Abstract
Background:
Genomic medicine has the potential to improve care by tailoring treatments to the individual. There is consensus in the literature that pharmacogenomics (PGx) may be an ideal starting point for real-world implementation, due to the presence of well-characterized drug-gene interactions. Clinical Decision Support (CDS) is an ideal avenue by which to implement PGx at the bedside. Previous literature has established theoretical models for PGx CDS implementation and discussed a number of anticipated real-world challenges. However, work detailing actual PGx CDS implementation experiences has been limited. Anticipated challenges include data storage and management, system integration, physician acceptance, and more.
Methods:
In this study, we analyzed the experiences of ten members of the Electronic Medical Records and Genomics (eMERGE) Network, and one affiliate, in their attempts to implement PGx CDS. We examined the resulting PGx CDS system characteristics and conducted a survey to understand the unanticipated implementation challenges sites encountered.
Results:
Ten sites have successfully implemented at least one PGx CDS rule in the clinical setting. The majority of sites elected to create an Omic Ancillary System (OAS) to manage genetic and genomic data. All sites were able to adapt their existing CDS tools for PGx knowledge. The most common and impactful delays were not PGx-specific issues. Instead, they were general IT implementation problems, with top challenges including team coordination/communication and staffing. The challenges encountered caused a median total delay in system go-live of approximately two months.
Conclusions:
These results suggest that barriers to PGx CDS implementations are generally surmountable. Moreover, PGx CDS implementation may not be any more difficult than other healthcare IT projects of similar scope, as the most significant delays encountered were not unique to genomic medicine. These are encouraging results for any institution considering implementing a PGx CDS tool, and for the advancement of genomic medicine.
Keywords: Clinical decision support, genomic medicine, personalized health care, pharmacogenomics, precision medicine
INTRODUCTION
There is a substantial body of literature highlighting the potential benefits of genomic medicine[1,2,3,4] and a general acceptance that clinical decision support (CDS) tools will play a significant role in its application.[2,5,6,7] Pharmacogenomics (PGx) is one area where decision support tools may be particularly effective, due to the existence of well-characterized drug-gene interactions.[2,6,8,9] Although many reports have discussed the use of general decision support tools in healthcare,[10,11,12,13] literature detailing the real-world use of genome-driven decision support for drug prescribing has been limited.
The Electronic Medical Records and Genomics (eMERGE) Network is a National Consortium funded by the National Human Genome Research Institute to link genomic data to electronic health records (EHR).[14] Since its initial phase starting in 2007, member sites have investigated numerous uses and challenges for such systems, including genome- and phenome-wide association studies,[15] return of results,[16] and ethical concerns.[17,18] In its second phase, eMERGE has focused on integrating genetic information into clinical care.[19] This includes a clinical PGx program that is sequencing 84 pharmacogenes in about 9000 individuals.[20] The network's EHR Integration (EHRI) workgroup has subsequently worked to bring such data to the clinical setting through pilot implementations of PGx CDS systems.
Previous publications from eMERGE have discussed the need for systems to manage the large amounts of omic data (genomic, proteomic, etc.,) that can now be collected, as well as the need to extract clinically relevant meaning from that data. We proposed an idealized model for an omic ancillary system (OAS) to manage, analyze, and integrate genomic data into the EHR,[5] which is a model several organizations have subsequently adopted.[21] We also discussed a conceptual model for filtering large amounts of genomic data into actionable results – with a key component being EHR integration (see elsewhere in this issue).
Our idealized model for integrating genomic data into clinical care identified five different paths through which genomic information could enter an EHR.[5] Two paths do not require an OAS and are commonly used today – human-readable text reports and simple structured data received directly from Clinical Laboratory Improvement Amendments-certified laboratories. The other three paths rely on an OAS and are less common today – human interpretation of actionable genomic information calculated in the OAS, direct import of actionable genomic information calculated in the OAS, and automated queries of CDS systems embedded in the OAS.
With these concepts in mind, member and affiliate sites of the eMERGE-EHRI workgroup have implemented systems and processes to incorporate genetic information into their EHRs and provide PGx CDS capabilities to prescribing clinicians. Participating sites are focusing their PGx CDS efforts on well-characterized, actionable variants[21] affecting primarily clopidogrel, warfarin, or simvastatin prescribing.[19] Data sources range from single-gene tests to multi-gene panels, to whole genome sequencing.
To inform future PGx CDS implementation efforts, we sought to understand and report the experiences of the eMERGE sites. We first examined the implementation characteristics of the participating sites to determine how they varied from the idealized model. We then identified challenges encountered in the implementation process. This approach allowed us to examine variability in real-world PGx CDS implementations and to identify valuable lessons learned from those implementations. We first report the various approaches sites took toward system infrastructure and the resulting heterogeneous implementation characteristics. We then describe lessons learned, which are based on a survey of challenges that each site encountered when first implementing PGx CDS.
METHODS
The study population included ten eMERGE-EHRI workgroup member sites and one affiliate site, which are large, research-oriented academic centers or health systems. These organizations have extensive technology infrastructures already in place, including a variety of EHR systems. Due to their participation in the eMERGE Network, all sites have prior interest and expertise in working with genetic and genomic data.
To evaluate the PGx CDS implementation characteristics, we collected free-text narratives from designated representatives at each organization in the workgroup, describing their respective setups. We performed a manual review to map those descriptions to the various features of the idealized model. The results of the mapping process were then circulated to the site representatives via E-mail for validation.
To evaluate challenges faced, we conducted a formal survey of the same organizations. The survey was conducted online via the REDCap platform.[22] It collected information about the challenges each site encountered and the lengths of the delays associated with those challenges. To ensure the final survey included options that fully covered the experiences of each site, we conducted an initial survey of the workgroup to solicit a list of delays the sites had faced. Each site, via a free-text online form, submitted any delay-inducing challenges they recalled encountering. We then reviewed, categorized, consolidated, and re-phrased the responses to be generalizable to the entire group. The edited responses were comprehensive and represented all of the initial responses. We then developed a draft survey and presented this to workgroup members for face validity, looking for accuracy, completeness, and clarity. Minor changes were made to the survey based on this feedback before it was finalized and distributed. Each site identified a single respondent that was directly involved in that site's implementation effort, resulting in eleven submissions. Any incomplete or ambiguous poll responses were clarified with the respondent via E-mail correspondence.
The final survey included seven categories of challenges, encompassing 26 different types of delays. Those categories were: Insufficient staffing, implementation team coordination and communication, institutional priorities, institutional approval, third party vendor software issues, difficulty translating knowledge to software alerts, and other [Table 1]. Each delay was presented in the poll with seven options to indicate the severity of delay that they caused, ranging from “none” (indicating no delay was encountered), through “unable to implement.”
Table 1.
Categorized delay reasons
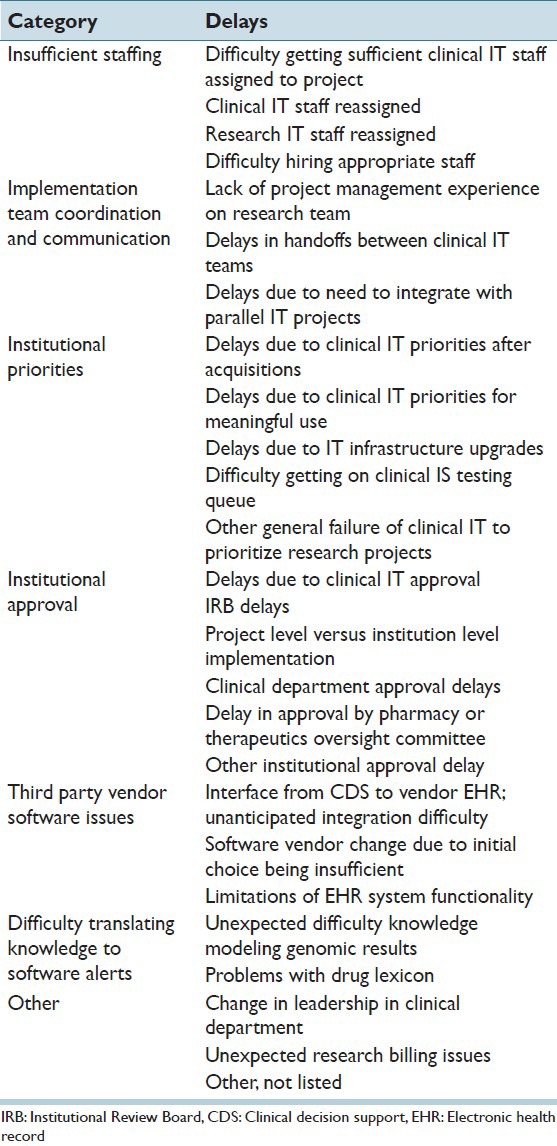
For analysis, individual delay reasons were categorized as low or high frequency and low or high impact. We determined the threshold between low and high frequency according to how often each delay reason was encountered. Delay reasons encountered less than the mean frequency were classified as low frequency and those at least the mean were classified as high frequency. We determined the threshold between low and high impact according to the median delay time reported for each delay reason (due to the ordinal nature of the response data). Those below the overall median delay time were classified as low impact and those at least the median were classified as high impact.
To understand how go-live dates were affected by the reported delays, we also collected a set of initially planned go-live dates and actual go-live dates from each site, via E-mail correspondence. We were aware that many sites deployed PGx CDS rules in a phased implementation plan, meaning that some delays reported in the survey might have occurred during later implementation phases after some rules were already “live” (i.e., triggered in the EHR in clinical practice). In addition, many of the delays could be encountered concurrently. For these reasons, it would not be accurate to simply sum the reported delays from each site to provide a single “go-live delay time.” In cases where sites implemented PGx CDS rules in phases, we focused this analysis on the first go-live date.
RESULTS
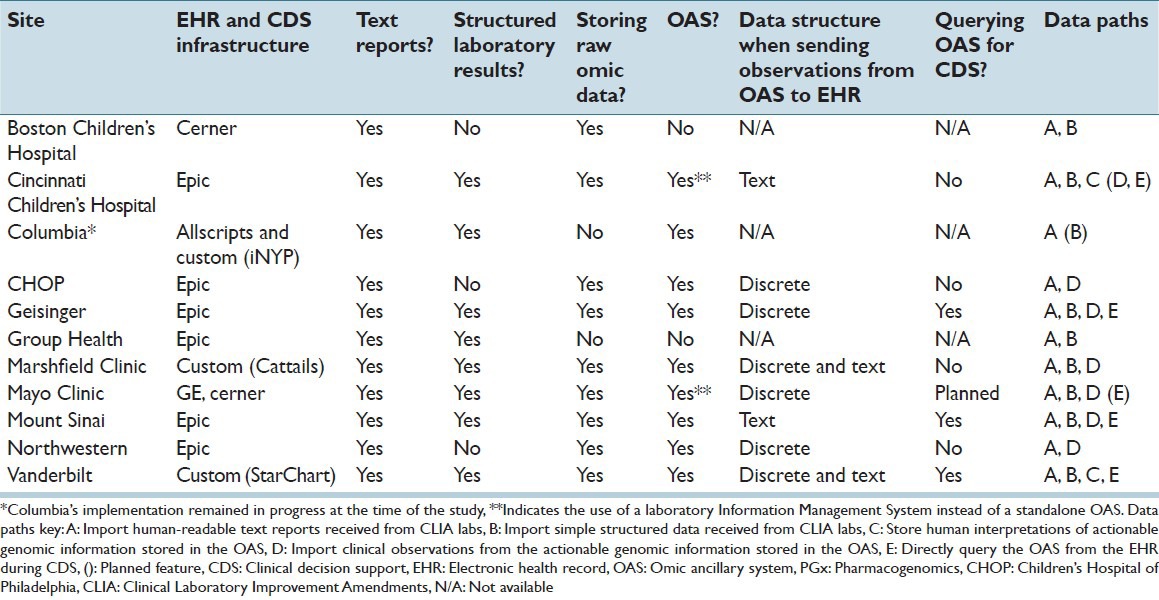
Most sites implemented an ancillary system of some kind to store raw omic data, which is consistent with earlier literature discussing the difficulties of integrating sequence data with the EHR.[5,23,24] One site that did not implement an OAS reported that their system was “interim” to achieve initial buy-in by users, with long-term plans for a more robust system. Of sites with an OAS, the stage of development varied, with some sites not yet incorporating the OAS into clinical workflows, or relying on human interpretation of information in the OAS. However, most sites were importing actionable genomic information directly into the EHR from the OAS. At the time of this assessment, three sites were querying an OAS directly for decision support and other sites planned to do so in the future. All sites used the CDS tools supplied by their existing EHR to implement PGx-related rules. Table 2 breaks down the characteristics of PGx CDS infrastructures across sites.
Table 2.
PGx decision support infrastructure characteristics
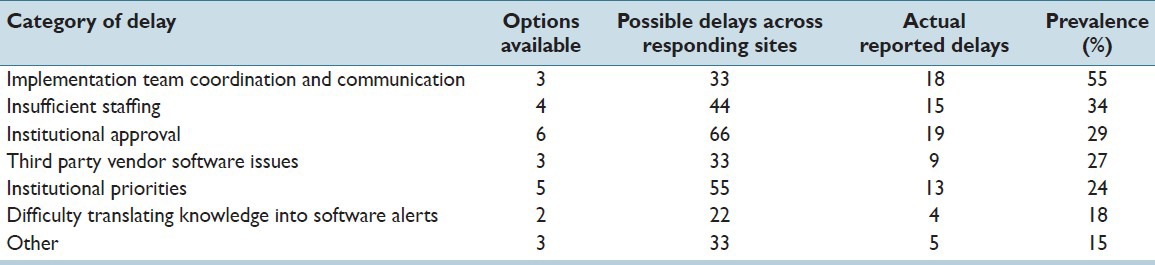
Representatives from all eleven sites in the eMERGE-EHRI workgroup responded to the challenges survey, reporting their experiences with the 26 possible delays. Ten of the eleven sites reported at least one delay in their implementation, including one site that was unable to implement prior to the study date. Overall, 83 delays were reported across the eleven sites. The “Institutional Approval” category had the most reported delays, with 19. However, this category also had the most possible types of delays, with six. Adjusting for the number of options in each category by examining prevalence instead of raw counts showed “Implementation Team Coordination and Communication” as the most frequent delay category, with 55% of all possible instances of delay being encountered [Table 3].
Table 3.
Delay prevalence, by category of delay, across 11 responding sites
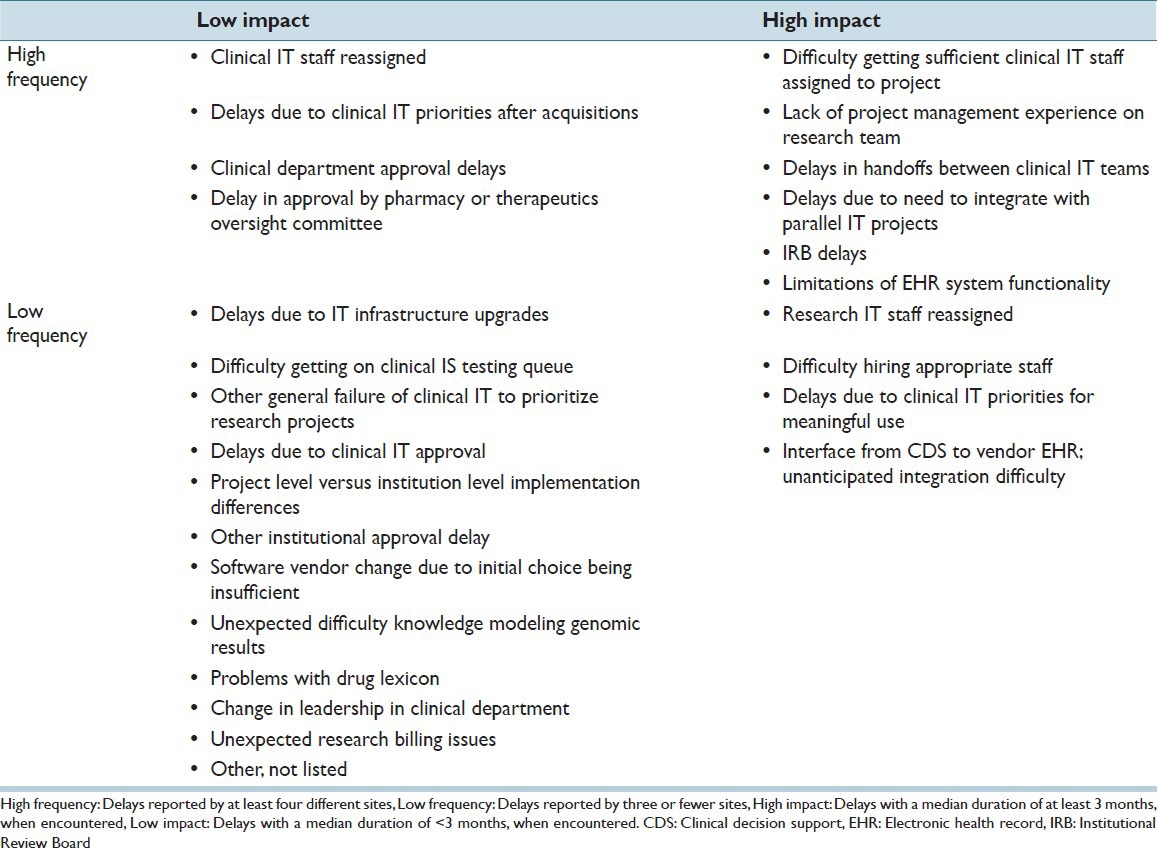
Each individual type of delay was encountered by a mean of 3.2 sites. Therefore, any delay encountered by three or fewer sites was categorized as low frequency, whereas any delay encountered by four or more sites was categorized as high frequency. The median individual delay time was approximately 3 months. Therefore, any delay with a median impact of less than 3 months was classified as low impact, whereas any delay with a median impact of at least 3 months was classified as high impact.
Under these criteria, 16 of the 26 tracked delay reasons were low frequency and 10 were high frequency. Similarly, 16 were low impact and 10 were high impact. Of the 83 individually reported delay incidents, 41 were low impact and 42 were high impact. Table 4 provides the complete breakdown of delay reasons, by impact and frequency.
Table 4.
Delay reasons, by impact and frequency
Sites reported a median total delay time of 67.5 days for their first go-lives, or just over 2 months (mean: 126 days). The longest delay was 323 days and the shortest was 0 days (i.e., no delay). Three sites reported no delay in their first go-live. Excluding those sites, the median delay time was 209 days, or approximately 7 months (mean: 180 days), and the minimum delay time was 37 days. One affiliate site reported an “unable to implement” delay in the challenges survey, due to “need to integrate with parallel IT projects,” and had no go-live date. This site was excluded from the total delay time analysis, though they continue to work toward implementation at a later date.
CONCLUSIONS
For practical reasons, each site took a different approach toward importing, storing, and applying the genomic information that was used in CDS. Reflecting the first-generation nature of PGx CDS systems, some sites created interim solutions to provide rapid functionality and evaluate user needs. Other institutional characteristics that led to differences in implementation included the type of EHR used, clinical workflow characteristics, and expectations for the role of technology in patient care. Consequently, implementation traits that differed across sites include whether laboratory results were stored in text reports or structured data, whether raw omic data were collected, and whether an OAS was used. Many sites plan to expand their system capabilities. Further development will be required to build robust, mature systems. All sites were able to make use of their existing CDS tools to implement PGx-related rules, suggesting that current tools are sufficient when genetic data are adequately processed.
Despite previous literature describing the complexities of genomic medicine, the eMERGE experience demonstrates that PGx CDS systems may not be any more difficult to implement than other healthcare IT initiatives of similar scope. Common themes in the genomic medicine literature included concerns about identifying actionable variants, lack of CDS infrastructure, difficulty integrating genetic data with existing IT systems, ethical issues, and concerns about physician education and adoption.[2,25,26,27] Ten eMERGE sites were able to overcome these anticipated barriers and successfully implement PGx CDS systems. Most unanticipated challenges encountered were not specific to genomic medicine, but were general IT implementation issues, such as team communication and difficulty getting appropriate staffing. “Difficulty translating knowledge into software alerts” was the only category in our survey that contained PGx-specific delays (“unexpected difficulty knowledge modeling genomic results” and “problems with drug lexicon”). Those delays were reported only four times and were ultimately classified as both low frequency and low impact.
Based on the results of this study, organizations planning a future PGx CDS system should prioritize efforts to mitigate the challenges that were classified in this report as both high impact and high frequency, as these are most likely to affect implementation timelines. Organizations should proactively plan to reduce the effects of these challenges. Additionally, organizations should be prepared for the other high impact or high frequency issues, if they appear particularly relevant to their institution. Any challenges classified as both low impact and low frequency should be monitored and addressed as they arise, but are less likely to require up-front planning.
The survey focused on the delays that sites encountered, meaning that we report only on unanticipated challenges. If a site accurately anticipated a major challenge with sufficient time allocated in the implementation plan, then it is not reported here. For example, one site has previously reported that implementation efforts included physician education plans, focus groups, and infrastructure updates to handle a new class of information.[28] However, sites participating in this study did not report frequent or impactful delays in these types of efforts.
The eMERGE sites that participated in this study are all large, research-oriented institutions with genomic expertise available to the project, so the results may not generalize to other facilities without the same local genomic and informatics expertise. Such organizations may encounter different types of delays, resulting in either greater or lesser total implementation delay.
This study did not investigate the effectiveness of PGx CDS post-implementation, although the eMERGE-EHRI workgroup is working with the eMERGE-PGx workgroup to capture data on process outcomes at each site. Analysis of effect on physician behavior and patient outcomes is ongoing, but similar systems implemented at other locations have shown promise.[29,30]
This study demonstrates some of the decisions that must be made when implementing a PGx CDS system. It also suggests that implementing PGx CDS rules may not result in significant unanticipated PGx-specific delays. Instead, most unexpected challenges are similar to other healthcare IT projects. These are encouraging results for any organization considering a PGx CDS tool and for genomic medicine in general. Future studies are needed to assess the effectiveness of these tools in altering physician behavior and improving patient outcomes.
Acknowledgments
The eMERGE Network was initiated and funded by NHGRI through the following grants: U01HG006828 (Cincinnati Children's Hospital Medical Center/Boston Children's Hospital); U01HG006830 (Children's Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative/University of Washington); U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai/Columbia University Medical Center); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); and U01HG006385 (Vanderbilt University Medical Center Serving as the Coordinating Center); T15LM007079 (Columbia University). Additional support was provided by the Mayo Clinic Center for Individualized Medicine (Mayo Clinic).
Financial Support and Sponsorship
Nil.
Conflicts of Interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2015/6/1/50/165999
REFERENCES
- 1.Green ED, Guyer MS. National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–13. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.Ginsburg GS, Willard HF. Genomic and personalized medicine: Foundations and applications. Transl Res. 2009;154:277–87. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Feero WG, Guttmacher AE, Collins FS. Genomic medicine – An updated primer. N Engl J Med. 2010;362:2001–11. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 4.Guttmacher AE, Collins FS. Realizing the promise of genomics in biomedical research. JAMA. 2005;294:1399–402. doi: 10.1001/jama.294.11.1399. [DOI] [PubMed] [Google Scholar]
- 5.Starren J, Williams MS, Bottinger EP. Crossing the omic chasm: A time for omic ancillary systems. JAMA. 2013;309:1237–8. doi: 10.1001/jama.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, et al. Implementing genomic medicine in the clinic: The future is here. Genet Med. 2013;15:258–67. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch BM, Eilbeck K, Del Fiol G, Meyer LJ, Kawamoto K. Technical desiderata for the integration of genomic data with clinical decision support. J Biomed Inform. 2014;51:3–7. doi: 10.1016/j.jbi.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–8. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 9.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: A systematic review. JAMA. 2001;286:2270–9. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 10.Garg AX, Adhikari NK, McDonald H, Rosas-Arellano MP, Devereaux PJ, Beyene J, et al. Effects of computerized clinical decision support systems on practitioner performance and patient outcomes: A systematic review. JAMA. 2005;293:1223–38. doi: 10.1001/jama.293.10.1223. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: A systematic review of trials to identify features critical to success. BMJ. 2005;330:765. doi: 10.1136/bmj.38398.500764.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuperman GJ, Bobb A, Payne TH, Avery AJ, Gandhi TK, Burns G, et al. Medication-related clinical decision support in computerized provider order entry systems: A review. J Am Med Inform Assoc. 2007;14:29–40. doi: 10.1197/jamia.M2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bates DW, Kuperman GJ, Wang S, Gandhi T, Kittler A, Volk L, et al. Ten commandments for effective clinical decision support: Making the practice of evidence-based medicine a reality. J Am Med Inform Assoc. 2003;10:523–30. doi: 10.1197/jamia.M1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shameer K, Denny JC, Ding K, Jouni H, Crosslin DR, de Andrade M, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133:95–109. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fullerton SM, Wolf WA, Brothers KB, Clayton EW, Crawford DC, Denny JC, et al. Return of individual research results from genome-wide association studies: Experience of the Electronic Medical Records and Genomics (eMERGE) Network. Genet Med. 2012;14:424–31. doi: 10.1038/gim.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clayton EW, Smith M, Fullerton SM, Burke W, McCarty CA, Koenig BA, et al. Confronting real time ethical, legal, and social issues in the electronic medical records and genomics (eMERGE) consortium. Genet Med. 2010;12:616–20. doi: 10.1097/GIM.0b013e3181efdbd0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuire AL, Basford M, Dressler LG, Fullerton SM, Koenig BA, Li R, et al. Ethical and practical challenges of sharing data from genome-wide association studies: The eMERGE Consortium experience. Genome Res. 2011;21:1001–7. doi: 10.1101/gr.120329.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: Past, present, and future. Genet Med. 2013;15:761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, Almoguera B, Basford MA, Bielinski SJ, et al. Design and anticipated outcomes of the eMERGE-PGx project: A multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther. 2014;96:482–9. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny JC. Surveying recent themes in translational bioinformatics: Big data in ehrs, omics for drugs, and personal genomics. Yearb Med Inform. 2014;9:199–205. doi: 10.15265/IY-2014-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) – A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louie B, Mork P, Martin-Sanchez F, Halevy A, Tarczy-Hornoch P. Data integration and genomic medicine. J Biomed Inform. 2007;40:5–16. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Kho AN, Rasmussen LV, Connolly JJ, Peissig PL, Starren J, Hakonarson H, et al. Practical challenges in integrating genomic data into the electronic health record. Genet Med. 2013;15:772–8. doi: 10.1038/gim.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinnon RA, Ward MB, Sorich MJ. A critical analysis of barriers to the clinical implementation of pharmacogenomics. Ther Clin Risk Manag. 2007;3:751–9. [PMC free article] [PubMed] [Google Scholar]
- 26.Scheuner MT, Sieverding P, Shekelle PG. Delivery of genomic medicine for common chronic adult diseases: A systematic review. JAMA. 2008;299:1320–34. doi: 10.1001/jama.299.11.1320. [DOI] [PubMed] [Google Scholar]
- 27.Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, Obeng AO, et al. Clinical pharmacogenetics implementation: Approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166C:56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, et al. Operational implementation of prospective genotyping for personalized medicine: The design of the vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, Baker DK, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2014;21:e93–9. doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldspiel BR, Flegel WA, DiPatrizio G, Sissung T, Adams SD, Penzak SR, et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. J Am Med Inform Assoc. 2014;21:522–8. doi: 10.1136/amiajnl-2013-001873. [DOI] [PMC free article] [PubMed] [Google Scholar]


