energy of tumor cell proliferation, ATP hydrolysis will be significant, therefore, the intracellular ATP level will be decreased significantly. Meanwhile, ADP and AMP, as the products of ATP hydrolysis, will accumulate in cells, so their levels will be up-regulated. The variation in trends of the three nucleotides is consistent with the abnormal metabolic ways of the tumor cell, which indicates that antibiotic drugs playing their roles of anticancer agents are not by way of interfering with tumor cell energy metabolism. On the other hand, the levels of ATP, CTP and UTP are all decreased. Their shortages will inhibit RNA synthesis and interfere with the transcription process, which is the same as the mechanism of drug action, since they are substrates for RNA synthesis. In summary, as the two best potential biomarkers for antibiotic anticancer drugs according to the ROC curve, ATP levels indicates that the energy metabolism of tumor cell is abnormal, while UTP may suggest some biological relevance of drug action. Even though all the changes in nucleotides cannot be explained thoroughly, the results show that the metabolism of nucleotides is closely related to mechanisms of drug action.
3.3. Distinguishing the mechanism of antitumor drug action
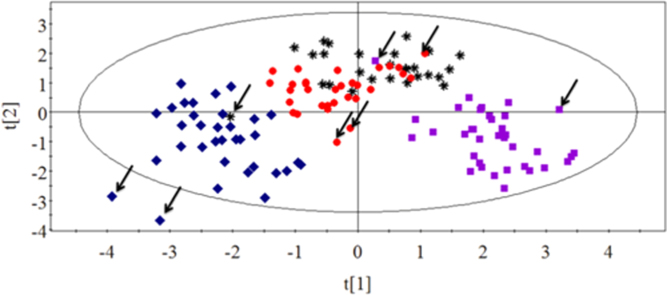
So far, we have investigated the effects of 12 kinds of chemotherapeutic drugs on nucleotides in cultured tumor cells and found some potential biomarkers associated with drugs but failed to develop a model to distinguish nine chemotherapeutic drugs individually. Because similar characteristics of drugs have the same mechanism of action, in this pilot study, 12 kind of chemotherapeutic drugs was classified by mechanisms of drug action into four groups: antimetabolic agents (5-fluorouracil, methotrexate and cytarabine), agents directly acting on DNA (mitomycin C, cisplatin and etoposide), antimitotic agents (taxol, vincristine and homoharringtonine) and antibiotic agents (DNR, EPI and ACD). The MTT assay was used to determine the antitumor activity of drugs. IC50 values represented the levels of cell sensitivity to antitumor drugs and the lower value meant more sensitivity. Four categories of drugs were investigated by PCA to find the link between nucleotide levels and different mechanisms of drug action. Points represented by the same mechanism of drug action showed a similar trend. As shown in Fig. 5, several points marked by an arrow were far away from their groups. Interestingly, these outliers had the very largest IC50 values relative to others of their group. Maybe the cell sensitivity to antitumor drug was too low, which resulted in the differential metabolism of nucleotides in cells. In consequence, the different cell sensitivity to drugs may influence the metabolism of nucleotides and the greater the difference the greater the impact on distinguishing mechanisms of drug action. Unfortunately, we have not found any common statistical correlation between outliers and IC50 values. Even so, except for the points marked by the arrow, data points grouped into three areas, of which, antimitotic agents and antibiotic agents could be separated well while the other two groups partially overlapped.
Figure 5.
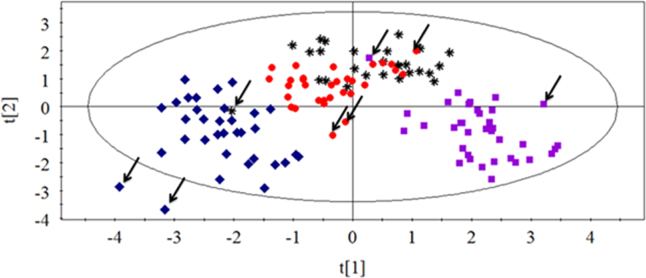
PCA score plot of cells in drug-treated groups with four classes of mechanisms of action ( Antimetabolic agents;
Antimetabolic agents;  agents directly acting on DNA;
agents directly acting on DNA;  antimitotic agents;
antimitotic agents;  antibiotic agents). Points marked by arrow were outliers which had the very largest IC50 values relative to the others of their group.
antibiotic agents). Points marked by arrow were outliers which had the very largest IC50 values relative to the others of their group.
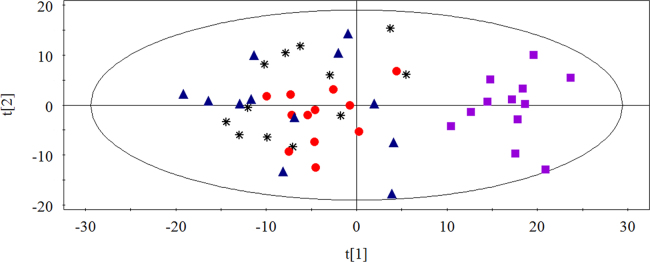
PCA analysis was used to find the metabolic differences between antimetabolic agents and agents directly acting on DNA. As shown in Fig. 6, two groups separated from each other. Antimetabolic agents function as antitumor drugs by combining with metabolic enzymes, which is different from the agents that directly act on DNA, but they all ultimately inhibit the synthesis of DNA in cells to achieve the antitumor results, which is similar to the mechanism of agents directly acting on DNA44, 45, 46, 47, 48, 49. According to the results above, the 12 antitumor drugs could be separated well based on their mechanisms of drug action, which may provide a theoretical basis and a novel insight for developing a model to predict a possible mechanism of action of an antitumor compound in the future.
Figure 6.
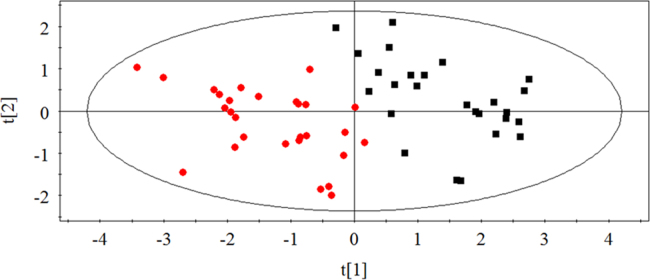
PCA score plot of cells in antimetabolite agents and agents directly acting on DNA ( Agents directly acting on DNA;
Agents directly acting on DNA;  antimetabolic agents). Two groups could be separated from each other well.
antimetabolic agents). Two groups could be separated from each other well.
4. Conclusions
In conclusion, an ion-pair RP-HPLC method was developed to analyze 12 nucleotides in mammalian tumor cells simultaneously for the targeted metabolomics analysis of antibiotic antitumor drugs (DNR, EPI and ACD), in order to find potential biomarkers. Following HPLC and data analysis, ATP and UTP were verified by ROC curve analysis as the most appropriate biomarkers for this class. Moreover, four categories of drugs with different mechanisms of action were analyzed by PCA to develop a discriminatory model. Combined with IC50 values, points away from their groups were found to have the very largest IC50 values (the lowest sensitivity).
Metabolomics can elucidate the metabolic characteristics of nucleotides in tumor cells treated with antitumor drugs, which could be used to study the correlation between metabolites and mechanisms of drug action. A more global, powerful and metabolomics-based approach to study the correlation between changes in metabolites and mechanisms of drug action may become an innovative method to predict the mechanism of an antitumor compound. Hence, the following approaches will be pursued. Firstly, the database should be extended: only 12 nucleotides and 12 chemotherapeutic drugs were analyzed, so more kinds of metabolites and antitumor drugs should be detected to get a more global metabolic characteristic and develop a more effective model. Secondly, more advanced metabolomics assay platforms should be used, such as MS and NMR, to increase the sensitivity and accuracy of the predictive model. Meanwhile, the different assay platforms used together can validate and complement each other, which can provide more information on the connection between metabolites and antitumor drugs. Thirdly, mechanisms of drug action should be further studied. Finding the potential general characteristics and differences may make the model more complete, as well as increase the ability to predict the mechanism of an antitumor-compound action. Lastly, more chemotherapeutic drugs of known mechanism should be used to validate our model.
To the best of our knowledge, this is the first development of a discriminatory model of mechanisms of antitumor-drug action based on metabolomics. However, to develop a more powerful model further work is needed. The method based on metabolomics, in this paper, may provide the theoretical basis of developing a recognition model to predict a possible mechanism of an antitumor-compound action, and also offer a novel approach to explore the connection between metabolites in cells and mechanisms of drug action.
Acknowledgments
This work was supported financially by the Natural Science Foundation of Liaoning Province, China (No. 201102210), and the Program for Liaoning Innovative Research Team in University (No. LH2012018).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.apsb.2015.03.010.
Appendix A. Supplementary materials
Supplementary data
References
- 1.Grune T., Siems W.G., Gerber G., Uhlig R. Determination of the ultraviolet absorbance and radioactivity of purine compounds separated by high-performance liquid chromatography. Application to metabolic flux rate analysis. J Chromatogr. 1991;553:193–199. doi: 10.1016/s0021-9673(01)88488-0. [DOI] [PubMed] [Google Scholar]
- 2.Frickenschmidt A., Fröhlich H., Bullinger D., Zell A., Laufer S., Gleiter C.H. Metabonomics in cancer diagnosis: mass spectrometry-based profiling of urinary nucleosides from breast cancer patients. Biomarkers. 2008;13:435–449. doi: 10.1080/13547500802012858. [DOI] [PubMed] [Google Scholar]
- 3.Woo H.M., Kim K.M., Choi M.H., Jung B.H., Lee J., Kong G. Mass spectrometry based metabolomic approaches in urinary biomarker study of women׳s cancers. Clin Chim Acta. 2009;400:63–69. doi: 10.1016/j.cca.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Qiu Y.P., Cai G.X., Su M.M., Chen T.L., Zheng X.J., Xu Y. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res. 2009;8:4844–4850. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]
- 5.Hirayama A., Kami K., Sugimoto M., Sugawara M., Toki N., Onozuka H. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 6.Sigoillot F.D., Sigoillot S.M., Guy H.I. Breakdown of the regulatory control of pyrimidine biosynthesis in human breast cancer cells. Int J Cancer. 2004;109:491–498. doi: 10.1002/ijc.11717. [DOI] [PubMed] [Google Scholar]
- 7.Madsen R., Lundstedt T., Trygg J. Chemometrics in metabolomics – a review in human disease diagnosis. Anal Chim Acta. 2010;659:23–33. doi: 10.1016/j.aca.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 8.Kell D.B. Systems biology, metabolic modelling and metabolomics in drug discovery and development. Drug Discov Today. 2006;11:1085–1092. doi: 10.1016/j.drudis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Hall R.D. Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol. 2006;169:453–468. doi: 10.1111/j.1469-8137.2005.01632.x. [DOI] [PubMed] [Google Scholar]
- 10.Gibney M.J., Walsh M., Brennan L., Roche H.M., German B., van Ommen B. Metabolomics in human nutrition: opportunities and challenges. Am J Clin Nutr. 2005;82:497–503. doi: 10.1093/ajcn.82.3.497. [DOI] [PubMed] [Google Scholar]
- 11.van der Werf M.J., Overkamp K.M., Muilwijk B., Coulier L., Hankemeier T. Microbial metabolomics: toward a platform with full metabolome. Anal Biochem. 2007;370:17–25. doi: 10.1016/j.ab.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Duarte I.F., Lamego I., Marques J., Merques M.P., Blaise B.J., Gil A.M. Nuclear magnetic resonance (NMR) study of the effect of cisplatin on the metabolic profile of MG-63 osteosarcoma cells. J Proteome Res. 2010;9:5877–5886. doi: 10.1021/pr100635n. [DOI] [PubMed] [Google Scholar]
- 13.Nyaga S.G., Jaruga P., Lohani A., Dizdaroglu M., Evans M.K. Accumulation of oxidatively induced DNA damage in human breast cancer cell lines following treatment with hydrogen peroxide. Cell Cycle. 2007;6:1472–1478. [PubMed] [Google Scholar]
- 14.Clayton T.A., Lindon J.C., Cloarec O., Antti H., Charuel C., Hanton G. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W., Tan S., Paintsil E., Dutschman G.E., Gullen E.A., Chu E. Analysis of deoxyribonucleotide pools in human cancer cell lines using a liquid chromatography coupled with tandem mass spectrometry technique. Biochem Pharmacol. 2011;82:411–417. doi: 10.1016/j.bcp.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokoloski J.A., Beardsley P.G., Sartorelli A.C. Induction of HL-60 leukemia cell differentiation by tetrahydrofolate inhibitors of de novo purine nucleotide biosynthesis. Cancer Chemother Pharmacol. 1991;28:39–44. doi: 10.1007/BF00684954. [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki H., Huang P., Keating M.J., Plunkett W. Differential incorporation of ara-C, gemcitabine, and fludarabine into replicating and repairing DNA in proliferating human leukemia cells. Blood. 1997;90:270–278. [PubMed] [Google Scholar]
- 18.van Moorsel C.J.A., Bergman A.M., Veerman G., Voorn D.A., Ruiz V.W.T., Haperen V. Differential effects of gemcitabine on ribonucleotide pools of twenty-one solid tumour and leukaemia cell lines. Biochim Biophys Acta. 2000;1474:5–12. doi: 10.1016/s0304-4165(99)00209-3. [DOI] [PubMed] [Google Scholar]
- 19.Cohen S., Megherbi M., Jordheim L.P., Lefebvre I., Perigaud C., Dumontet C. Simultaneous analysis of eight nucleoside triphosphates in cell lines by liquid chromatography coupled with tandem mass spectrometry. J Chromatogr B. 2009;877:3831–3840. doi: 10.1016/j.jchromb.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C.C., Liu Z., Liu X., Wei L., Liu Y.J., Yu J. Targeted metabolic analysis of nucleotides and identification of biomarkers associated with cancer in cultured cell models. Acta Pharm Sin B. 2013;3:254–262. [Google Scholar]
- 21.Liu X., Zhang C.C., Liu Z., Wei L., Liu Y.J., Yu J. LC-based targeted metabolomics analysis of nucleotides and identification of biomarkers associated with chemotherapeutic drugs in cultured cell models. Anticancer Drugs. 2014;25:690–703. doi: 10.1097/CAD.0000000000000096. [DOI] [PubMed] [Google Scholar]
- 22.Mansat-de M.V., Bezombes C., Quillet-Mary A., Bettaïeb A., D’orgeix A.D., Laurent G. Implication of radical oxygen species in ceramide generation, c-Jun N-terminal kinase activation and apoptosis induced by daunorubicin. Mol Pharmacol. 1999;56:867–874. doi: 10.1124/mol.56.5.867. [DOI] [PubMed] [Google Scholar]
- 23.Kebieche M., Lakroun Z., Lahouel M., Bouayed J., Meraihi Z., Soulimani R. Evaluation of epirubicin-induced acute oxidative stress toxicity in rat liver cells and mitochondria, and the prevention of toxicity through quercetin administration. Exp Toxicol Pathol. 2009;61:161–167. doi: 10.1016/j.etp.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Bock J., Szabó I., Jekle A., Gulbins E. Actinomycin D-induced apoptosis involves the potassium channel Kv1.3. Biochem Biophys Res Commun. 2002;295:526–531. doi: 10.1016/s0006-291x(02)00695-2. [DOI] [PubMed] [Google Scholar]
- 25.Masquelier M., Zhou Q.F., Gruber A., Vitols S. Relationship between daunorubicin concentration and apoptosis induction in leukemic cells. Biochem Pharmacol. 2004;67:1047–1056. doi: 10.1016/j.bcp.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 26.Lo Y.L., Ho C.T., Tsai F.L. Inhibit multidrug resistance and induce apoptosis by using glycocholic acid and epirubicin. Eur J Pharm Sci. 2008;35:52–67. doi: 10.1016/j.ejps.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Kleeff J., Kornmann M., Sawhney H., Korc M. Actinomycin D induces apoptosis and inhibits growth of pancreatic cancer cells. Int J Cancer. 2000;86:399–407. doi: 10.1002/(sici)1097-0215(20000501)86:3<399::aid-ijc15>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 28.Shepelevtseva N.G. Effect of the anthracycline antineoplastic antibiotics, rubomycin and carminomycin, on the glycogen and nucleic acid content in the myocardium of white mice. Antibiotiki. 1977;22:553–557. [PubMed] [Google Scholar]
- 29.Zhang H., Gao Y.G., van der Marel G.A., van Boom J.H., Wang A.H. Simultaneous incorporations of two anticancer drugs into DNA. The structures of formaldehyde-cross-linked adducts of daunorubicin-d(CG(araC)GCG) and doxorubicin-d(CA(araC)GTG) complexes at high resolution. J Biol Chem. 1993;268:10095–10101. [PubMed] [Google Scholar]
- 30.Meriwether W.D., Bachur N.R. Inhibition of DNA and RNA metabolism by daunorubicin and Adriamycin in L1210 mouse leukemia. Cancer Res. 1972;32:1137–1142. [PubMed] [Google Scholar]
- 31.Zunina F., Gambetta R., Di Marco A. The inhibition in vitro of DNA polymerase and RNA polymerases by daunomycin and adriamycin. Biochem Pharmacol. 1975;24:309–311. doi: 10.1016/0006-2952(75)90300-7. [DOI] [PubMed] [Google Scholar]
- 32.Bachur N.R., Yu F., Johnson R., Hickey R., Wu Y., Malkas L. Helicase inhibition by anthracycline anticancer agents. Mol Pharmacol. 1992;41:993–998. [PubMed] [Google Scholar]
- 33.Bachur N.R., Gordon S.L., Gee M.V., Kon H. NADPH cytochrome P-450 reductase activation of quinone anticancer agents to free radicals. Proc Natl Acad Sci USA. 1979;76:954–957. doi: 10.1073/pnas.76.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidal R.F., Eksborg S., Sundberg M., Carlberg M., Elfsson B., Andersson B.S. Doxorubicin- and daunorubicin-induced energy deprivation and nucleotide degradation in isolated cardiomyocytes. Toxicology. 1996;114:1–10. doi: 10.1016/s0300-483x(96)03410-5. [DOI] [PubMed] [Google Scholar]
- 35.Benderra Z., Trussardi A., Morjani H., Villa A.M., Doglia S.M., Manfait M. Regulation of cellular glutathione modulates nuclear accumulation of daunorubicin in human MCF7 cells overexpressing multidrug resistance associated protein. Eur J Cancer. 2000;36:428–434. doi: 10.1016/s0959-8049(99)00288-9. [DOI] [PubMed] [Google Scholar]
- 36.Cao X., Fang L., Gibbs S., Huang Y., Dai Z., Wen P. Glucose uptake inhibitor sensitizes cancer cells to daunorubicin and overcomes drug resistance in hypoxia. Cancer Chemother Pharmacol. 2007;59:495–505. doi: 10.1007/s00280-006-0291-9. [DOI] [PubMed] [Google Scholar]
- 37.Zhong L.L., Shen H.L., Huang C.F., Jing H.W., Cao D.L. AKR1B10 induces cell resistance to daunorubicin and idarubicin by reducing C13 ketonic group. Toxicol Appl Pharmacol. 2011;255:40–47. doi: 10.1016/j.taap.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charak S., Jangir D.K., Tyagi G., Mehrotra R. Interaction studies of Epirubicin with DNA using spectroscopic techniques. J Mol Struct. 2011;1000:150–154. [Google Scholar]
- 39.Coukell A.J., Faulds D. Epirubicin. Drugs. 1997;53:453–582. doi: 10.2165/00003495-199753030-00008. [DOI] [PubMed] [Google Scholar]
- 40.Sun W.L., Chen J., Wang Y.P., Zheng H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7:1035–1044. doi: 10.4161/auto.7.9.16521. [DOI] [PubMed] [Google Scholar]
- 41.Cantoni O., Sestili P., Cattabeni F., Geroni C., Giuliani F. Comparative effects of doxorubicin and 4′-epi-doxorubicin on nucleic acid metabolism and cytotoxicity in a human tumor cell line. Cancer Chemother Pharmacol. 1990;27:47–51. doi: 10.1007/BF00689275. [DOI] [PubMed] [Google Scholar]
- 42.Scott E.V., Jones R.L., Banville D.L., Zon G., Marzilli L.G., Wilson W.D. Proton and phosphorus-31 NMR investigations of actinomycin D binding selectivity with oligodeoxyribonucleotides containing multiple adjacent d(GC) sites. Biochemistry. 1988;27:915–923. doi: 10.1021/bi00403a012. [DOI] [PubMed] [Google Scholar]
- 43.Atkinson D.E. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry. 1968;7:4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- 44.Longley D.B., Harkin D.P., Johnston P.G. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 45.Galmarini C.M., Mackey J.R., Dumontet C. Nucleoside analogues and nucleobases in cancer treatment. Lancet Oncol. 2002;3:415–424. doi: 10.1016/s1470-2045(02)00788-x. [DOI] [PubMed] [Google Scholar]
- 46.Kremer J.M. Toward a better understanding of methotrexate. Arthritis Rheum. 2004;50:1370–1382. doi: 10.1002/art.20278. [DOI] [PubMed] [Google Scholar]
- 47.Tomasz M., Mitomycin C. small, fast and deadly (but very selective) Chem Biol. 1995;2:575–579. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 48.Fuertes M.A., Castilla J., Alonso C., Pérez J.M. Cisplatin biochemical mechanism of action: from cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Curr Med Chem. 2003;10:257–266. doi: 10.2174/0929867033368484. [DOI] [PubMed] [Google Scholar]
- 49.Hande K.R. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur J Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data