Abstract
Background
Extended spectrum β-lactamases (ESBLs), a group of enzymes conferring resistance to third generation cephalosporins have rapidly increased in Enterobacteriacae and pose a major challenge to human health care. Resistant isolates are common in domestic animals and clinical settings, but prevalence and genotype distribution varies on a geographical scale. Although ESBL genes are frequently detected in bacteria isolated from wildlife samples, ESBL dissemination of resistant bacteria to the environment is largely unknown. To address this, we used three closely related gull species as a model system and collected more than 3000 faecal samples during breeding times in nine European countries. Samples were screened for ESBL-producing bacteria, which were characterized to the level of ESBL genotype groups (SHV, TEM), or specific genotypes (CTX-M).
Results
ESBL-producing bacteria were frequently detected in gulls (906 of 3158 samples, 28.7 %), with significant variation in prevalence rates between countries. Highest levels were found in Spain (74.8 %), The Netherlands (37.8 %) and England (27.1 %). Denmark and Poland represented the other extreme with no, or very few positive samples. Genotyping of CTX-M isolates identified 13 different variants, with blaCTX-M-1 and blaCTX-M-14 as the most frequently detected. In samples from England, Spain and Portugal, blaCTX-M-14 dominated, while in the rest of the sampled countries blaCTX-M-1 (except Sweden where blaCTX-M-15 was dominant) was the most frequently detected genotype, a pattern similar to what is known from studies of human materials.
Conclusions
CTX-M type ESBLs are common in the faecal microbiota from gulls across Europe. The gull ESBL genotype distribution was in large similar to published datasets from human and food-production animals in Europe. The data suggests that the environmental dissemination of ESBL is high from anthropogenic sources, and widespread occurrence of resistant bacteria in common migratory bird species utilizing urban and agricultural areas suggests that antibiotic resistance genes may also be spread through birds.
Keywords: ESBL, CTX-M, Wildlife, Birds, Gulls, Antibiotic resistance, E. coli, Europe
Background
Nosocomial infections (in human clinics) with bacteria harboring extended spectrum beta-lactamase (ESBL) genes started to appear in Europe in the mid-1980s, and has since then constituted an increasing everyday challenge in European clinics [1, 2]. A shift in the prevalence and genotypes of ESBLs in Europe occurred around 2000, when the CTX-M type ESBLs became the dominating class, with much greater penetration into the Escherichia coli populations than TEM or SHV type ESBLs, and is now found in human outpatient settings [3]. Today more than 170 different CTX-M genotypes are described (Bush K, Palzkill T, Jacoby G. Lahey Clinic. http://www.lahey.org/studies/, accessed September 2015), broadly divided in five groups (CTX-M group 1, 2, 8, 9 and group 25 [4]). In humans the most common CTX-M variants are blaCTX-M-14 and blaCTX-M-15 [3]. In food production animals ESBL-producing bacteria mainly occur in poultry, but are also reported with lower prevalence rates in cattle and swine, with blaCTX-M-1, blaCTX-M-14, blaTEM-52 and blaSHV-12 as the currently most frequently reported genotypes [5]. By comparing population genetic relationships between E. coli from humans and poultry it has been shown that antibiotic-resistant E. coli isolates are more frequently related than antibiotic-susceptible isolates are [5]. Further, a coherence between poultry and human ESBLs has been shown by sequence typed E. coli comparisons [6], illustrating transmission of resistant strains between sources. Consequently, both the clonality of E. coli and specific resistance genotypes are important study topics for fully understanding how transfer of resistance may occur. This is also concluded by Lazarus et al. in a systematic review, finding that food production animals, more apparent for poultry, may constitute a source of human extraintestinal ESCR-EC (expanded-spectrum cephalosporin-resistant Escherichia coli) infections. [7].
Recently, the occurrence of ESBL-producing bacteria in the environment has started to receive more attention and it has repeatedly been shown that they are regularly found in both wildlife and the environment [8–12]. The level of ESBL-producing bacteria in the environment seems to be higher in areas with high human densities, but are also found in seemingly remote areas, including Escherichia coli of O25b-ST131 clone harboring blaCTX-M-15 in gulls sampled at the isolated Commander Islands in Russia and 37 % ESBL harboring isolates in gulls sampled in Barrow, Alaska [13, 14]. Thus, wildlife has been pointed out as a potential reservoir for resistant bacteria [9, 10], and especially species that live close to humans [15–17]. In recent years, gulls (Laridae) have become particularly studied since they have many characteristics which make them suitable for resistance dissemination studies [11, 12, 18, 19]. Several gull species have large breeding distributions, are common in man-made environments, and to a large extent feed on human and food-animal waste. High carriage rates of ESBL-producing bacteria in gulls have been reported, for instance in a study from France where 9.4 % of the sampled Yellow-legged gulls (Larus michaelis) harbored ESBL-producing bacteria [11], and two studies of gulls in Portugal where 19.3 and 32 % of the gulls, respectively, carried ESBL-producing bacteria [19, 20]. A problem when addressing resistance occurrence in wildlife bacteria is that studies have generally been small, or not utilized standardized methodologies [10]. In order to gain a broader view of the resistance situation in the environment we undertook a standardized study where >3000 fecal samples from gulls in nine European countries were screened for ESBL-producing bacteria, with primary focus on prevalence of CTX-M genotypes. Specifically, we wanted to describe the occurrence and spatial variation in ESBL prevalence and blaCTX-M genotype distribution in Europe. If the occurrence of ESBL is primarily driven by transfer of resistant genotypes from humans and food-production animals into wildlife, we hypothesize that ESBL prevalence and genotype distributions would conform to patterns seen in bacterial populations from these sources.
Methods
Studied avian species, sampling sites and sampling methodology
Fecal samples were collected from Herring gulls (Larus argentatus) and Lesser-Black backed gulls (Larus fuscus) in northern Europe, and Yellow-legged gulls (Larus michaelis) in southern Europe (Table 1; Fig. 1). Field sampling was conducted from mid-June to early July 2009 in nine European countries. From each of the 14 sampling sites, 101–323 samples with bacterial growth were used (sample viability ensured by growth on non-selective media). Samples were collected from adult and subadult birds in or around breeding colonies. By collecting the samples during the late breeding season we limited the risk of sampling non-stationary birds. The sampling areas were chosen to be as similar as possible according to human activities, but different human density, suitable sampling locations, etc. made it impossible to have completely identical sites. To avoid that multiple samples were collected from one bird, samples were collected from separated flocks, and with fewer samples than individuals present at each site.
Table 1.
Description of sampling sites and gull species
| Country | Location | Speciesa | Samples (n)b | Description |
|---|---|---|---|---|
| Denmark | Blavand | Larus fuscus (60 %) | 158 | Coastline close to Roskilde |
| Larus argentatus (40 %) | ||||
| England | Bristol | Larus argentatus | 133 | Central Bristol |
| Ireland | Howth | Larus argentatus | 266 | Suburb of Dublin close to fishing habour |
| Latvia | Daugava | Larus argentatus | 323 | Close to central Riga |
| Kaltene | Larus argentatus | 101 | Countryside 60 km SV Riga | |
| Netherlands | Borsele | Larus argentatus | 280 | Industry habour |
| Rotterdam | Larus fuscus (90 %) | 280 | Industry habour | |
| Larus argentatus (10 %) | ||||
| Poland | Wladyslawowo | Larus argentatus | 280 | Coastline close to Gdansk |
| Portugal | Lagos | Larus michaellis | 314 | Fish market in Lagos |
| Portimao | Larus michaellis | 111 | Coastline close to Portimao | |
| Spain | Emporda | Larus michaellis | 199 | Citydump outside Emporda |
| Mazarron | Larus michaellis | 321 | Breeding colonie close to Mazarron | |
| Malaga | Larus michaellis | 175 | Harbour in Malaga | |
| Sweden | Hudiksvall | Larus argentatus | 217 | Archipelago outside Hudiksvall |
aPercentage (%) represents the frequency of a gull species in the sampled flocks when there was more than one species present in the flock
bNumber of viable samples from each sampling site
Fig. 1.
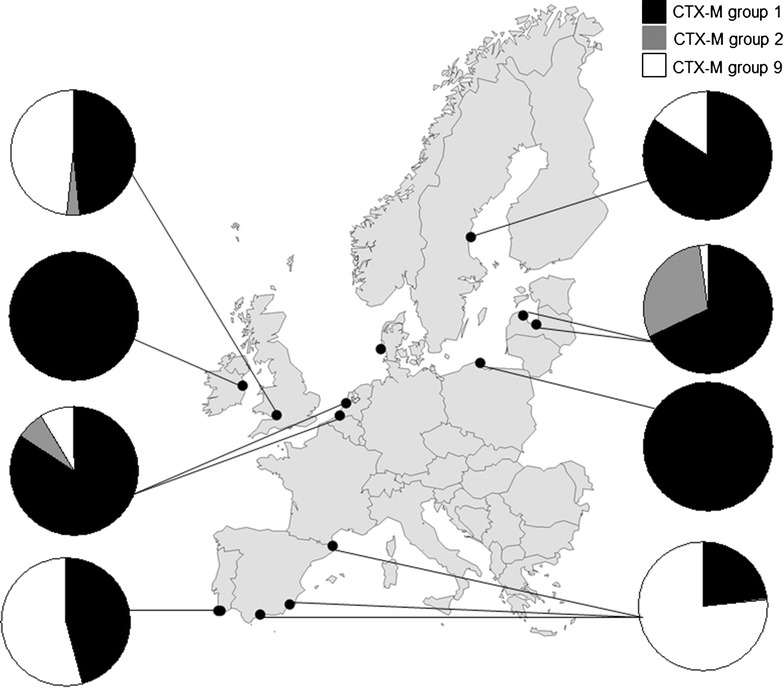
Distribution of CTX-M groups in E. coli from gulls in the nine European countries. In Spain (Emporda) one sample included a CTX-M group 8 (bla CTX-M-8) harboring isolate
Sterile cotton wool swabs were swirled in freshly deposited fecal material on the ground where gulls had recently been roosting. Swabs were put in tubes containing bacterial freeze medium [Luria broth; BD, Sparks, USA, phosphate buffered saline containing 0.45 % Na-citrate, 0.1 % MgSO4, 1 % (NH4)2SO4, and 4.4 % glycerol] and immediately frozen in liquid nitrogen until arrival at the laboratory in Sweden where they were subsequently stored in −80 °C.
Isolation of ESBL-producing bacteria
In total, 3158 fecal samples with bacterial growth (101–323 samples/sampling site) were available for analysis (Table 1). All samples were enriched in brain heart infusion broth (Becton–Dickinson, Franklin Lakes, NJ, USA), supplemented with 16 mg/L vancomycin, for 18–24 h in 37 °C, and subsequently inoculated on a ChromID™ ESBL plates (bioMérieux, Solna, Sweden), according to the manufacturer’s instructions. Presumptive ESBL producing colonies were isolated and species identity was confirmed by biochemical testing (standard fermentation analysis such as lactose ferm., ONPG, Voges-Proskauer, spot indole etc.) and MALDI/TOF (Bruker Daltonik GmbH, Bremen Germany). ESBL plates with bacterial growth of more than one bacterial species each presumable species was isolated. ESBL-production was confirmed with the cefpodoxime/cefpodoxime + clavulanic acid double-disc test (MAST Diagnostics, Bootle, UK). Samples were regarded as ESBL producing and further analyzed when zone diameter around cefpodoxime was ≥5 mm than the zone diameter around cefpodoxime + clavulanic acid, according to manufacturer’s instructions (MAST Diagnostics, Bootle, UK).
Genetic determination of ESBL variants
The presence of blaCTX-M genotype was detected using a previously described multiplex real-time PCR protocol [21], allowing designation of CTX-M subgroups (CTX-M group 1, 2, 9 and 8/25) [21] using StepOnePlus™ real-time PCR system (Applied biosystems). Positive isolates were sequenced using specific primers and protocols described previously [22, 23]. Sequencing was performed by Eurofins Genomics (Ebersberg, Germany).
The presence of blaTEM and blaSHV was detected using previously described primers [24] and a SYBR Green-based real-time PCR protocol [12].
Results
Prevalence of ESBL producing bacteria
ESBL producing bacteria were detected in 906 (28.7 %) of the 3158 samples. Since 44 samples contained more than one ESBL-producing bacterial species, the total number of ESBL producing bacteria was 950 (Table 2). The vast majority of the isolated ESBL were E. coli (902 isolates, 94.9 %) followed by 42 Klebisella (41 K. pneumoniae and one K. oxytoca) and only six isolates from other bacterial genera: Acinetobacter spp 1 (blaTEM,), Citrobacter spp 2 (blaTEM+blaSHV), Enterobacter spp 2 (blaSHV) and Proteus spp 1 (blaCTX-M-2).
Table 2.
Description of ESBL harboring samples and bla CTX-M variants found in investigated countries
| CTX-M group | Denmark | England | Ireland | Latvia | Poland | Portugal | Spain | Sweden | Netherlands | |
|---|---|---|---|---|---|---|---|---|---|---|
| Samples (n) | 158 | 133 | 266 | 424 | 280 | 425 | 695 | 217 | 560 | |
| ESBL genotypes (n)a | 0 | 50 | 12 | 98 | 2 | 74 | 572 | 74 | 280 | |
| ESBL samples(%)b | (0) | (27.1) | (4.5) | (17.4) | (0.7) | (12.7) | (74.8) | (20.7) | (37.8) | |
| 1 | ||||||||||
| CTX-M-1 | 5.3c | 2.6 | 4.2 | 0.4 | 1.4 | 3.2 | 2.3 | 21.3 | ||
| CTX-M-3 | 0.8 | 0.2 | 0.1 | 0.2 | ||||||
| CTX-M-15 | 4.5 | 0.4 | 3.5 | 0.5 | 1.3 | 13.8 | 2.1 | |||
| CTX-M-32 | 0.4 | 0.5 | 0.4 | 0.7 | 2.9 | 0.7 | ||||
| CTX-M-55 | 0.2 | |||||||||
| 2 | ||||||||||
| CTX-M-2 | 1.5 | 3.3 | 2.1 | |||||||
| 8 | ||||||||||
| CTX-M-8 | 0.1 | |||||||||
| 9 | ||||||||||
| CTX-M-9 | 0.2 | 2.7 | 0.2 | |||||||
| CTX-M-14 | 8.3 | 2.4 | 16.8 | 0.9 | 1.4 | |||||
| CTX-M-14b | 1.5 | 0.2 | 1.2 | 0.9 | 0.4 | |||||
| CTX-M-24 | 1.4 | |||||||||
| CTX-M-27 | 0.8 | |||||||||
| CTX-M-65 | 0.2 | |||||||||
| TEMd | 3.0 | 0.7 | 6.6 | 26.2 | 3.6 | |||||
| SHVd | 12.0 | 1.1 | 10.1 | 5.6 | 27.8 | 14.7 | 17.9 | |||
aTotal number of ESBL genotypes (n) in each country. Note that some bacterial strains carried more than one ESBL genotype
bPercentage (%) of total number of samples carrying ESBL
cPercentage (%) of the total n the total number of ESBL in each country umber of ESBL in each country belonging to a certain genotype
dPercentage (%) of TEM and SHV of the total number of ESBL in each country. However, note that a bla TEM and bla SHV genotype present in the same bacterial strain as another ESBL genotype could be bla TEM-1, bla TEM-2, or bla SHV-1 which are not considered true ESBL genotypes, but the original genes from which later genotype variants are derived from
The levels of ESBL positive samples varied significantly between countries. In Spain 74.8 % of the total number of samples carried ESBL. Also, The Netherlands, England and Sweden had high levels of ESBL producing bacteria (37.8, 27.1 %, respectively 20.7 %), while only 0.8 % of the samples in Poland, and in Denmark no ESBL positive samples were detected at all (Table 2).
Distribution of ESBL variants
CTX-M
In total, 602 (66.4 %) of the ESBL harboring isolates carried CTX-M type ESBL. The number of detected blaCTX-M variants was 13, distributed on five CTX-M group 1 variants, one variant of CTX-M group 2, one variant of CTX-M group 8/25, and six variants of CTX-M group 9 (Table 2). Group 1 (52.9 % of the total number of isolated CTX-M) and group 9 (42.2 %) dominated, and only a few isolates were designated group 2 (4.7 %) and group 8/25 (0.2 %) (Table 2). The most common genotypes were blaCTX-M-1 and blaCTX-M-14, but there was a large variation between blaCTX-M variants between countries (Table 2). In Spain, Portugal and England blaCTX-M-14 was the most common CTX-M genotype. In The Netherlands there was a large dominance of blaCTX-M-1 (76.6 % of all CTX-M) while in Sweden blaCTX-M-15 (71.2 % of all CTX-M) was the most common. The CTX-M group 2 (blaCTX-M-2) was only found in three countries, Latvia, The Netherlands and England, and corresponded to 32.5, 7.8 and 3.4 % of the total number of CTX-M type ESBL, respectively. In Spain, one E. coli carried a blaCTX-M-8 while no sample carried genotypes from CTX-M group 25. A number of ESBL positive samples carried two different blaCTX-M genotypes and a great part of those that carried blaCTX-M were also positive for both blaTEM and/or blaSHV (data not shown).
TEM and SHV
ESBL positive isolates were screened for blaTEM and blaSHV genotypes, but not sequenced for specific genotype variants. In total 222 isolates were blaTEM positive and 372 blaSHV positive. Of these, 216 (80 %) blaTEM and 242 (65 %) blaSHV were present in the same isolate as another ESBL genotype. In those isolates it is not possible to exclude the commonly appearing variants blaTEM-1, blaTEM-2 and blaSHV-1 which are not true ESBLs. Thus, the level of true ESBLs could be lower for these variants.
Discussion
Having investigated several of the European countries, particularly in the western parts, we can report a wide difference in ESBL prevalence, starting with none or very low ESBL prevalence in Denmark (0 %), Poland (0.7 %) and Ireland (4.5 %), to intermediate in Portugal (12.7 %), Latvia (17.4 %) and Sweden (20.7 %), and increasingly higher in England (27.1 %), The Netherlands (37.8 %), and Spain (74.8 %). Compared to current human clinical or veterinary data, most countries investigated show a high level of acquired ESBL resistance. Despite that on a national level prevalence rates could vary considerably, to our knowledge, the rates of ESBL producing bacteria reported in this study are greater than any previous study of clinically relevant bacteria from a wildlife source.
Standardized European studies covering prevalence of ESBLs in E. coli among community isolates from healthy humans are limited and not always including genotype details [25]. For European coverage EARS-net has a yearly report presenting levels of bacteria with resistance to third generation cephalosporins from clinical isolates [26]. In our study all countries except Denmark and Poland had ESBL levels higher then presented for clinical E. coli isolates from Europe in the corresponding countries, although AmpC was included in the Ears-net report and also the sample populations are very different. In countries such as Sweden and Spain, the levels of ESBL positive samples were considerably higher than reported from samples of healthy humans in separate studies in corresponding country [27–29]. Denmark and Poland were the only two countries that had lower prevalence in comparison to the data presented on EARS-net. From Denmark more recent data from 2011 presented by the DANMAPS yearly report show low but increasing levels of ESBLs in clinical samples and ESBLs were found in almost 8 % of blood culture samples from hospitalized humans [30]. Also in Portugal the levels of ESBL from our gull samples were low in comparison to the neighboring country Spain and a previously performed study on gulls from Portugal where ESBL was found in 32 % of the sampled gulls [19].
Spain showed extremely high levels of ESBLs, and in Sweden which is known to have low level of ESBLs in humans [31], 20.7 % of the gulls carried ESBL. This is much higher than previously found in gull studies performed in Sweden [12, 32]. The Netherlands have in coherence with Sweden low antibiotic usage in human clinical settings, but in gulls the levels were second highest when the studied countries are compared (37.8 %) [33]. However, The Netherlands have far higher antibiotic consumption in food-production [34].
Food-production animals are suggested to be an important source in the environmental dissemination of resistant bacteria, and the total consumption of antibiotics is much higher than what is used by human medicine [35]. The most comprehensive data for antibiotic resistance levels in food-production animals are presented for poultry, pigs and cattle in a yearly report by the European Food Safety Authority (EFSA). Compared to our study ESBL levels from seven of the countries are included (Denmark, England, Ireland, Latvia, Netherlands, Spain and Sweden). The levels of third generation cephalosporins resistance from 2008 were very low (<1 %) in cattle and pigs for all included countries. In poultry the levels of resistant E. coli varied between 0 and 26 % [36]. The overall trends corresponded, with highest levels of ESBLs in E. coli found in Spain from poultry and from our sampled gulls. Further ESBL was lacking in Danish poultry as well as the gull samples. Although trends should be the same, resistance levels may differ due to the randomized sampling approach used in the EFSA material.
In our gull material significant differences in genotype distributions could be seen between countries. In Spain, Portugal and England blaCTX-M-14 was dominant, while blaCTX-M-1 and blaCTX-M-15 was most frequently detected in the other surveyed countries (Table 2). This is in coherence with the human situation, where blaCTX-M-14 followed by blaCTX-M-15 are the most common genotypes in humans while blaCTX-M-1 and blaCTX-M-14 are most frequently detected in domestic animals [36, 37]. The pattern seen in gull isolates is to large extent in coherence with the pattern of blaCTX-M genotype distribution in human isolates in Europe [3]. Livermore et al. [3] have elegantly summarized CTX-M data from humans in Europe in a review article and here blaCTX-M-14 is noted as one of the most frequent genotypes in Spain which is in coherence with results. Further, a study on E. coli isolated at Spanish hospitals showed high similarity in genotype frequency with our results (52 % of all CTX-M was blaCTX-M-9 and 39 % blaCTX-M-14) [38]. Also in Portugal and England there was a dominance of blaCTX-M-14 in isolates from gulls which is not the situation in human isolates. Separate publications from humans in UK and Portugal, and also a previous study on gulls in Portugal, found that blaCTX-M-14 was scarce in these [19, 39, 40]. This contrast could be explained by sampling site deviation from the “general picture”, or possibly dissemination from other sources as for example food-production animals. Studies show that blaCTX-M-14 genotype is frequent in Spanish and Portuguese poultry [41–43].
In England, Ireland and Sweden, included in our study, Livermore et al. [3] reports blaCTX-M-15 as the most frequently occurring genotype in isolates from humans and blaCTX-M-1 is also frequently occurring. This is in coherence with our results where blaCTX-M-1 was dominant in each country except Sweden which showed a large dominance of blaCTX-M-15. Interestingly, blaCTX-M-15, which is the dominating variant in humans in many parts of Europe, was not common in samples from gulls, except in those sampled in Sweden. This is another example of overlapping patterns between observations in wild birds and in humans, as blaCTX-M-15 is also the most common CTX-M variant in humans in Sweden [3, 44].
In contrast, The Netherlands where the human antibiotic consumption in humans are comparable to Sweden but far higher in food-production animals there was a total dominance of blaCTX-M-1 [34]. This is considered as a CTX-M genotype most often found in poultry and other food-production animals in Europe [45, 46] and the poultry production in The Netherlands is the most intense in Europe [47]. High similarities between ESBL variants in humans and poultry in the Netherlands have been seen with a dominance of blaCTX-M-1 [6].
The CTX-M group 2 is unevenly distributed in Europe, with different variants mainly spread in Russia and Eastern Europe [2]. This pattern was seen in the gull material where 32 % of the CTX-M isolated from E. coli in Latvia belonged to blaCTX-M-2. It is also noteworthy that one blaCTX-M-8 was isolated from a gull E. coli in Spain, to our knowledge the first record of blaCTX-M-8 from wildlife [9].
Conclusions
Despite low availability of easily comparable data from human and veterinary settings, levels of ESBL are seemingly higher in wild gulls in some regions of Europe. Certainly, the diet of gulls make them exposed to a variety of bacteria from different sources. Gulls are gregarious, especially during breeding, which could contribute to rapid spread of ESBL between individuals in a local population. Spread between gulls could also be mediated by the environment, as shown for Salmonella which can be maintained in a breeding colony of gulls between breeding seasons [48]. Since many gull species migrate, sometimes even between continents, there is a risk that these birds will contribute to the global spread of antibiotic resistance genes. The results from this study are therefore remarkable and the high environmental ESBL levels, as seen in gull fecal samples, alarming.
Authors’ contributions
JS, MD, JW, BMcM, BO and JB planned and designed the project. JS, JH and BMcM performed the fieldwork. JS, MD, CT and JH did all the culturing of bacteria and sequencing. JS wrote the initial draft of the manuscript and edited it together with MD. All authors read and approved the final manuscript.
Acknowledgements
We would like to extend our thanks to all people that made it possible to sample large parts of Europe in this study: A. Eriksson, B. Hasan, G. Norevik, A. Steen, M. Stervander and P. Österman. We also got invaluable help from local ornithologists in the different countries: M. Axbrink, R. J. Buijs, P. Feliu, P. Andersen Harild, S. Garcia, P. Rock, D. Rodrigues, A. Cama Torell and S. Wates. Financial support was received from the Swedish Research Council FORMAS (2008-326), The Karin Korsner’s Foundation and The Olle Engkvist Byggmästare Foundation.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Johan Stedt, Email: johan.stedt@lnu.se.
Jonas Bonnedahl, Email: jonas.bonnedahl@ltkalmar.se.
Jorge Hernandez, Email: Jorge.hernandez@lnu.se.
Jonas Waldenström, Email: jonas.waldenstrom@lnu.se.
Barry J. McMahon, Email: barry.mcmahon@ucd.ie
Conny Tolf, Email: conny.tolf@lnu.se.
Björn Olsen, Email: bjorn.olsen@medsci.uu.se.
Mirva Drobni, Email: mirva.drobni@regionjh.se.
References
- 1.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canton R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, Coque TM. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14(Suppl 1):144–153. doi: 10.1111/j.1469-0691.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- 3.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.EFSA Panel on Biological Hazards (BIOHAZ). Scientific opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J 2011;9:2322. doi:10.2903/j.efsa.2011.2322. http://www.efsa.europa.eu/efsajournal.
- 6.Leverstein-van Hall MA, Dierikx CM, Stuart JC, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011;17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 7.Lazarus B, Paterson D, Mollinger J, Rogers B. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis. 2015;60(3):439–452. doi: 10.1093/cid/ciu785. [DOI] [PubMed] [Google Scholar]
- 8.Kummerer K. Resistance in the environment. J Antimicrob Chemother. 2004;54:311–320. doi: 10.1093/jac/dkh325. [DOI] [PubMed] [Google Scholar]
- 9.Guenther S, Ewers C, Wieler LH. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. 2011;2:246. doi: 10.3389/fmicb.2011.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 11.Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus A, Kahlmeter G, Waldenstrom J, Johansson A, Olsen B. Dissemination of Escherichia coli with CTX-M type ESBL between humans and Yellow-legged gulls in the south of France. PLoS One. 2009;4:Article No.:e5958. [DOI] [PMC free article] [PubMed]
- 12.Bonnedahl J, Drobni P, Johansson A, Hernandez J, Melhus A, Stedt J, Olsen B, Drobni M. Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J Antimicrob Chemother. 2010;65:1939–1944. doi: 10.1093/jac/dkq222. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez J, Bonnedahl J, Eliasson I, Wallensten A, Comstedt P, Johansson A, Granholm S, Melhus A, Olsen B, Drobni M. Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring bla(CTX-M-15), found in Glaucous-winged gull at remote Commander Islands, Russia. Environ Microbiol Rep. 2010;2:329–332. doi: 10.1111/j.1758-2229.2010.00142.x. [DOI] [PubMed] [Google Scholar]
- 14.Bonnedahl J, Hernandez J, Stedt J, Waldenström J, Olsen B, Drobni M. Extended-spectrum β-lactamases in Escherichia coli and Klebsiella pneumoniae in gulls, Alaska, USA. Environ Microbiol Rep. 2014;20(5):897–899. doi: 10.3201/eid2005.130325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilliver MA, Bennett M, Begon M, Hazel SM, Hart CA. Enterobacteria—antibiotic resistance found in wild rodents. Nature. 1999;401:233–234. doi: 10.1038/45724. [DOI] [PubMed] [Google Scholar]
- 16.Souza V, Rocha M, Valera A, Eguiarte LE. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl Environ Microbiol. 1999;65:3373–3385. doi: 10.1128/aem.65.8.3373-3385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skurnik D, Ruimy R, Andremont A, Amorin C, Rouquet P, Picard B, Denamur E. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J Antimicrob Chemother. 2006;57:1215–1219. doi: 10.1093/jac/dkl122. [DOI] [PubMed] [Google Scholar]
- 18.Dolejska M, Cizek A, Literak I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from Black-headed Gulls in the Czech Republic. J Appl Microbiol. 2007;103:11–19. doi: 10.1111/j.1365-2672.2006.03241.x. [DOI] [PubMed] [Google Scholar]
- 19.Simoes RR, Poirel L, Da Costa PM, Nordmann P. Seagulls and Beaches as reservoirs for multidrug-resistant Escherichia coli. Emerg Infect Dis. 2010;16:110–112. doi: 10.3201/eid1601.090896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poeta P, Radhouani H, Igrejas G, Goncalves A, Carvalho C, Rodrigues J, Vinue L, Somalo S, Torres C. Seagulls of the Berlengas Natural Reserve of Portugal as carriers of fecal Escherichia coli harboring CTX-M and TEM extended-spectrum beta-lactamases. Appl Environ Microbiol. 2008;74:7439–7441. doi: 10.1128/AEM.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birkett CI, Ludlam HA, Woodford N, Brown DFJ, Brown NM, Roberts MTM, Milner N, Curran MD. Real-time TaqMan PCR for rapid detection and typing of genes encoding CTX-M extended-spectrum beta-lactamases. J Med Microbiol. 2007;56:52–55. doi: 10.1099/jmm.0.46909-0. [DOI] [PubMed] [Google Scholar]
- 22.Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47:3724–3732. doi: 10.1128/AAC.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother. 2006;57:14–23. doi: 10.1093/jac/dki398. [DOI] [PubMed] [Google Scholar]
- 24.Pitout JDD, Thomson KS, Hanson ND, Ehrhardt AF, Moland ES, Sanders CC. beta-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob Agents Chemother. 1998;42:1350–1354. doi: 10.1128/aac.42.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coque TM. Baquero F. Euro Surveill Canton R: Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe; 2008. p. 13. [PubMed] [Google Scholar]
- 26.European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2009. Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). Stockholm. 2010. doi 10.2900/35994.
- 27.Valverde A, Grill F, Coque TM, Pintado V, Baquero F, Canton R, Cobo J. High rate of intestinal colonization with extended-spectrum-beta-lactamase-producing organisms in household contacts of infected community patients. J Clin Microbiol. 2008;46:2796–2799. doi: 10.1128/JCM.01008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tangden T, Cars O, Melhus A, Lowdin E. Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum beta-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother. 2010;54:3564–3568. doi: 10.1128/AAC.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vinue L, Saenz Y, Martinez S, Somalo S, Moreno MA, Torres C, Zarazaga M. Prevalence and diversity of extended-spectrum beta-lactamases in faecal Escherichia coli isolates from healthy humans in Spain. Clin Microbiol Infect. 2009;15:954–957. doi: 10.1111/j.1469-0691.2009.02803.x. [DOI] [PubMed] [Google Scholar]
- 30.DANMAP. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. 2011. ISSN 1600-2032. http://www.danmap.org.
- 31.Tangden T, Cars O, Melhus A, Lowdin E. Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum beta-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother. 2010;54:3564–3568. doi: 10.1128/AAC.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallensten A, Hernandez J, Ardiles K, Gonzalez-Acuna D, Drobni M, Olsen B. Extended spectrum beta-lactamases detected in Escherichia coli from gulls in Stockholm, Sweden. Infect Ecol Epidemiol. 2011. doi:10.3402/iee.v1i0.7030. [DOI] [PMC free article] [PubMed]
- 33.Cars O, Molstad S, Melander A. Variation in antibiotic use in the European Union. Lancet. 2001;357:1851–1853. doi: 10.1016/S0140-6736(00)04972-2. [DOI] [PubMed] [Google Scholar]
- 34.Grave K, Torren-Edo J, Mackay D. Comparison of the sales of veterinary antibacterial agents between 10 European countries. J Antimicrob Chemother. 2010;65:2037–2040. doi: 10.1093/jac/dkq247. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. Antimicrobial resistance—global report on surveilllance. WHO Press. 2014. Available on the WHO website http://www.who.int/.
- 36.European Food Safety Authority. The community summary report on antimicrobial resistance in zoonotic and indicator bacteria from animals and food in the European Union in 2008. EFSA J. 2010;8:1658. doi:10.2903/j.efsa.2010.1658; Available online: http://www.efsa.europa.eu/efsajournal.
- 37.Bush K. Proliferation and significance of clinically relevant beta-lactamases. Antimicrob Ther. 2013;1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez JR, Martinez-Martinez L, Canton R, Coque TM, Pascual A. Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum beta-lactamases in Spain. Antimicrob Agents Chemother. 2005;49:2122–2125. doi: 10.1128/AAC.49.5.2122-2125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, Johnson AP, Pike R, Warner M, Cheasty T, et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J Antimicrob Chemother. 2004;54:735–743. doi: 10.1093/jac/dkh424. [DOI] [PubMed] [Google Scholar]
- 40.Mendonca N, Leitao J, Manageiro V, Ferreira E, Canica M. Spread of extended-spectrum beta-lactamase CTX-M-producing Escherichia coli clinical isolates in community and nosocomial environments in Portugal. Antimicrob Agents Chemother. 2007;51:1946–1955. doi: 10.1128/AAC.01412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa D, Vinue L, Poeta P, Coelho AC, Matos M, Saenz Y, Somalo S, Zarazaga M, Rodrigues J, Torres C. Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli isolates in faecal samples of broilers. Vet Microbiol. 2009;138:339–344. doi: 10.1016/j.vetmic.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 42.Brinas L, Moreno MA, Zarazaga M, Porrero C, Saenz Y, Garcia M, Dominguez L, Torres C. Detection of CMY-2, CTX-M-14, and SHV-12 beta-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob Agents Chemother. 2003;47:2056–2058. doi: 10.1128/AAC.47.6.2056-2058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanc V, Mesa R, Saco M, Lavilla S, Prats G, Miro E, Navarro F, Cortes P, Llagostera M. ESBL- and plasmidic class C beta-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet Microbiol. 2006;118:299–304. doi: 10.1016/j.vetmic.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Helldal L, Karami N, Floren K, Welinder-Olsson C, Moore ER, Ahren C. Shift of CTX-M genotypes has determined the increased prevalence of extended-spectrum beta-lactamase-producing Escherichia coli in south-western Sweden. Clin Microbiol Infect. 2013;19:E87–E90. doi: 10.1111/1469-0691.12086. [DOI] [PubMed] [Google Scholar]
- 45.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, et al. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis. 2011;17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Girlich D, Poirel L, Carattoli A, Kempf I, Lartigue MF, Bertini A, Nordmann P. Extended-spectrum beta-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl Environ Microbiol. 2007;73:4681–4685. doi: 10.1128/AEM.02491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Global Livestock Production and Health Atlas. http://kids.fao.org/glipha.
- 48.Literak Ivan AC, Smola J. Survival of salmonellas in a colony of common Black-headed gulls Larus ridibundus between two nesting periods. Colon Waterbirds. 1996;19:268–9.


