Significance
Clinical use of the currently available antibiotics is severely compromised by the increasing resistance to them, acquired by the natural bacterial capability to manipulate their genomes. Many existing antibiotics target the fundamental process of protein biosynthesis, mainly by paralyzing the ribosome. Although antibiotics’ modes of action are similar across most eubacteria, species specificity has been detected. We determined the structures of the large ribosomal subunit from Staphylococcus aureus, a pathogenic bacterial species with a known capacity to become multiresistant, and of its complexes with known antibiotic compounds, as well as with a novel potential pleuromutilin derivative. Our new insights provide unique chemical tools for enhanced distinction between pathogens and the useful benign microbiome, as well as for suggesting novel sites for potential future antibiotics.
Keywords: antibiotic resistance, potential advanced pleuoromutilin, species specificity, protein biosynthesis
Abstract
The emergence of bacterial multidrug resistance to antibiotics threatens to cause regression to the preantibiotic era. Here we present the crystal structure of the large ribosomal subunit from Staphylococcus aureus, a versatile Gram-positive aggressive pathogen, and its complexes with the known antibiotics linezolid and telithromycin, as well as with a new, highly potent pleuromutilin derivative, BC-3205. These crystal structures shed light on specific structural motifs of the S. aureus ribosome and the binding modes of the aforementioned antibiotics. Moreover, by analyzing the ribosome structure and comparing it with those of nonpathogenic bacterial models, we identified some unique internal and peripheral structural motifs that may be potential candidates for improving known antibiotics and for use in the design of selective antibiotic drugs against S. aureus.
The emergence of bacterial resistance to antibiotics threatens to cause regression to the preantibiotic era, as the treatment of infections with the available arsenal of clinically used antibiotics has been badly affected by the appearance of multidrug-resistant strains (1–3). Consequently, today many infections are caused by highly resistant bacterial strains of Staphylococcus aureus. These Gram-positive, versatile, and potentially aggressive strains are among the most worrisome pathogenic bacteria (3, 4).
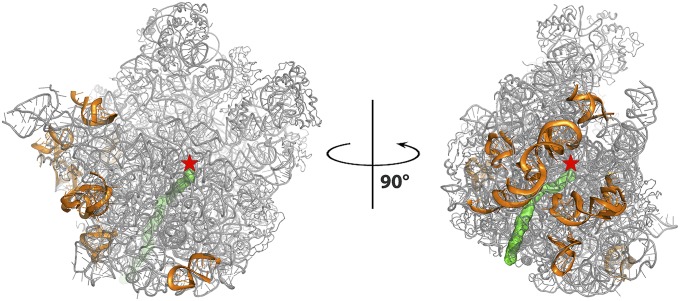
A large number of antibiotics paralyze ribosomes, the universal cellular multicomponent RNA/PROTEIN particles that translate the genetic code into proteins in all living cells. Previous structural studies of the modes of action of antibiotics that inhibit bacterial ribosomes, performed using their complexes with then-available crystallizable ribosomal particles, yielded high-resolution crystal structures. These include the large ribosomal subunit of Deinococcus radiodurans (D50S) and the whole ribosome of Thermus thermophilus (T70S), as well as Escherichia coli (E70S) (5–7). These studies provided useful clues about the common traits of inhibitory mechanisms of antibiotics, namely binding at ribosomal functional sites, such as the peptidyl transferase center (PTC) or the protein exit tunnel (Fig. 1); illuminated the structural basis for the distinction between pathogenic bacteria and their mammalian hosts despite the high conservation of the ribosomal functional sites; shed light on antibiotic synergism; and highlighted the general principles of resistance and cross-resistance to antibiotics.
Fig. 1.
The structure of the large ribosomal subunit of SA50S is shown in gray, the polypeptide exit tunnel is shown in green, and the PTC location is marked by a red star. The rRNA regions with fold variability compared with all other known structures (see below) on the SA50S are shown in orange and are rotated 90° to one another.
Based on their similar ribosomal RNA (rRNA) and ribosomal protein sequences, the structures of ribosomes from pathogens resemble those of ribosomes from other eubacteria. Nevertheless, species specificity in clinically relevant properties, particularly the modes of acquiring antibiotic resistance, have been identified (8). Given the knowledge that small structural differences between bacterial species can affect drug binding (9), for the progress of structure-based drug design, it is imperative to have a high-resolution crystal structure of the ribosome and its subunits from the pathogenic bacterial species. As a first step toward this goal, we determined the structure of large ribosomal subunit of the pathogen S. aureus (SA50S).
Infections caused by S. aureus have been treated with various drugs, including the ribosomal antibiotics oxazolidinones, pleuromutilins, macrolides, and ketolides, which bind to the large ribosomal subunit. Eubacteria possess several copies of rRNA operons; thus, acquisition of single-nucleotide mutations and/or posttranscription modifications in the 23S rRNA, which are among the common resistance mechanisms with respect to ribosomal antibiotics that enter the cells, should occur after relatively long periods (10). Nevertheless, the epidemiology of S. aureus is increasing (11). Resistance mutations in S. aureus are also associated with ribosomal protein (rProtein) uL3, located in proximity to the PTC, as well as with uL4 and uL22 rProteins, whose segments are exposed in the exit tunnel (12–20).
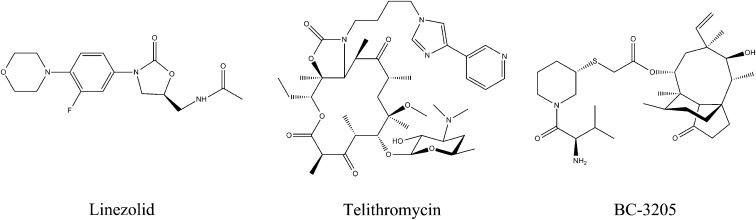
Linezolid (Fig. 2), a synthetic antibiotic drug of the oxazolidinone class, binds at the PTC (21). It was approved by the Food and Drug Administration in 2000 to treat Gram-positive infections. As a synthetic drug, linezolid had no known preexisting resistance mechanisms, and resistance to it was expected to emerge rather slowly (22). Despite these expectations, however, S. aureus-acquired linezolid resistance was detected within a relatively short time. This resistance is conferred by a specific 23S rRNA point mutation (G2576U) (E. coli rRNA numbering is used throughout) (23). Additional resistant strains were identified later, including strains with rRNA mutations in U2500A (24), A2503G, U2504C, and G2447U (25), as well as those that have acquired a transmittable gene of rRNA methyltransferase cfr targeting A2503 (26). Within 10 years, linezolid resistance was detected in >1% of S. aureus clinical isolates (16).
Fig. 2.
Chemical structures of the oxazolidinone linezolid, the ketolide telithromycin, and the pleuromutilin BC-3205.
Telithromycin (Fig. 2), a ketolide antibacterial drug that is structurally related to the macrolides, was developed specifically to provide optimal therapy for respiratory tract infections. All ketolides have two structural modifications compared with macrolides of previous generations: a C3-keto group and an additional long alkyl-aryl arm linked in telithromycin to the C11, C12-carbamate cycle. Importantly, telithromycin shows potent in vitro activity against Streptococcus pneumoniae, including strains resistant to macrolide-lincosamide-streptogramin B-ketolide (MLSBK). Crystal structures of telithromycin in complex with the bacterial 50S subunits or 70S ribosomes reveal that telithromycin binds to the large ribosomal subunit at the macrolide-binding pocket in the ribosome’s exit tunnel (27–30), and that in different species its flexible alkyl-aryl arm is stacked in various modes to different sites in the nascent protein exit tunnel.
Pleuromutilin (Fig. 2 and Fig. S1) is a natural product of the fungus Pleurotus mutilus (now called Clitopilus scyphoides) (31), which serves as a base for the synthesis of several semisynthetic antibacterial agents (32). All pleuromutilins consist of a tricyclic mutilin core, a C21 carboxyl group essential for antimicrobial activity (32), and various substituents at their C14 position, most of which are extensions with diverse chemical natures. Some pleuromutilins currenty in clinical use in humans and veterinary medicine have elevated activity over a broad spectrum of pathogens, including multidrug-resistant staphylococci and streptococci. Among the group of C14-sulfanyl-acetate derivatives, retapamulin is a topical antibiotic that was approved for clinical use in 2007. All strains of S. aureus and Streptococcus pyogenes were susceptible to retapamulin with low minimum inhibitory concentrations, <0.5 µg/mL (33, 34). Other C14-sulfanyl-acetate derivatives, valnemulin and tiamulin, are approved for veterinary clinical use.
Fig. S1.
Chemical formulas of selected pleuromutilin derivatives. Typically, the tricyclic mutilin core (Right) is conserved among pleuromutilin derivatives. Note the variability of the C14 extension (R1). Chemical formulas for all mentioned pleuromutilins are shown.
The crystal structures of complexes of the large ribosomal subunit from a nonpathogenic bacterium, D50S, with various pleuromutilin compounds (namely tiamulin, retapamulin, SB-264128, and SB-571519) reveal that the pleuromutilins are bound to the large ribosomal subunit at the PTC (35). In all cases, their cores are placed in a similar fashion at the A-site and their C14 extensions are pointing toward the P-site; thus, they directly inhibit peptide bond formation. Because the PTC is almost fully conserved, the clinical efficacy of the pleuromutilins stems from their efficient inhibitory modes, which are attained by structural diversity in the second or third shells around the PTC, by exploiting the ribosomal intrinsic functional flexibility for induced fit and remote conformational rearrangements that result in a tightening up of the binding pockets (35, 36).
Recent advances in pleuromutilin chemistry have yielded several new potential drugs, among which are BC-3205 (Fig. 2 and Fig. S1) and lefamulin (BC-3781). These novel semisynthetic pleuromutilin derivatives were developed by Nabriva Therapeutics (Vienna, Austria) for i.v. or oral treatment of community-acquired bacterial pneumonia and skin infections. Being 16- to 32-fold more potent than linezolid against S. aureus, BC-3205 exhibits clinically relevant potent antibacterial activity (37). Nabriva’s lead product, Lefamulin, is about to enter phase 3 of clinical trials on community-acquired bacterial pneumonia (38, 39).
Here we present the crystal structure of the large ribosomal subunit from S. aureus alongside the structures of its complex with linezolid, telithromycin, and BC-3205. By analyzing these structures and their comparisons with ribosomes from eubacteria in complex with various oxazolidinones, ketolides, and pleuromutilins, we have elucidated specific traits that may govern S. aureus inhibition. We have also provided structural definitions of ribosomal features that could be selectivity used in designing novel antibiotic drugs to target the S. aureus ribosome.
Results and Discussion
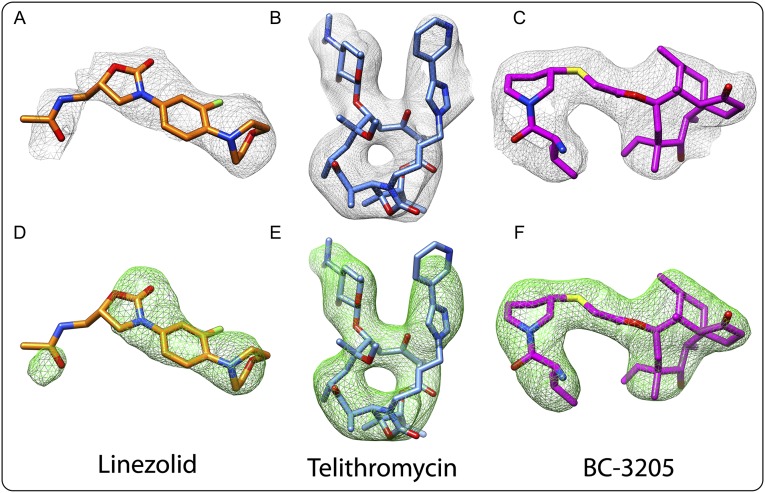
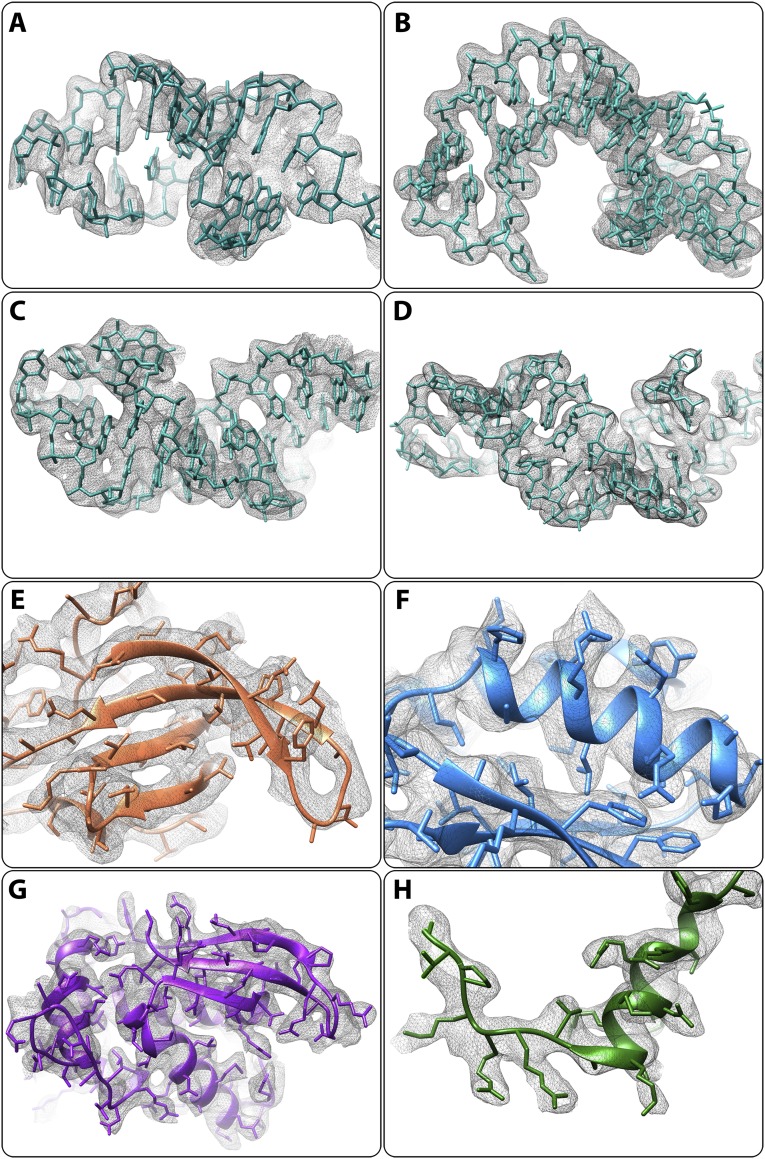
We determined the crystal structures of the large ribosomal subunit from S. aureus (SA50S) and of its complexes with linezolid, telithromycin, and BC-3205 at near-atomic resolution (Table 1). Most of the amino acid residues of 26 of the SA50S rProteins and 93% of the rRNA 50S nucleotides were reliably traced in the electron density map. Electron density maps around linezolid, telithromycin, and BC-3205 are shown in Fig. S2 A–F. In the vicinity of the rRNA, we also detected an electron density that may be interpreted as hydrated ions, presumably of Mg2+ and Mn2+.
Table 1.
Crystallographic data and refinement statistics of SA50S (native 50S), SA50S in complex with linezolid (SA50Slin) SA50S in complex with telithromycin (SA50Stel), and SA50S in complex with BC-3205 (SA50S-BC3205)
| Statistic | SA50S | SA50Slin | SA50Stel | SA50S-BC3205 |
| Crystal information | ||||
| Space group | P6522 | P6522 | P6522 | P6522 |
| a = b, Å | 279.8 | 279.9 | 282.7 | 280.9 |
| c, Å | 872.7 | 870.6 | 877.0 | 875.6 |
| α, β, γ, ° | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Diffraction data statistics | ||||
| X-ray source (ESRF) | ID23-2, ID23-1, ID23-1 | ID23-1, ID23-1 | ID23-1 | ID23-2 |
| Wavelength, Å | 0.873, 0.972, 1.000 | 0.972, 1.000 | 0.973 | 0.873 |
| Number of crystals | 11 | 8 | 11 | 20 |
| Resolution, Å* | 50–3.51 (3.57–3.51) | 200–3.4 (3.46–3.4) | 50–3.45 (3.51–3.45) | 50–3.43 (3.47–3.43) |
| Observed reflections | 2,813,593 | 1,392,748 | 1,792,405 | 2,109,298 |
| Unique reflections | 236,855 | 246,474 | 257,382 | 253,918 |
| Redundancy | 12.0 (6.0) | 5.7 (2.8) | 7.0 (5.0) | 8.3 (3.4) |
| Completeness, % | 96.3 (84.3) | 89.4 (64.8) | 98.5 (96.5) | 92.1 (78.9) |
| <I>/<σ(I)> | 7.40 (1.43) | 8.45 (1.16) | 8.84 (1.21) | 10.51 (1.21) |
| R-merge, % | 25.3 (97.8) | 15.6 (77.6) | 16.5 (92.6) | 15.2 (74.4) |
| Refinement statistics | ||||
| R-factor, % | 20.18 | 20.18 | 19.19 | 20.42 |
| R-free (5%), % | 24.65 | 24.28 | 23.15 | 24.20 |
| rmsd bonds, Å | 0.006 | 0.006 | 0.006 | 0.006 |
| rmsd angles, ° | 1.161 | 1.086 | 1.106 | 1.099 |
ESRF, European Synchrotron Radiation Facility.
Values for the highest-resolution shells are in parentheses.
Fig. S2.
Electron density of linezolid, telithromycin, and BC-3205 within their complexes with SA. (A–C) Weighted 2Fo-Fc electron density maps contoured at 1.0 σ. (D–F) Weighted Fo-Fc electron density maps contoured at 3.0 σ.
Main Features of S. aureus rRNA and rProteins.
Sequence alignment of the 23S rRNA shows 81%, 76%, and 73% identity between S. aureus and D. radiodurans, E. coli, and T. thermophilus, respectively. Despite the high sequence conservation, local variability was detected. To highlight the unique features of SA50S, we compared it with the available eubacterial ribosome structures D50S (40) and the large ribosomal subunits in the T70S (6) and E70S (7) ribosomes (29, 41).
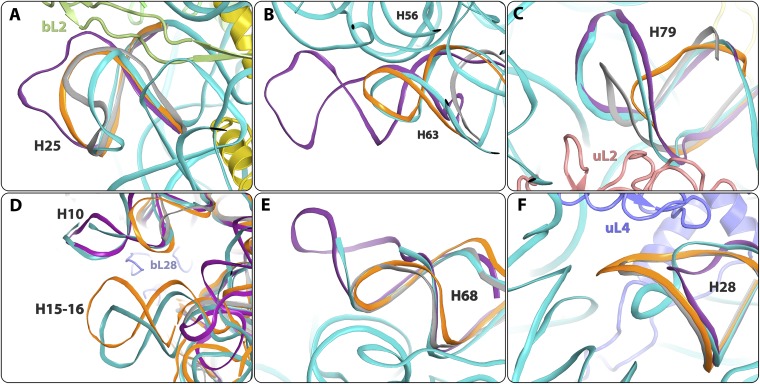
As expected, we found significant similarity in the rRNA folds of all high-resolution structures examined here, yet also identified several prominent differences on the surface of the 50S subunit. These differences are located mainly at the subunit surface (Fig. 1), and some at the interface with the small subunit within the active ribosome. Most of these differences are not involved in crystal contacts (all being located >5 Å from their crystal neighbors), and thus seem to originate from sequence variability or from the difference between isolated and bound large subunits. Examples include H25 (uppercase H is used throughout to refer to specific 23S rRNA helices), which has different folds in each of the four structures (Fig. 3A and Fig. S3B). Similarly, H63, which is located in proximity to the intersubunit bridge B5 (42), has a different length in each of the four structures (longest in E70S, shorter in SA50S and T70S, and shortest in D50S) (Fig. 3B and Fig. S3D). Helices H10 and H79, which interact with each other and with rProteins bL28 and uL2, respectively, are extended in the pathogen’s SA50S and E70S compared with their conformation in T70S and D50S (Fig. 3 C and D). H16 of SA50S and E70S are longer than H16 in T70S and D50S (Fig. 3D). Of interest, H15 shows small structural diversity between SA50S and T70S that may result from different interactions with a symmetry-related molecule owing to crystal packing, indicating its flexibility. It could not be traced in D50S electron density maps and does not exist in E70S (Fig. 3D). Also of note, H68, which is involved in the binding of rProtein uL1, elongation factor-G, and ribosomal recycling factor, and belongs to the intersubunit bridge B7a (42) with the 30S subunit, is longer in SA50S than in T70S and D50S; however, it is not fully traced in the SA50S electron density map (Fig. 3E). Additional fold variability was observed for H28, which has a different orientation in SA50S compared with the other three structures (Fig. 3F and Fig. S3C).
Fig. 3.
Details of structural variability in the rRNA backbone among the four known eubacteria structures. SA50S is in teal, E. coli (E70S) is in purple, D. radiodurans (D50S) is in gray, and T. thermophilus (T70S) is in orange. (A) H25. (B) H63. (C) H79. (D) H10 and H15-16. (E) H68. (F) H28.
Fig. S3.
(A–D) Electron density of 23S rRNA. Weighted 2Fo-Fc electron density maps contoured at 1.2 σ. (E–H) Electron density of rProteins. Weighted 2Fo-Fc electron density maps contoured at 1.0 σ. (A) H9. (B) H25. (C) H28. (D) H63. (E) uL3. (F) uL16. (G) bL17. (H) bL32 N terminus.
Sequence alignment of the large ribosomal subunit proteins showed an overall identity of ∼50% between S. aureus and E. coli, T. thermophilus, and D. radiodurans. Consequently, the rProteins show a greater diversity in their detailed structures compared with the differences observed for the rRNA structure. Typically, the main features of the rProtein folds, such as the globular domains and the secondary structural elements that interact with the rRNA, are rather conserved; however, several SA50S rProteins contain extensions that were not seen in any other published ribosome structure. (We use S. aureus rProtein residues numbering throughout, to minimize confusion owing to the significant variability in the rProtein sequences and folds of the four organisms.)
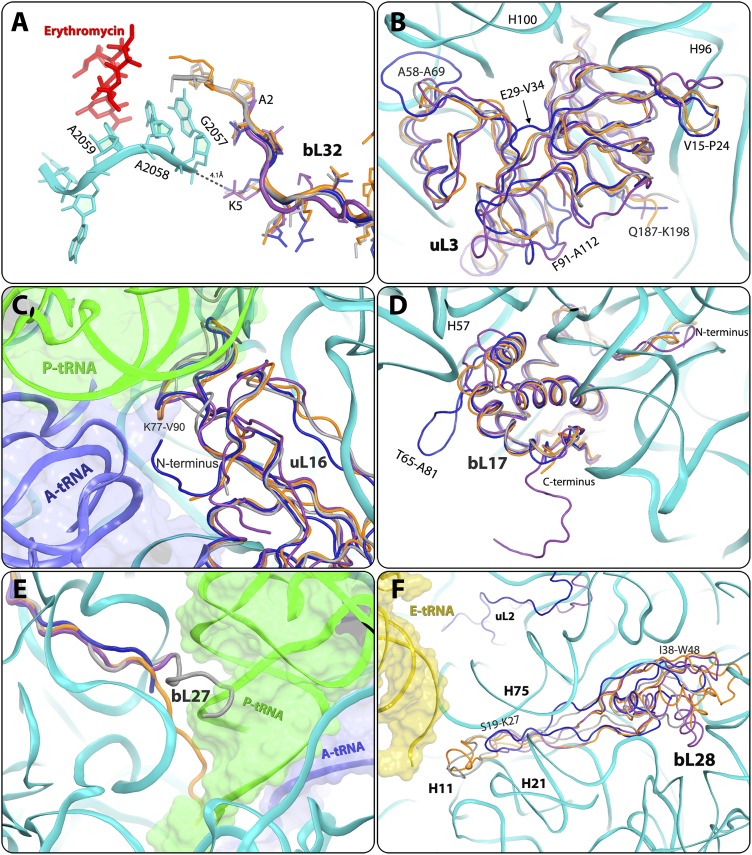
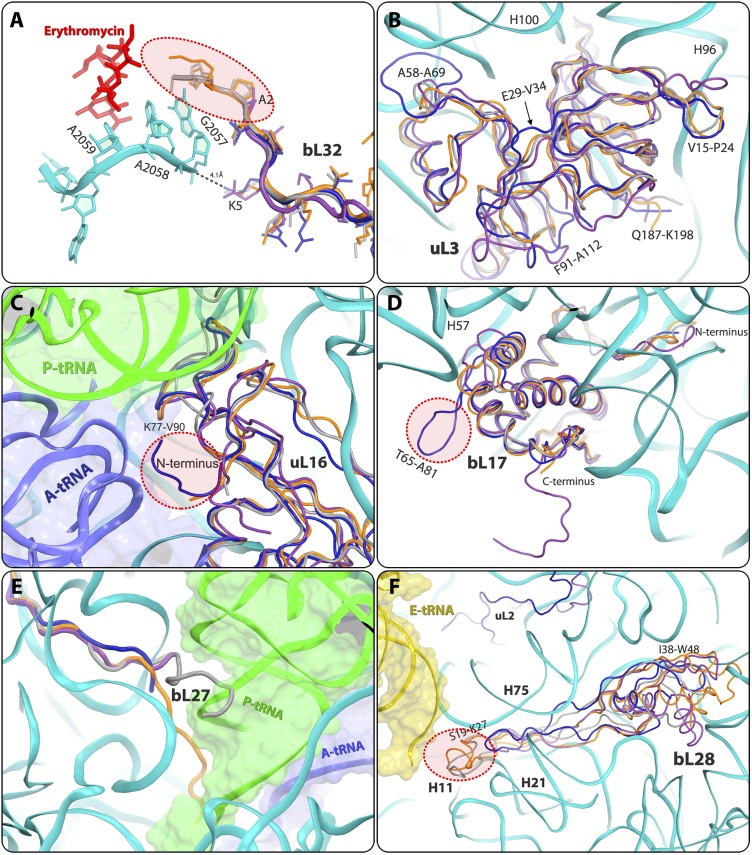
Important differences in S. aureus were detected in the N-terminal region of bL32 rProtein, resulting in the creation of a unique void at the rims of the macrolide- binding site. Thus, in D50S and T70S, the N-terminal tails of bL32 reach the erythromycin-binding pocket, whereas in the ribosome from SA50S and E70S, they are distal to this pocket (Fig. 4A and Figs. S3H and S4A). Sequence alignment of bL32 to other 721 eubacterial species (43) confirms that the elongated N-terminal end of bL32 is unique to Deinococcus and Thermus species. In addition, in all known structures of eubacterial ribosome nucleotide A2058 is stacked to G2057, which interacts with residue 5 of the bL32 N-terminal tail. The sequence and structure variability of this residue (Lys in SA50S, T70S, and D50S and Gln in E70S) creates different environments around G2057. Of interest, this protein does not exist in the archaeon Haloarcula marismortui or in eukaryotes.
Fig. 4.
Structural differences in rProteins. SA50S is shown in navy, E. coli (E70S) is in purple, D. radiodurans (D50S) is in gray, and T. thermophilus (T70S) is in orange. (A) The N-terminal end of protein bL32 that resides in the second shell around the erythromycin-binding pocket is shorter in S. aureus (SA50S, navy) and E. coli (E70S, purple) compared with in D. radiodurans (D50S, gray) and T. thermophilus (T70S orange), and thus may provide a space for a specific extension of erythromycin (Fig. S5). (B) SA50S rProtein bL3 has a unique extended A58-A69 loop compared with D50S, T70S, and E70S. (C) SA50S uL16 points opposite to E70S and thus may provide a potential specific binding site (Fig. S5) for a compound that may interfere with binding of the A-site tRNA acceptor stem (blue). P-site tRNA (green) interacts with the loop K77-V90, which has some structural variability. (D) SA50S bL17 has an extended (T65-A81) loop that is unique to SA50S. Because it is exposed on the surface of the SA50S subunit, it may provide a potential specific binding site (Fig. S5) for a compound that may interfere with the two subunits association. Its C terminus is ∼10 aa longer in E70S relative to the other three organisms. (E) In D50S, bL27 reaches the position of the acceptor stem of the P-site tRNA (green), whereas in T70S it reaches the PTC, in the proximity of the CCA-end of the P-site tRNA. (F) bL28 is located near the 50S surface, close to the position of the CCA 5′ of E-site tRNA (yellow). in D50S and T70S, it has a 15-residue extended loop (S19-K27) This loop is shorter in SA50S and E70S and does not exist in D50S and T70S, and thus may provide a potential site for a compound that may interfere with the binding of the E-site tRNA acceptor stem (yellow) (Fig. S5).
Fig. S4.
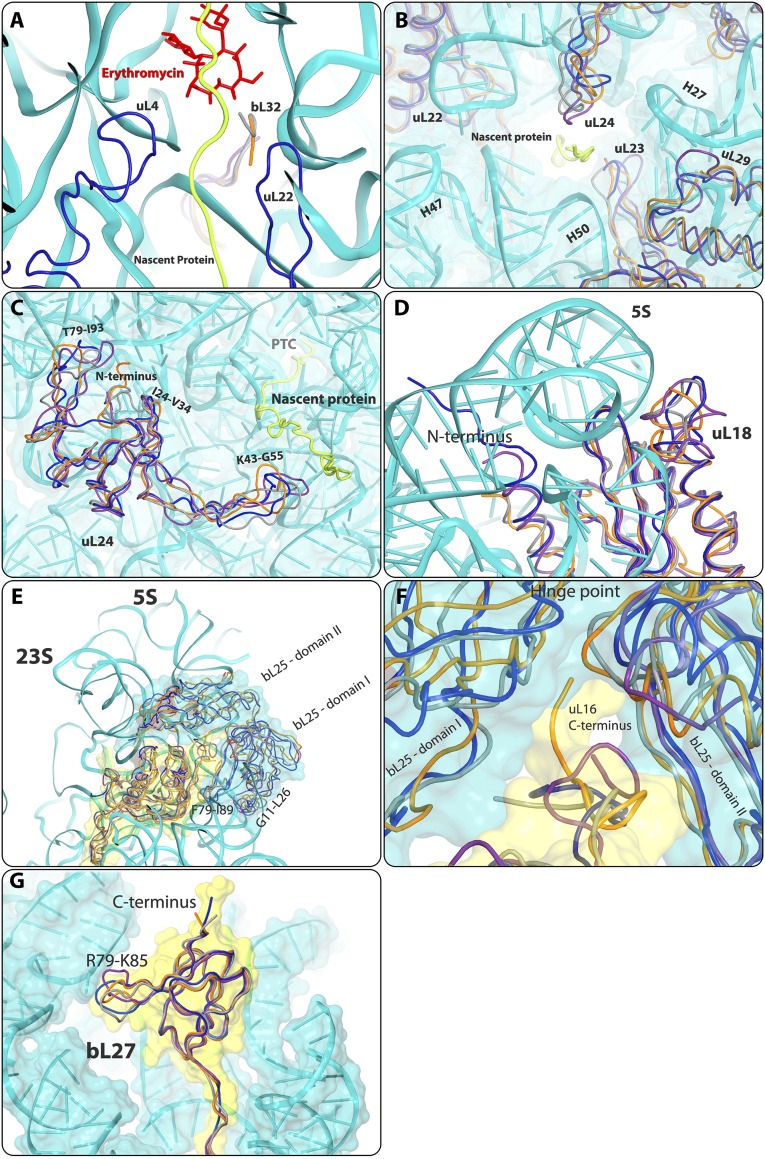
Additional structural differences identified in rProteins. A possible path of the backbone of a modeled nascent protein chain is indicated in yellow. The SA50S structure is shown in teal, the E. coli (E70S) structure is in purple, the D. radiodurans (D50S) structure is in gray, and the T. thermophilus (T70S) structure is in orange. (A) The bL32 N terminus in D50S and T70S is elongated relative to that in SA50S and E70S, reaching the rims of the erythromycin-binding site. (B) View into the exit tunnel opening, showing structural variability of the rProteins uL23, uL24, and uL29. The polypeptide chain was modeled in the tunnel for clarity (lime). (C) uL24 in all four structures. (D) The uL18 N-terminal domain is elongated in S. aureus relative to D50S, T70S, and E70S reaching the other side of the central protuberance. (E) bL25 domains, as well as structural variability, in the fold of G11-L26 and F79- I86 loops. The uL16 C terminus is longer in T70S than in SA50S, D50S, and E70S. (F) Zoom-in view of the hinge between the bL25 domains. The uL16 C terminus is longer in T70S than in SA50S, D50S, and E70S. It penetrates into the proximity of bL25, thereby changing the angle between bL25 domains. (G) bL27 R79-K85 loop and C-terminal fold variability among the four structures.
SA50S rProtein uL3 has an extended (A58-A69) loop next to h100 of 23S rRNA. Sequence alignment of uL3 to other 756 eubacterial species (43) confirms that this loop is unique to staphylococci and thus may serve as a new antibiotic target site against staphylococci ribosomes. In addition, variability in the SA50S uL3 backbone fold compared with D50S, T70S, and E70S was observed. Among these, E29-V34 and F91-A112 and loops V15- P24, S142-P151, G152-P170, and Q187-K198 have a different fold and orientation in SA50S than in T70S and E70S (Fig. 4B and Fig. S3E).
uL16 protein is positioned in the intersubunit surface and interacts with the A- and P-site transfer RNAs (tRNAs) (superimposed on PDB ID code 2WDK) (6). The N terminus of uL16 SA50S is pointing in the opposite direction compared with E70S, and thus may form a unique network of interactions with the acceptor stem of A-site tRNA (Fig. 4C and Figs. S3F and S5C).
Fig. S5.
Structural differences in rProteins. SA50S is shown in navy, E. coli (E70S) is in purple, D. radiodurans (D50S) is in gray, and T. thermophilus (T70S) is in orange. (A) The N-terminal end of protein bL32 that resides in the second shell around the erythromycin-binding pocket is shorter in S. aureus (SA50S, navy) and E. coli (E70S, purple) compared with that in D. radiodurans (D50S, gray) and T. thermophilus (T70S, orange), thus may provide a space for a specific extension of erythromycin (encircled in pink). (B) SA50S rProtein bL3 has a unique extended A58-A69 loop compared with that in D50S, T70S, and E70S. (C) SA50S uL16 is pointing opposite compared with E70S thus may provide a potential specific binding site (encircled in pink) for a compound that may interfere with binding of the A-site tRNA acceptor stem (blue). P-site tRNA (green) interacts with the loop K77-V90 that has some structural variability. (D) SA50S bL17 has an extended (T65-A81) loop that is unique to SA50S. Because it is exposed on the surface of the SA50S subunit, it may provide a potential specific binding site (encircled in pink) for a compound that may interfere with the two subunits association. Its C-terminal is ∼10 aa longer in E70S relative to the other three organisms. (E) bL27 in D50S reaches the position of the acceptor stem of the P-site tRNA (green), whereas in T70S it reaches the PTC, in the proximity of the CCA-end of the P-site tRNA. (F) bL28 is located near the 50S surface; close to the position of the CCA 5′ of E-site tRNA (yellow). In D50S and T70S, it has a 15-residue extended loop (S19–K27). This loop is shorter in SA50S and E70S and does not exist in D50S and T70S, and thus may provide a potential site for a compound that may interfere with the binding of the E-site tRNA acceptor stem (yellow) (encircled in pink).
rProtein bL17 of SA50S has a unique extended (T65-A81) loop. Sequence alignment of bL17 to other 788 eubacterial species (43) confirms that this loop has a unique conserved sequence in staphylococci and thus may provide a potential specific binding site, because it may interfere with subunit association or with nonribosomal factor interactions. The N terminus of SA50S bL17, which is located in proximity to H100-H101, H61, and H96, is four residues shorter relative to that in the other three structures. Its C terminus is ∼10 aa longer in E70S relative to the other three organisms (Fig. 4D and Figs. S3G and S5D).
The bL27 N-terminal has various conformations in the four known structures. In SA50S, it is traced only from residue 19 (based on sequence alignment with PDB ID code 2ZJR) (Fig. 4E). It was recently reported that in S. aureus, bL27 has an extended N terminus that is cleaved posttranslationally after amino acid 9 by a specific essential protease, before or concurrent with ribosome assembly (44). In D50S, it reaches the acceptor stem of the P-site tRNA (40), whereas in T70S, it reaches the PTC and interacts with the CCA 3′ end of the P-site tRNA (6). In the E70S structure, its five N-terminal residues are missing in the electron density map, and the N-terminal tail is shorter than in the other large subunits. In E. coli, rProtein bL27 appears to be essential for cell survival and for peptide bond formation (45), and the variations in its length and orientation apparently do not hamper its functional key role.
bL28 rProtein has a 15-residue extended loop (S19-K27) that reaches H11 of the 23S rRNA and interacts with the CCA 5′ of E-site tRNA in the T70S and D50S structures. This loop is shorter in SA50S and E70S (Fig. 4F) and, although it interacts with H21 and H75, it does not reach H11 or the E-site tRNA. In addition, the core of bL28, which is situated in proximity to the outer surface of the large subunit, is folded differently in the four structures. Moreover, the bL28 I38-W48 loop adopts a different orientation in SA50S than in E70S, T70S, and D50S (Fig. 4F and Fig. S5F).
A detailed comparative structural analysis presenting the main structural differences found in SA50S rProteins and rRNA is summarized in Tables S1 and S2 and shown in Figs. S4, S6, and S7.
Table S1.
Main structural differences in rProteins
| rProtein | Residues | Difference | Ribosome site | Fig. |
| uL2 | N terminus | Variation in length and fold. Thirty residues are missing from the D50S structure. In the current SA50S structure, it is traced from the third residue. | Intersubunit surface, next to H34 | |
| uL3 | V15-P24 | Different fold and orientation compared with T70S and E70S | Subunit surface, next to uL14 and bL19 | 4C |
| uL3 | E29-V34 | Variability in backbone folds in all four structures | Subunit surface within its globular domain | 4C |
| uL3 | A58-A69 | SA50S extended loop next to H100 of 23S rRNA compared with E70S, D50S, and T70S | Subunit surface, next to H100 | 4C |
| uL3 | F91-A112 | Variability in backbone folds in all four structures | Subunit surface within its globular domain | 4C |
| uL3 | H134-P170 | Different fold and orientation compared with T70S and E70S | Subunit core, in between h61, H35, and H90 | 4C |
| uL3 | Q187-K198 | Different fold and orientation compared with T70S and E70S | Subunit surface within its globular domain | 4C |
| uL4 | N terminus | Different backbone folds in the four structures | Subunit surface within its globular domain, next to H29 | S4C |
| uL4 | V6-I17 | Extended surface loop compared with this loop in E70S and D50S. This loop adopts a different orientation in the T70S structure. | Subunit surface within its globular domain | S4A |
| uL4 | W65 | W65 of SA50S (W/W/Y in E70S, T70S, and D50S) points into the exit tunnel and has a different orientation relative to the other structures. | Protein exit tunnel entrance | 6F |
| uL4 | K66 | K66 in SA50S (R/G/P in E70S, D50S, and T70S) interacts with nucleotide U790 in the 23S rRNA that is pointing into the exit tunnel. | Protein exit tunnel entrance | 6 E and F |
| UL4 | K68 | Conserved among all four structures. Situated in proximity to the PTC nucleotides A2060 and U2061, but exhibits a different orientation in each of the four structures. | Protein exit tunnel entrance | 6F |
| uL4 | R72 | R72 in SA50S (R/R/N in E70S, T70S, and D50S) points into the exit tunnel in different orientations in the four structures. | Protein exit tunnel entrance | 5E |
| uL4 | Q119-E132 | Different backbone fold in the four structures | Subunit surface, close to the H19–H20 junction | S4C |
| uL4 | C terminus | Different structure and orientation in the four structures | Subunit surface within its globular domain, next to H28 | S4B |
| uL5 | M38-A53 | High structural variability observed among the four structures | Part of intersubunit bridge B1b | S3E |
| uL5 | G126-D144 | High structural variability observed among the four structures. | Part of intersubunit bridge B1b | S3E |
| uL14 | L25-S28 | Extended loop in D50S compared with SA50S, E70S, and T70S, forming a unique interaction with H95 in addition to the common interaction with H90 | Subunit core in between h90, H92 and H95 | |
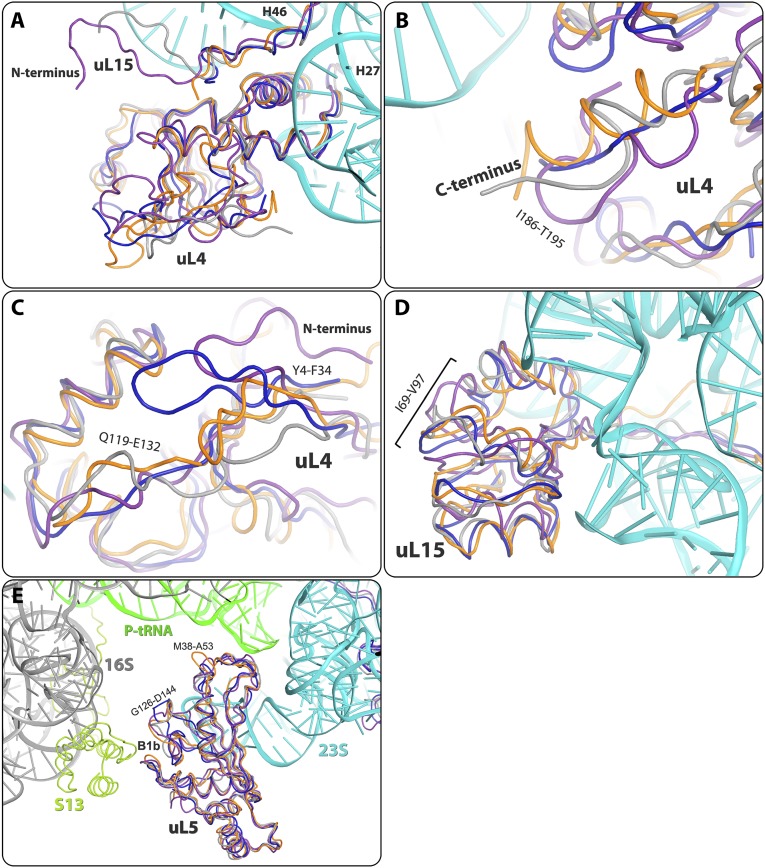
| uL15 | N terminus | The uL15 N-terminal is a long extended strand with fold variation among the four structures that begins at the subunit surface and penetrates into the core of the 23S rRNA. In SA50S, this loop is the shortest. | Reaches subunit surface, next to uL4 | S4A |
| uL15 | I69-V97 | Different structure of this loop in the four structures | Subunit surface within its globular domain, close to H28 | S4D |
| uL16 | N terminus | The N terminus of uL16 SA50S points in the opposite direction compared with that in E70S, and thus may form a unique interaction network with the acceptor stem of A-site tRNA. | Intersubunit surface, between H38 and H89, A-site tRNA binding site | 4D |
| uL16 and bL25 | C terminus | The T70S C terminus of uL16 is longer, penetrating deeper between bL25 domains; thus, the angle between the bL25 domains is larger compared with that in D50S and SA50S bL25 structures. | Central protuberance, subunit surface | S3F |
| bL17 | N terminus | The SA50S N terminus is four residues shorter relative to other three structures, and it folds differently. | Subunit core in between H96, H91 and H100 | 4E |
| bL17 | T65-A81 | This loop is most extended in SA50S. | Subunit surface between H47, H59 and H57 | 4E |
| bL17 | C terminus | ∼10 aa longer in E70S compared with the other three structures, and thus extended at the subunit surface | Subunit surface within its globular domain, close to H101 | 4E |
| uL18 | N terminus | The SA50S N terminus is longer than the E70S tail, reaching protein uL5 across the 5S rRNA. In D50S and T70S, this N terminus has been traced only from amino acids 8 and 11, respectively. | Central protuberance, subunit surface | S3D |
| uL18 | T53-A73 | Variable fold among the four structures | Central protuberance, subunit surface | S3D |
| bL19 | N terminus | The SA50S bL19 N terminus interacts with H101, whereas the D50S bL19 N terminus interacts with H100 and uL3. The T70S and E70S N termini are shorter. | Subunit surface next to H101 | |
| bL21 | N terminus | The SA50S backbone fold is slightly different from the backbone fold in the other three structures. | Subunit surface within its globular domain next to bL20 | |
| bL21 | K24-D31 | The SA50S backbone fold is slightly different from the backbone folds in the other three structures. | Subunit surface next to H45 | |
| bL21 | G43-V58 | The SA50S backbone fold is slightly different from the backbone folds in the other three structures. | Subunit surface within its globular domain next to bL20 | |
| uL22 | N terminus | The D50S N terminus is 22 residues longer than the N termini in SA50S, T70S, and E70S, resulting in a rather different surface of the D50S subunit at this face. | Subunit surface next to H24 | |
| uL22 | K83 | This Lys side chain in SA interacts with nucleotide C1261 (H26), adopting a different orientation compared with the E70S, T70S, and D50S structures. | Protein exit tunnel entrance | |
| uL22 | R84 | R84 points in different direction in SA50S, E70S, and T70S than in D50S. | Protein exit tunnel entrance | |
| uL22 | S85-F94 | Hydrophobic loop that folds differently in SA50S, D50, and T70S structures compared with the E70S structure | Protein exit tunnel entrance | 6A |
| uL22 | R86 | Only in SA50S does structure residue R86 (I/L/M in D50S, T70S, and E70S) reach nucleotide C1325 (h50) U/A/A in E70S, D50S, and T70S. | Protein exit tunnel entrance | 6A |
| uL22 | Q90 | Q in SA and R in D50S and T70S, pointing into the exit tunnel. The glutamine side chain is shorter than the arginine side chain, allowing a wider passage in the tunnel. Because E. coli has a different backbone fold in this region, the aligned residue is K and is in a different position. | Protein exit tunnel entrance | 6A |
| uL22 | R92 | R/S/R in T70S, D50S, and E70S. Orientation is different in all the four structures. | Protein exit tunnel entrance | 6A |
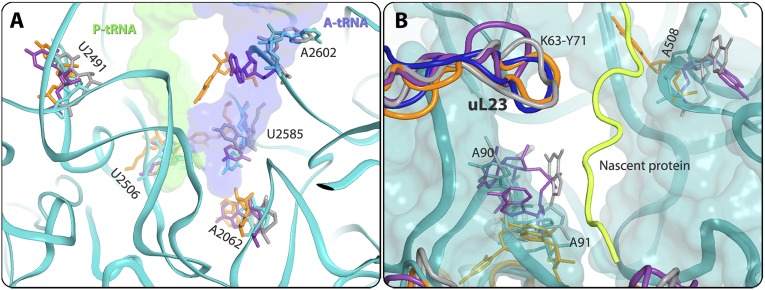
| uL23 | K63-Y71 | Pointing into the tunnel. Exhibits different orientations in SA50S/T70S, D50S, and E70S, especially, residue R67 (Q in E70S), which points into the tunnel in different directions. | Protein exit tunnel | S3B |
| uL23 | C terminus | The SA50S C-terminal tail is similar in length to that in D50S and T70S. It folds different from that in E70S, which exhibits an extended C-terminal tail. Consequently, this part of the subunit surface, which interacts with the ER membrane, is altered among the four structures. | Protein exit tunnel end (surface) | |
| uL24 | N terminus | Reaches the outer subunit surface via interactions with H7 and the junction of H18, H19, H20 of the 23S rRNA. In D50S, the N terminus of uL24 is 10–13 residues longer than that in the other three structures. The SA50S and T70S N termini are shorter (the SA50S N terminus is partly untraced, owing to uninterruptable density). The E70S N terminus is the shortest. | Subunit surface via interactions with the H7 and the junction H18, H19, H20 | S3C |
| uL24 | I24-V34 | The E70S loop folds different from in SA50S, D50S, and T70S. | Surface next to H7 | S3C |
| uL24 | K43- G55 | This loop has a different orientation and a slightly different fold in all four structures. | Protein exit tunnel end (surface) next to H24 and H7 | S3C |
| uL24 | T79-I93 | Highly divergent among the four structures | Surface of the subunit between H7 and uL18 | S3C |
| uL24 | C terminus | Highly divergent among the four structures | Surface, close to H7 | |
| bL25 | Domains | bL25 of E70S is a single-domain protein, whereas in SA50S and T70S,, bL25 is composed of two domains,, and in D50S,b, L25 is composed of three domains. | Central protuberance, subunit surface | S3E |
| bL25 | G11-L26 | SA50S and E70S have an extended loop that interacts with 5S rRNA. | Central protuberance, subunit surface | S3E |
| bL25 | I49-T69 | Variable backbone fold and orientation in all 4 structures. | Central protuberance, subunit surface | S3E |
| bL25 | F79-I89 | Variable backbone fold and orientation in all 4 structures. | Central protuberance, subunit surface | S3E |
| bL27 | N terminus | S. aureus bL27 has an extended N- terminus which is cleaved posttranslationally after the ninth aa by a specific essential protease, before or concurrent with ribosome assembly. bL27 N-terminal has various conformations in the four known structures. In SA50S it is traced only from residue 19 (based on sequence alignment with 2ZJR) and is located differently in all known structures. | Intersubunit surface next to the CCA 3′ P-site tRNA | 4F |
| bL27 | R79-K85 | Variable fold among the four structures | Central protuberance, subunit surface | S3G |
| bL27 | C terminus | Structural variations, as reported previously for all known structures (45) | Central protuberance, subunit surface | S3G |
| bL28 | Entire protein | The T70S bL28 has a different fold compared with the E70S, D50S, and SA50S structures. | Intersubunit surface | 4H |
| bL28 | S19-K27 | In D50S and T70S, bL28 has an extended loop compared with E70S and SA50S, which reaches H11 of the 23S rRNA and should interact with the CCA 5′ of E-site tRNA. | Intersubunit surface, between H11 and H21 | 4G |
| bL28 | I38-W48 | Adopts a different orientation in SA50S compared with E70S, T70S, and D50S structures. | Intersubunit surface, between H79 and H10 | 4H |
| uL29 | N terminus | Extended in T70S and E70S compared with SA50S and D50S N-terminal structures. | Subunit surface, next to H7 | |
| uL29 | L37-T40 | The E70S loop is more extended than in the three other structures. | Protein exit tunnel exit | S3B |
| uL29 | C terminus | Variable folds among the four structures | Subunit surface next to H18 | |
| uL30 | N terminus | Pointing in different directions in SA50S and T50S structures compared with E70S. In D50S, the N terminus is shorter. | Subunit surface close to its C terminus and H38 | |
| uL30 | C terminus | Longer in T70S compared with in the other three structures. | Subunit surface close to its N terminus and H38 | |
| bL32 | N terminus | D50S and T70S are elongated relative to SA50S and E70S, thus penetrating deeper into the tunnel wall. | Protein exit tunnel entrance | 4 A and B |
| S3A | ||||
| bL32 | V24-Q37 | Structural variability among the four structures | Surface between uL22 globular domains h101 and h99 |
Table S2.
Main structural differences in rRNA
| rRNA | Difference | Ribosome site | Fig. |
| H10 | Elongated in SA50S and E70S structures relative to T70S and D50S structures | Interacting with H79 and with proteins bL28 | 3D |
| H15-H16 | In SA50S, H15 and H16 fold differently than in E70S and H16 in D50S. Interestingly, H15 is missing in the E70S structure and could not be traced in D50S, presumably owing to its flexibility. | Surface, in vicinity of bL28 | 3D |
| H25 | Different fold among the four structures compared | Surface, in vicinity of bL21 | 3A |
| H28 | Different fold among the four structures compared | Next to the surface of rProtein uL4 globular domain | 3F |
| H63 | Variable lengths in all four structures, longest in E70S, shorter in SA50S and T70S, and shortest in D50S | In proximity to intersubunit bridge B5 | 3B |
| H68 | Not fully traced in SA50S structure, because it is longer than H68 of T70S and D50S structures | Involved in the binding of rProtein uL1, EF-P, and ribosomal recycling factor, and part of the intersubunit bridge B7a | 3E |
| H79 | Elongated in SA50S and E70S relative to T70S and D50S | Interacting with H10 and with proteins uL2 | 3C |
| U2506, U2504, U2609, U2585, A2062, A2602 (H93), U2491 (H89) | Flexible nucleotides | PTC | S1A |
| A508, A90-A91 (H7) | Flexible nucleotides | Exit tunnel exit, on the surface of the subunit | S1B |
Fig. S6.
(A) Flexible nucleotides at the PTC and at the exit tunnel in U2506, U2585, A2062, A2602, and U2491, where the P-site tRNA (green surface) and A-site tRNA (blue surface) should bind. S. aureus 23S RNA backbone and nucleotides are shown in teal. D. radiodurans, T. thermophilus, and E. coli nucleotides are shown in gray, orange, and purple, respectively. (B) Flexible nucleotides toward the tunnel opening. A90, A91, and A508 are located in the ribosomal exit tunnel, and were detected with different conformations in all four structures. A possible path of the backbone of a modeled nascent polyalanine chain is shown in yellow.
Fig. S7.
Structural differences in rProteins at the subunit surface. (A) Length variations detected in the V6-I17 surface loop of protein uL4 and the N terminus of uL15 located in its vicinity. Most extend in SA50S and less in E70S and D50S, and have a different orientation in T70S. Shown are the uL15 N termini in E70S, D50S (traced from the fourth amino acid), T70S (traced from the fifth amino acid), and SA50S (the shortest, traced from the first amino acid). (B and C) The C and N terminals of uL4 have different backbone folds in the four eubacterial structures. (D) SA50S L15 loops I69-T89 and T89-V97 have different structures compared with the corresponding loops in D50S, T70S, and E70S. (E) Structural variability in the M38-A53 and G126-D144 loops of uL5 that participate in the B1b intersubunit bridge with the 30S subunit (16S in gray) with rProtein uS13 (dark green).
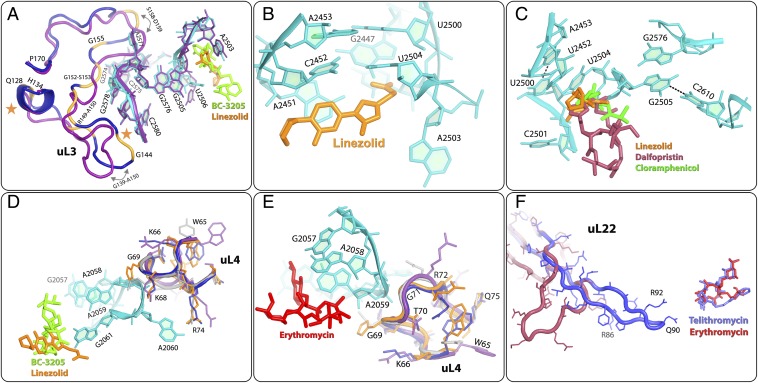
Structure of the SA50S-Linezolid Complex.
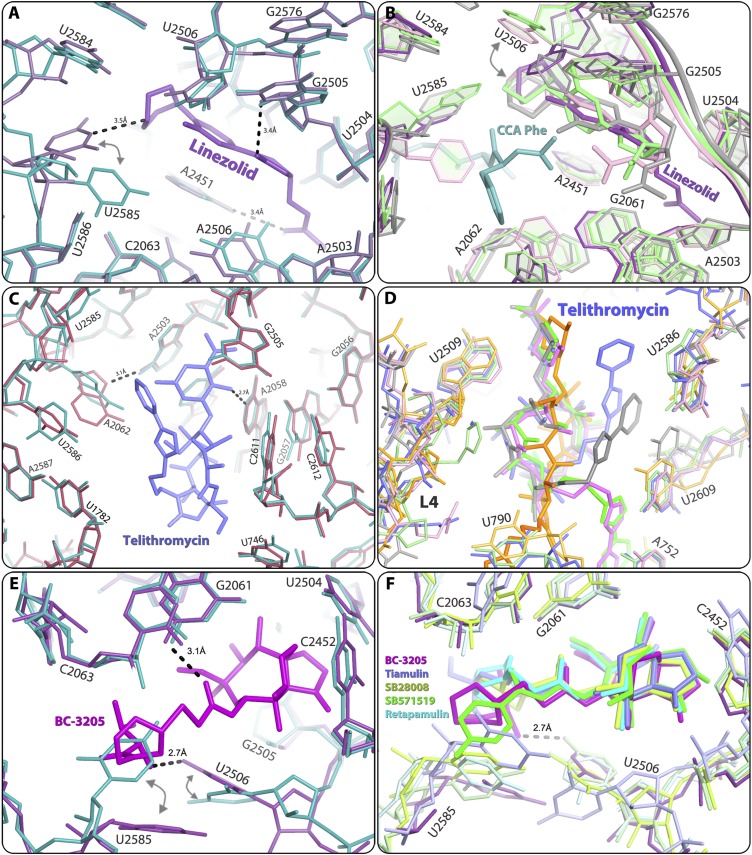
The SA50Slin crystal structure indicates that linezolid is bound at the PTC, blocking the A-site in an orientation clearly similar to that observed in other ribosome linezolid complexes with a D50S (46), H50S (47), and E70S linezolid model (48). However, in the SA50Slin complex, the flexible nucleotide U2585 (49) undergoes significant rotation and forms a hydrogen bond with the O4 of the linezolid morpholino ring, leading to a nonproductive conformation of the PTC (Fig. 5 A and B).
Fig. 5.
The linezolid-binding site as identified in the SA50Slin complex structure (A and B), with hydrogen bonds between the bound drugs and 23S rRNA shown in black dashes, the telithromycin-binding site as identified in the SA50Stel complex structure (C and D), and the BC3205-binding site as identified in the SA50S-BC3205 complex structure (E and F). (A) Comparison of native SA50S PTC (teal) and SA50Slin complex (pale purple) structures. (B) Overlay of the structures of various ribosome-linezolid complexes, including SA50Slin (pale purple; this study), H50Slin (green) + CCA-Phe substrate analog (teal) (PDB ID code 3CPW) (47), and D50Slin (gray) (PDB ID code 3DLL) (46), and of the model of E70Slin (pink) (48). The color coding of the rRNA components of the various linezolid-binding pockets is the same as that of the corresponding linezolid molecules. (C) Comparison of the structures of native SA50S PTC (teal) and SA50Stel complex (red). (D) Structural overlay of telithromycin conformations observed in various ribosome-telithromycin complex structures: SA50Stel (slate; this study), D50Stel (orange) (PDB ID ID code 1P9X) (27), H50Stel (gray) (PDB ID code 1YIJ) (28), E70S-tel (pink) (PDB ID code 3OAT) (29), and T70Stel (green) (PDB ID code 3OI3) (30). The color coding of the rRNA components of the various telithromycin-binding pockets is in a brighter tone than the corresponding telithromycin molecules. (E) Comparison of the PTC structure in native SA50S (teal) and in SA50S-BC3205 complex (purple). The arrows indicate the movements of nucleotides U2585 and U2506 in the bound vs. native structure. (F) Structural overlay of various pleuromutilins in their binding pockets: SA50S-BC3205 (violet; this study), D50S-SB571519 (green) (PDB ID code 2OGM), D50S-retapamulin (cyan) (PDB ID code 2OGO), D50S-tiamulin (slate) (PDB ID code 1XBP), and D50S-SB280080 (lemon) (PDB ID code 2OGN). Only one hydrogen bond between BC-3205 (violet) and 23S rRNA is shown here (as black dashes) for increased clarity.
The linezolid 1,3-oxazolidin-2-one moiety and acetamide group form additional hydrogen bonds with G2505 and A2451, respectively. The fluorophenyl moiety of linezolid is located in a heteroaromatic crevice formed by the PTC residues C2451 and C2452, the so-called “A-site cleft.” All of its other interactions with rRNA nucleotides G2061, C2501, U2504, U2506 and G2447 are either van der Waals or hydrophobic.
Comparing the SA50Slin structure (Fig. 5A) with other available crystal structures or models of large ribosomal subunits in complex with linezolid, namely of H50S (H50Slin) (47), D50S (D50Slin) (46) and E70S (E70Slin) (48) (Fig. 5B) revealed that linezolid binds in the same pocket in all structures. However, there are subtle differences between the conformations of the linezolid acetamide group in SA50Slin and in the other structures. These include a 100°–120° rotation of the acetamide group, which in SA50Slin enables fixation of A2451 by a hydrogen bond. Of note, the H50Slin complex was crystallized in the presence of CCA-Phe, a tRNA-3′ end substrate analog, assuming that in such an environment it should interact strongly with the 50S subunit P-site (47). Indeed, it seems that this P-site analog altered linezolid conformation compared with other linezolid complex structures that were determined with an empty P-site. Of importance, a hydrogen bond between the morpholino ring of linezolid and nucleotide U2585 was observed in D50Slin and in SA50Slin, but not in the complex of the archaeal H50S with linezolid.
Structure of the SA50S-Telithromycin Complex.
The SA50Stel structure shows that telithromycin is bound at the MLSBK-binding site and forms the typical ketolide (and macrolide) hydrogen bond between its desosamine sugar and A2058. At this position, it is partially blocking the protein exit tunnel, as has been found in other ribosome-MLSBK complex structures (5, 27, 50). In the SA50Stel complex, the flexible nucleotide A2062 is rotated compared with its conformation in native SA50S, and its conformational range is minimized by the hydrogen bond with A2503 (Fig. 5C). All of the other interactions of telithromycin with the rRNA nucleotides G2505, A2059, C2611, and U746 are either hydrophobic or based on van der Waals distances.
Comparing the SA50Stel structure with other available crystal structures of ribosomal particles in complex with telithromycin (27–30) revealed that in all structures, the drug is bound at the same pocket but with distinct differences in orientations of the alkyl-aryl moiety. The orientation of this moiety was similar in SA50Stel and in H50Stel. In both, the arm is folded back over the macrolactone ring, creating a rather compact structure of the drug; however, in SA50Stel, the alkyl-aryl arm reaches the center of the tunnel ∼5 Å closer to the PTC compared with its location in H50Stel, and may block nascent protein progression at an earlier stage. In this orientation, the alkyl-aryl arm is almost overlapping the location of the cladinose sugar of erythromycin in its complex with D50S (Fig. 6F) (5). In contrast, in T70Stel and E70Stel, this arm is stacked to A752 and U2609, a base pair located on the tunnel wall further away from the PTC, so that it can block nascent protein progression ∼10 Å away from the point of blockage observed in H50Stel. In D50Stel, the alkyl-aryl arm is extended, and thus it blocks the tunnel by interacting with U790 located across the tunnel, creating a barrier located 15 Å further along the tunnel compared with SA50Stel (Fig. 5D). It has been suggested that the structure of E70Stel reflects the telithromycin binding mode to the ribosomes of medically relevant (namely pathogenic) eubacteria species, because the A752-U2690 base pair is conserved among all eubacteria (29). In addition, telithromycin partially protects A752 from chemical modification (29), and the telithromycin-resistant S. pneumoniae ΔA752 mutant has been isolated (51). Nevertheless, in SA50Stel, the alkyl-aryl arm of telithromycin does not interact with the A752-U2690 base pair. This observation clearly demonstrates that the general description of the overall antibiotic-binding properties, which is the main outcome from the previous structures, cannot be extrapolated safely to other species.
Fig. 6.
Resistance and cross-resistance mechanisms in S. aureus. SA50S rRNA and rProteins are colored in navy. D50S (gray), E70S (purple), and T70S (orange) structures are superimposed on the SA50S structure for comparative analysis. rRNA nucleotides of S. aureus are shown only in regions in which they can be well aligned with the corresponding nucleotides in all other structures used for the comparisons. (A) SA50S rRNA nucleotides of the linezolid-binding sites (orange) and BC-3205–binding sites (green) are superimposed on the corresponding E70S rRNA (purple). G2576 is located in the second shell around the linezolid- and BC-3205–binding sites, in proximity to the first shell nucleotides G2505 and U2506; thus, its mutations may cause alterations in them. The locations of the uL3 mutations acquiring resistance to S. aureus are marked on the protein chain (yellow). For comparison, the structure of E. coli uL3 (purple) is superposed on SA50S uL3 (navy). Key structural differences are marked by arrows. The orange stars indicate deletions. (B and C) Linezolid (orange), chloramphenicol (green), and dalfopristin (streptogramins A, in raspberry) are shown in the SA50S rRNA-binding pocket. (D) uL4 rProtein is in the vicinity of S. aureus linezolid-binding (orange) and S. aureus BC-3205–binding (green) pockets. The structural variability of its loop (W65-Q75) in four species is shown. (E) SA50S rRNA A2058 and A2059 are main binding determinants of MLSBK family of antibiotics, represented here by erythromycin (PDB ID code 3OFR) (red). uL4 and bL32 rProteins form a second shell around the erythromycin-binding pocket next to A2058 and A2059. Structural variability of uL4 among D50S, E70S, T70S, and SA50S is shown. (F) S. aureus telithromycin (slate) conformation within its S. aureus complex, where its alkyl-aryl arm is folded such that it overlaps the desosamine sugar of erythromycin (red) in its complex with D50S. SA50tel uL22 (blue) is superimposed on the same rProtein in H. marismortui large ribosomal subunit with a deletion mutation Δ82–84 in uL22 (red) that allows protein progression even though it binds erythromycin (PDB ID code 1YJ9).
Structure of the SA50S-BC3205 Complex.
In the SA50S-BC3205 crystal structure, BC-3205 is bound at the PTC, so that the tricyclic mutilin core is blocking the A-site, and its C14 extension is pointing into the P-site, thus perturbing A- and P-site tRNA accommodation, as was seen in the ribosome-pleuromutilin complexes with D50S (35, 36). In the SA50S-BC3205 complex, the conformation of the flexible nucleotide U2585 is different from that of the unbound SA50S, and its conformational range is reduced because of partial overlap by the BC-3205 (Fig. 5E). In addition, U2506 is shifted toward the walls of the binding pocket, forming a hydrogen bond between its O5 carbonyl and the NH2 of the valyl moiety of BC-3205. An additional hydrogen bond is formed between the acetyl carbonyl and the NH2 of G2061. All of its other interactions with the rRNA nucleotides A2063, A2503, U2504, G2505, A2451, C2452, and U2585 are either hydrophobic or based on van der Waals forces.
Comparing the crystal structure of SA50S-BC3205 with those of D50S in complex with various pleuromutilins (35, 36) (Fig. 5F) revealed that all pleuromutilins bind to the same pocket, albeit by creating somewhat different interaction networks, including a shift of U2585. This shift is larger in SA50S-BC3205 compared with all other pleuromutilin complexes, so that the base of U2585 is located ∼6 Å away from its position in unbound SA50S. Similarly, a movement of U2506 toward the bound drug was observed in pleuromutilin complexes with D50S. Notably, in the SA50S-BC3205 complex, U2506 forms a hydrogen bond with the NH2 of the valyl moiety of the compound, and thus its shift is the largest. Consequently, the two sides of the BC-3205 molecule are held within its binding pocket by hydrogen bonds, compared with the single hydrogen bond created in the other pleuromutilin complexes with G2061, indicating a better fit of BC-3205 to its binding site. This additional interaction of BC-3205 seems to account for its greater potency against methicillin-resistant S. aureus–resistant strains and its low IC50 value in a S. aureus cell-free translation system.
Structural Analysis of Known S. aureus Resistance Mechanisms.
S. aureus linezolid resistance is caused mainly by the G2576U mutation (23), a nucleotide that is ∼98% conserved throughout all kingdoms of life (52). The G2576U resistance mechanism is in accord with all crystal structures of ribosome-linezolid complexes, as well as with a model of such a complex (46–48). These structures indicate that linezolid binds to the PTC, and that linezolid blocks the A-site within the PTC. Of interest is the G2576U mutation, which is located in the second shell of the PTC and is also associated with linezolid resistance in the multidrug-resistant bacteria Enterococcus faecium, Enterococcus faecalis (53), and S. pneumoniae, as well as in Mycobacterium smegmatis, the species used in the diagnosis of Mycobacterium tuberculosis (54). G2576 is located in the second shell around the linezolid-binding pocket in the PTC, in the vicinity of the uL3 rProtein. Compared with E70S, the uL3 loop G152-P170 in SA50S is bent toward the minor groove of H72 away from G2576, thereby permitting more flexibility (Fig. 6A).
S. aureus linezolid resistance is also caused by rRNA mutations U2500A (24), A2503G, U2504C, and G2447U (25) of nucleotides that are ∼98% conserved throughout all kingdoms of life (52). In SA50S, D50S, and H50S linezolid complexes, linezolid interacts with U2504 (46, 47). Mutating it to C will abolish the C:U noncanonical base pair interactions (55) with C2452, and thus likely change the shape of the linezolid-binding pocket. G2447 interacts with A2451; thus, the mutated G2447U may form a new base pair with A2451, which may limit A2451 flexibility and thereby influence drug binding. U2500 is base-paired with A2453. Its mutation to A will abolish the base pair interactions and consequently change the PTC environment (Fig. 6C) and prevent drug binding (56). The G2447U mutation, which renders M. smegmatis, M. tuberculosis (57), and E. coli (58) linezolid-resistant, has been suggested to be involved in functional differences between Gram-positive and Gram-negative bacteria (59). Of interest, significant structural similarity was observed in this region of ribosomes from T70S, D50S, E70S, and SA50S, which represent both Gram-positive and Gram-negative species.
Cross-resistance to linezolid, chloramphenicol, and dalfopristin (the streptogramin A component of Synercid) is associated with the second shell nucleotide G2576U mutation (10). Linezolid, chloramphenicol, and streptogramin A bind at the PTC via the first shell nucleotide G2505, which is stacked to U2576. In addition, cross-resistance of S. aureus to linezolid and chloramphenicol, of which the binding pockets partially overlap, may be caused by G2505A and U2500A mutations (60), even though the conservation level of these nucleotides is ∼98% throughout all kingdoms of life (52). These mutations indicate that despite its high conservation, this nucleotide is not essential for ribosomal function. Nucleotide G2505 is base-paired to C2610, and its ribose and phosphate are located in the vicinity of the antibiotic. The mutation G2505A leads to a mismatch in this base pair, thereby altering the antibiotic binding pocket. Similarly, U2500 is base-paired with A2453 and has hydrophobic interactions with U2504. The mutation U2500A disrupts the base pair U2500-A2453, and thus may allow U2504 to tilt away from the linezolid binding pocket (56) (Fig. 6B).
S. aureus-resistant mutations are also associated with the region of the uL3 rProtein located in proximity to the PTC (12–17, 61). The fold of uL3 loop G139-A150 is similar in SA50S to its folds in D50S and T70S, all bending toward H90 and H62, but is different in E70S, where it bends toward the H61 major groove (Fig. 6A). All of the mutated amino acids found in clinically resistant isolates are located in the uL3 region, which is adjacent to the third shell of nucleotides around the PTC, and appear to reshape the antibiotic-binding pocket (marked in yellow in Fig. 6A). Mutations in the uL3 loops in the vicinity of G2578 and C2575, which are part of the second shell nucleotides, may affect G2576 and its interactions with nucleotides G2505-U2506, which are part of the pleuromutilin- and linezolid-binding pockets (35, 46).
The uL3 loop F127-P170 is also located in the vicinity of A2572, which interacts with the flexible nucleotide U2504. In D50S and S. aureus, this nucleotide is located in the first shell around the linezolid- and tiamulin-binding pockets, and interacts with them (Fig. 6A). The nucleotide A2572 has been previously implicated in PTC antibiotic resistance acquired by induced-fit alterations that, although not directly interacting with the bound drugs, reshape the binding pocket via networks of remote interactions, most of which occur through the flexible nucleotide U2504 (56). In another S. aureus linezolid-resistant strain, a deletion of F127-H146 occurs in the rplC gene, which encodes uL3 rProtein (12). In wild-type uL3, this loop is located within the area described above, in the vicinity of 23S rRNA H90 nucleotides G2576-G2580, which reside in the second shell around the linezolid-binding pocket. Loop deletion may eliminate or alter interactions of uL3 with its proximal rRNA, and thus may reshape the drug-binding pocket and hinder antibiotic binding (Fig. 6A). Similar mutations in locations somewhat distal from the actual antibiotic-binding pockets that confer resistance by allosteric rearrangements have been detected in other systems as well (54, 62, 63).
A2058 and A2059, known as the main determinants for binding MLSBK antibiotics by direct interaction (5, 64), are in direct contact with the W65-Q75 loop of uL4 rProtein, which has diverse sequences in SA50S, E70S, T70S, and D50S (Fig. 6 D and E). Mutations in uL4 rProtein, G69A, and T70P produce S. aureus resistance to linezolid (65), whereas Q67K, G69E, G69A, and T70P mutations produce S. aureus resistance to erythromycin (15). In addition, ΔK68 and ΔG69 are susceptible to linezolid, and K68Q mutation produces S. aureus resistance to linezolid, pleuromutilins, chloramphenicol, and tedizolid (17). Because the uL4 W65-Q75 loop is located in the vicinity of the phosphate of A2059, which belongs to the MLSBK-binding pocket, the aforementioned mutations may alter the W65-Q75 loop conformation, affecting A2059 conformation (Fig. 6 D and E). In E70S, T70S, and SA50S structures, residue 72 of uL4 is Arg, whereas in D50S it is Asn. Each of these four side chains of residue 72 point to a different direction, creating a different environment around nucleotide A2059 in the four crystal structures compared. Residue K68 of uL4 is located in the vicinity of the phosphate of nucleotide G2061 and the sugar-phosphate backbone of A2059, which are in direct contact with linezolid (46). It has a similar orientation in E70S, SA50S and T70S, but different from that in D50S (Fig. 6D). The mutation K68Q replaces a long, positively charged side chain with a shorter, uncharged side chain with no electrostatic interactions with G2061 and A2059. G69A and T70P point mutations in uL4 introduce hydrophobic residues, which may increase the flexibility of the binding pocket, thereby reducing the efficiency of binding and explaining how resistance to linezolid is gained in these cases. All available National Center for Biotechnology Information sequences of uL4 rProtein indicate that its residue 68 lysine is highly conserved; however, in H50S, the respective residue is serine, a short polar uncharged residue, which seems to reduce binding efficiency. This may be an additional feature of the weaker binding mode of linezolid to H50S (46, 47).
Point mutations, deletions, and insertions in the uL22 hairpin loop that is part of the exit tunnel wall in residues R80-S108 in S. aureus confer resistance to erythromycin, Synercid, and telithromycin, although its location is too far to allow direct chemical interactions with the bound drugs (18, 19). Importantly, similar resistance mutations were also observed in additional bacterial species. The crystal structure of one of these mutants of a H. marismortui large ribosomal subunit with a deletion mutation Δ82–84 in uL22 demonstrates that the deletion triggers a shift in the position of the uL22 hairpin loop and it is located deeper in the tunnel (28). It is conceivable that similar mechanisms play roles in acquiring resistance in S. aureus deletion/insertion mutants.
Methylation of the C8 position of A2503 in S. aureus by the Cfr enzyme leads to multidrug resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (PhLOPSA) (26, 66, 67). The A2503 nucleotide interacts with linezolid and is ∼98% conserved throughout all kingdoms (52); nevertheless, it can be modified without interfering with protein biosynthesis. A2503 methylation of C8 by Cfr, which increases its size, interferes with PhLOPSA drug binding, thereby conferring resistance to these drugs. In addition, hindering the C2 position of A2503 indigenous methylation by inactivating the gene of the methyltransferase RlmN renders resistance to linezolid, because a lack of methyl substitution may change its orientation with respect to the drug (25, 68) (Fig. 6 A and B).
Summary and Concluding Remarks.
The sequence and structure alignments of S. aureus large ribosomal subunit rRNA with other eubacteria indicate high conservation. Nevertheless, diversity was identified in several locations, some of which may serve as antibiotic target sites. Furthermore, structural characterization of this ribosomal particle and of its complexes with linezolid, telithromycin, and BC-3205 revealed minute but nonetheless significant differences. In this respect, the superiority of the new pleuromutilin compound BC-3205 over known veterinary pleuromutilins or retapamulin, as demonstrated by its strong binding to S. aureus ribosome and low minimum inhibitory concentration, is clearly supported by the structural elements of its binding mode.
Our structural analysis identified structural motifs within the particle core and on its periphery that do not belong to the known binding sites of antibiotics but may be candidates for the design of novel selective drugs against S. aureus, thus enriching the pool of potential antibiotic compounds. Several of these features are shown in Fig. 4 and highlighted in Fig. S5. Some of these features may provide inhibitory mechanisms similar to that of the periphery binding site of RsfS that inhibits protein synthesis by interfering with the association of the two subunits (69, 70).
Analysis of the structures of the unbound and drug-bound ribosomes vs. ribosome structures from other known eubacteria identified specific structural motifs that may indicate possible features involved in species-specific responses to antibiotics. Thus, our novel insights provide unique structural tools for discriminating between pathogenic bacteria and the microbiome, namely the useful bacterial species within the human body, mainly in the gut flora.
Methods
S. aureus Growth and Cell Wall Disruption.
S. aureus strain RN4220 (American Type Culture Collection 35556) (71) was grown overnight at 37 °C. Cells were harvested at an OD600 of ∼1.5. The bacterial culture was centrifuged three times at 5,000 rpm using a GS-3 rotor at 4 °C for 15 min, after which the supernatant was discarded. The pellet was resuspended and centrifuged in a tabletop centrifuge at 4,000 rpm using a GS-3 rotor at 4 °C for 10 min. The supernatant was then discarded, and the wet pellets (cells) were weighed. To disrupt the cell membranes, the samples were resuspended in 10 mM Tris-acetate buffer pH 8.0, 14 mM Mg-acetate, 1 M KCl, 1 mM DTT, and 50 µg/mL Lysostaphin, then incubated at 37 °C for 1 h and periodically inverted during incubation. The lysates were centrifuged at 16,000 rpm using a SS34 rotor, at 4 °C for 30 min to remove cell debris. The supernatants were incubated in 670 mM Tris-acetate buffer pH 8.0, 20 mM Mg-acetate, 7 mM DTT, 7 mM Na3-phosphoenolpyruvate, 5.5 mM ATP, 70 mM of each amino acid, and 1.9 mg of pyruvatkinase at 37 °C for 30 min and then dialyzed overnight at 4 °C against dialysis in 10 mM Tris-acetate buffer pH 8.0, 14 mM Mg-acetate, 60 mM K-acetate and 1 mM DTT. The extract was then flash-frozen and stored at −80 °C until use.
S. aureus Ribosome Purification.
Cell extract was layered on a 1.1 M sucrose cushion (72), H10M15N100K50βMe6 pH 8.0 (10 mM Hepes pH 8.0, 15 mM MgCl2, 100 mM NH4Cl, 50 mM KCl, and 6 mM β-mercaptoethanol), and ultracentrifuged twice, each time at 55,000 rpm using a Ti-70 rotor at 4 °C for 17 h. The supernatant was then discarded, and the pellet was dissolved in H10M15N150βMe6 buffer pH 8.0 at 4 °C. Ribosomal subunits were then separated by zonal ultracentrifugation, using a Ti-15 zonal rotor with a gradient of 8–40% sucrose, at low Mg2+ concentration (1 mM MgCl2) for 17.5 h at 27,000 rpm. After separation, the Mg2+ concentration was adjusted to 10 mM, and the ribosomal subunits fractions were concentrated using sequential centrifugations. The samples were kept in H10M10N60K15 at pH 7.6, brought to a final concentration not exceeding 1,000 A260 mL−1, and then flash-frozen for storage at −80 °C.
Ribosome Activity Assay.
S. aureus ribosome activity was determined in a bacterial-coupled transcription/translation assay system, which measures the expression of the luciferase gene (73).
Crystallization of the SA50S Subunit.
Crystals of SA50S were obtained at 20 °C by the hanging-drop vapor diffusion technique. The crystallization drop contained 0.166% MPD, 0.333% EtOH, H20M10N60K15 (pH range, 6.8–7.8), 5 mM spermidine, 0.5 mM MnCl2, and 1–1.6 mg/mL SA50S subunits. The reservoir solution contained 15% of 1:2 ethanol-MPD and H110M10N60K15 buffer (pH range, 6.8–7.8). The SA50S subunits were heat-activated for 30 min at 37 °C before crystallization. These conditions usually yield ∼60- to 300-µm hexagonal crystals, which appear as hexagons. High-resolution diffracting crystals were obtained by macro seeding, using crystals that had been fished out of the crystallization drop; washed in 10 µL of 7.5% EtOH, 7.5% MPD, H110M10N60K15 buffer, and 0.5 mM MnCl2; and seeded for 24 h in a preequilibrated crystallization drop. SA50S crystals were kept in stabilization solution consisting of 15% MPD, 15% EtOH, H110M10N60K15 buffer (pH range, 6.8–7.8), and 0.5 mM MnCl2.
Crystals of SA50S Complexes.
For obtaining SA50S antibiotic complexes, SA50S crystals were soaked in solutions containing 11.4–22.7 µg/mL BC-3205 in the stabilization solution for 3–6 h before flash-freezing and data collection.
Data Collection and Processing.
Before data collection, the crystals were immersed in cryoprotectant solution of 20% MPD, 15% EtOH, H110M10N60K15 buffer, and 0.5 mM MnCl2. Crystallographic data were collected at the ID23-1, ID23-2, and ID-29 beamlines at the European Synchrotron Radiation Facility in Grenoble, France. X-ray diffraction data were collected from the hexagonal crystals at 100K. Up to 15 crystals were needed to yield complete datasets of SA50S using 0.1° oscillations. Data were processed with HKL2000 (74) and the CCP4 package suite (75).
Map Calculation, Model Building, and Refinement.
The structures were determined using PHASER for molecular replacement, implemented in PHENIX, with the D50S structure (PDB ID code 2ZJR) used as a starting model. Once initial phases were obtained, rigid body and positional refinement were performed using Phenix.refine (76) and CNS (77). For R-free calculations during refinement cycles, a random 5% of the data were omitted during refinement cycles. Tracing the ribosomal RNA and remodeling the ribosomal proteins with S. aureus strain NCTC8325 sequence (78) according to the electron density maps was performed using Coot (79, 80), Rosetta ERRASER (81) was used to facilitate further building and to improve the quality of the rRNA geometry. Figures were generated using Pymol (82) and Chimera (83). Sequence alignments were performed using BLAST (84), and structure alignments were done using LSQMAN (85) and Coot.
SI Results and Discussion
Several Cases of Structural Variability in Bacterial rRNA Nucleotides.
Within the core of the ribosome, the rRNA fold is mostly conserved in the four eubacterial ribosome structures, including the previously identified flexible nucleotides (with variable conformations) at the PTC (U2506, U2585, A2062, A2602, and U2491) and at the exit tunnel (A508 and A90-91) (Fig. S6).
Several Cases of Structural Differences in rProteins.
Relative positions of the globular domains of the ribosomal proteins on the subunit surface are shown in Fig. S8. Note that almost all of the ribosomal proteins contain N- and/or C-terminal extensions, as well as extended internal loops (both types may reach 70–80 Å in length), which penetrate into the ribosome core and interact with and presumably stabilize the ribosomal RNA. These extensions contain most of the structural variability.
Fig. S8.
Surface of the SA50S structure indicating the locations of the globular regions of the rProteins. rRNA is shown in gray, and rProteins are shown in different colors. (Upper, Left) View from the SA50S intersubunit surface. (Upper, Right) View from the SA50S outer surface. (Lower) +900 and −900 vertical rotation of the intersubunit surface view.
The Nascent Protein Exit Tunnel.
The exit tunnel, approximately 100 Å long, is of nonuniform shape with a variable diameter. It contains a constriction located roughly five to seven peptide bonds away from its entrance, which is shaped at its far end by the hairpin loops of rProteins uL4 and uL22, which reach the tunnel walls in all four compared structures despite some slight sequence and structural variability (Fig. S4A). The uL4 loop of S. aureus is more hydrophobic compared with the other bacterial species that we evaluated. From the opposite tunnel wall, the uL22 hydrophobic hairpin loop has a similar fold in all structures compared; however, the orientations of R92 (R/S/R in T70S, D50S, and E70S) differ somewhat in the four structures. The macrolide-binding site is proximal to this constriction.
The N terminals of bL32 in D50S and T70S are elongated relative to those in SA50S and E70S (Fig. 3A and Fig. S4A and B), thus creating a void proximal to the macrolides binding site. This void may be exploited for potential new or improved drugs (i.e., extended macrolides).
In the vicinity of the tunnel opening, structural variations were observed in ribosomal proteins uL23, uL29, uL24, bL32, and uL22, creating different surfaces for the interactions of ribosomes with cellular factors, such as the translocon, endoplasmic reticulum (ER) membranes, chaperons, and so on. The uL23 K63-Y71 loop that is pointing into the tunnel displays a different conformation in SA50S and T70S than in E70S and D50S. Specifically, in the four compared structures, residue R67 (Q in E70S) points into the tunnel in different orientations. The tyrosine in D50S stacks to the A508 nucleotide, but A508 has a different conformation in each of the four structures, highlighting its flexibility (Fig. S6B). The uL23 C terminus of SA50S is similar in length to its mates in D50S and T70S, but its fold is different from that of E70S, which has an extended C-terminal tail. Consequently, this region of the subunit surface that interacts with other cellular components, such as the ER membrane or chaperons, is altered among the four structures examined (Fig. S4B).
The uL24 N terminus interacts with H7 and with the junction of H18, H19, and H20 of the 23S rRNA. The D50S N terminus is 10–13 residues longer than that in the other three structures. The SA50S and T70S N terminals are shorter (the SA50S N terminal’s first two amino acids could not be traced owing to an uninterpretable density) and the E70S N terminus is the shortest. The uL24 I24-V34 loop in E70S folds differently from that in SA50S, D50S, and T70S, whereas the K43-G55 loop in SA50S folds similar to that in T70S, exhibiting an extended loop different from that in E70S and D50S. In addition, the 5′ end of the uL24 T79-I93 loop and the C-terminal tail are highly divergent among the four structures (Fig. S4C). The L37-T40 loop of rProtein uL29 is extended in E70S compared with in the three other structures (Fig. S4B).
Subunit Core and the Central Protuberance.
The ribosomal proteins’ globular regions are located on the surface of the large ribosome subunit. Their long extended tails and loops penetrate into the subunit core and interact with the rRNA. Differences in proteins that interact with the rRNA may result in allosteric alterations in the nucleotides (36). Here we describe several substantial structural variabilities detected in ribosomal core proteins. uL14 loop L25-S28 is extended in D50S compared with in SA50S, E70S, and T70S and has a unique interaction with H95, in addition to the common interaction with H90. The eL15 C terminus is connected to a long extended strand with fold variation among the structures that we compared (with SA50S the shortest), which penetrates into the subunit’s core (Fig. S4D), interacting with protein bL35, which has a similar fold in the four structures compared.
The central protuberance lying on the surface of the 50S subunit is a subcomplex of the 5S rRNA with rProteins uL5, uL18, bL27, and bL25 (called TL5 in T50S and CTC in D50S). This rProtein interacts with the 5S rRNA. In E70S, rProtein bL25 has a single domain, in SA50S and T70S it has two domains, and in D50S the CTC protein contains three domains (86). The bL25 first domain of SA50S and E70S has an extended G11-L26 loop that interacts with 5S rRNA. The F79-I86 loops that interact with the 23S rRNA have different backbone folds and orientations in the four structures. The C terminus of uL16 R134-T141 reaches the bL25 domain’s interface and 5S rRNA. The C terminus of uL16 is longer in T70S and penetrates deeper into the domain’s interface; thus, the angle between the two TL5 domains is larger compared with that in D50S and SA50S (Fig. S4F). The uL18 N-terminal in SA50S the longest modeled relative to E70S, D50S, and T70S, and reaches rProtein uL5 which is located on the other side of the 5S rRNA (Fig. S4D). D50S and T70S N terminals have been traced only from residues 8 and 11, respectively. rProtein bL27 has a similar structure in all four structures apart from a variability in the fold of R79-K85 loop that reaches the 5S rRNA in all structures (Fig. S4G). Structural variations of the C terminus that reaches the subunit surface are observed (Fig. S4G), in addition to the other differences in the N-terminal tail mentioned in the main text.
The Intersubunit Interface and Subunit Outer Surface.
The fold of the N terminus of protein uL2 in SA50S is somewhat different from that in E70S, in which the uL2 of E. coli is longer. Of interest in the D50S structure, 32 residues of protein uL2 are not traced. rProtein uL5 participates in the B1 (42) intersubunit bridge with the 30S subunit, together with rProtein uS13. By superimposing the SA50S subunit on the corresponding part of the T70S structure, we identified structural variability in the M38-A53 and G126-D144 loops of uL5 that interact with rProtein uS13 in the 70S ribosome, embracing the uS13 interface helix in all four organisms (Fig. S7E).
Among the structural differences identified in the structures of the large ribosomal subunits located on their surfaces (Table S1) are the N and C terminals of uL29, which reach the subunit surface and have variable folds in the four structures. The N termini are extended in T70S and E70S compared with their folds in SA50S and D50S structures, whereas the C termini are of similar length but with variable folds.
The uL4 of SA50S has an extended V6-I17 surface loop compared with that in E70S and D50S, which in T70S adopts a different orientation (Fig. S7A). In addition, in SA50S the uL4 N-terminal has a different backbone fold, Q119-E132 (Fig. S7C), and a different structure and orientation of its C-terminal tail (I186-T195) (Fig. S7B) compared with E70S, T70S, and D50S.
The SA50S uL15 C-terminal domain is located on the surface of the 50S subunit. Its loops I69-T89 and T89-V97 have different structures compared with the corresponding loops of E70S, but are similar to the T70S and D50S C-terminal domains (Fig. S7D). This region is close to symmetry-related molecules within the crystal packing of SA50S, E70S, and T70S and thus may influence the loop conformation. Fig. S7A shows the surface shared by uL4 and uL15, and the overall structural differences among the four different crystal structures. The uL15 N terminus is the longest in E70S, shorter in D50S (traced from the fourth amino acid), and much shorter in T70S (traced from the fifth amino acid). It is the shortest in SA50S (traced from the first amino acid), but its 70-aa extended N-terminal tail intercalates differently with the 23S rRNA of the four structures compared (Fig. S7A).
Furthermore, although the bL19 N termini of SA50S and D50S are positioned differently, both interact with H101, whereas in D50S, the bL19 N terminus interacts with H100 and bL3, and in T70S and E70S, these N termini are shorter. In the SA50S bL21 N terminus, the K24-D31 and G43-V58 loops have a slightly different backbone fold than in the other three structures. The N terminus of D50S uL22 is 22 residues longer than that of SA50S, T70S, and E70S, resulting in a rather different surface of the D50S subunit. The uL30 N terminus located on the particle’s outer surface is positioned differently in SA50S and T50S structures compared with E70S, whereas the D50S N terminus is shorter. The uL30 C terminus, which also reaches the subunit outer surface, is longer in T70S compared with the other three structures.
Acknowledgments
We thank Moran Ben-Ami, Elinor Breiner-Goldshtein, Rufayda Murwat, Ophir Pick, Shoshana Tel- Or, Miriam Lachever, Maggie Kessler, and Yaacov Halfon for their interest and experimental support; Astrid Gruss for S. aureus ribosome preparation; Moshe Peretz, Linda Shimon, and Yael Diskin-Posner for skillful services; staff members at Beamlines ID23-1 and ID23-2 of the European Synchrotron Radiation Facility/European Molecular Biology Laboratory for their assistance during data collection. Funding was provided by European Research Council Grants 322581 (NOVRIB) and POC 632167 (PATRES), an Advanced Merieux Research Grant, and the Kimmelman Center for Macromolecular Assemblies. M.K. is a recipient of an Adams Fellowship. A.Y. holds the Martin S. and Helen Kimmel Professorial Chair at the Weizmann Institute of Science.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 4WCE, 4WFB, 4WF9, and 4WFA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517952112/-/DCSupplemental.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Kurosu M, Siricilla S, Mitachi K. Advances in MRSA drug discovery: Where are we and where do we need to be? Expert Opin Drug Discov. 2013;8(9):1095–1116. doi: 10.1517/17460441.2013.807246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization . Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; Geneva: 2014. [Google Scholar]
- 4.Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7(2):178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase center in eubacteria. Nature. 2001;413(6858):814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 6.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16(5):528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5-Å resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DN. On the specificity of antibiotics targeting the large ribosomal subunit. Ann N Y Acad Sci. 2011;1241(1):1–16. doi: 10.1111/j.1749-6632.2011.06192.x. [DOI] [PubMed] [Google Scholar]
- 9.Yonath A. Antibiotics targeting ribosomes: Resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 10.Besier S, Ludwig A, Zander J, Brade V, Wichelhaus TA. Linezolid resistance in Staphylococcus aureus: Gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob Agents Chemother. 2008;52(4):1570–1572. doi: 10.1128/AAC.01098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wada A, Ohta H, Kulthanan K, Hiramatsu K. Molecular cloning and mapping of 16S-23S rRNA gene complexes of Staphylococcus aureus. J Bacteriol. 1993;175(22):7483–7487. doi: 10.1128/jb.175.22.7483-7487.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long KS, Vester B. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother. 2012;56(2):603–612. doi: 10.1128/AAC.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller K, Dunsmore CJ, Fishwick CW, Chopra I. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob Agents Chemother. 2008;52(5):1737–1742. doi: 10.1128/AAC.01015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentry DR, Rittenhouse SF, McCloskey L, Holmes DJ. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob Agents Chemother. 2007;51(6):2048–2052. doi: 10.1128/AAC.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosowska-Shick K, et al. Single- and multistep resistance selection studies on the activity of retapamulin compared to other agents against Staphylococcus aureus and Streptococcus pyogenes. Antimicrob Agents Chemother. 2006;50(2):765–769. doi: 10.1128/AAC.50.2.765-769.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endimiani A, et al. Emergence of linezolid-resistant Staphylococcus aureus after prolonged treatment of cystic fibrosis patients in Cleveland, Ohio. Antimicrob Agents Chemother. 2011;55(4):1684–1692. doi: 10.1128/AAC.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Locke JB, Hilgers M, Shaw KJ. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700) Antimicrob Agents Chemother. 2009;53(12):5265–5274. doi: 10.1128/AAC.00871-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentry DR, Holmes DJ. Selection for high-level telithromycin resistance in Staphylococcus aureus yields mutants resulting from an rplB-to-rplV gene conversion-like event. Antimicrob Agents Chemother. 2008;52(3):1156–1158. doi: 10.1128/AAC.00923-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malbruny B, et al. Resistance to quinupristin-dalfopristin due to mutation of L22 ribosomal protein in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2200–2207. doi: 10.1128/AAC.46.7.2200-2207.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory ST, Dahlberg AE. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J Mol Biol. 1999;289(4):827–834. doi: 10.1006/jmbi.1999.2839. [DOI] [PubMed] [Google Scholar]
- 21.Kloss P, Xiong L, Shinabarger DL, Mankin AS. Resistance mutations in 23 S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J Mol Biol. 1999;294(1):93–101. doi: 10.1006/jmbi.1999.3247. [DOI] [PubMed] [Google Scholar]
- 22.Zurenko GE, et al. In vitro activities of U-100592 and U-100766, novel oxazolidinone antibacterial agents. Antimicrob Agents Chemother. 1996;40(4):839–845. doi: 10.1128/aac.40.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsiodras S, et al. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet. 2001;358(9277):207–208. doi: 10.1016/S0140-6736(01)05410-1. [DOI] [PubMed] [Google Scholar]
- 24.Meka VG, et al. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J Infect Dis. 2004;190(2):311–317. doi: 10.1086/421471. [DOI] [PubMed] [Google Scholar]
- 25.Livermore DM, Warner M, Mushtaq S, North S, Woodford N. In vitro activity of the oxazolidinone RWJ-416457 against linezolid-resistant and -susceptible staphylococci and enterococci. Antimicrob Agents Chemother. 2007;51(3):1112–1114. doi: 10.1128/AAC.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toh S-M, et al. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol. 2007;64(6):1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berisio R, et al. Structural insight into the antibiotic action of telithromycin against resistant mutants. J Bacteriol. 2003;185(14):4276–4279. doi: 10.1128/JB.185.14.4276-4279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121(2):257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci USA. 2010;107(40):17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci USA. 2010;107(40):17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kavanagh F, Hervey A, Robbins WJ. Antibiotic substances from Basidiomycetes, VIII: Pleurotus multilus (Fr.) Sacc. and Pleurotus passeckerianus Pilat. Proc Natl Acad Sci USA. 1951;37(9):570–574. doi: 10.1073/pnas.37.9.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egger H, Reinshagen H. New pleuromutilin derivatives with enhanced antimicrobial activity, II: Structure–activity correlations. J Antibiot (Tokyo) 1976;29(9):923–927. doi: 10.7164/antibiotics.29.923. [DOI] [PubMed] [Google Scholar]
- 33.Parish LC, et al. SB275833/032 Study Team Topical retapamulin ointment (1%, wt/wt) twice daily for 5 days versus oral cephalexin twice daily for 10 days in the treatment of secondarily infected dermatitis: Results of a randomized controlled trial. J Am Acad Dermatol. 2006;55(6):1003–1013. doi: 10.1016/j.jaad.2006.08.058. [DOI] [PubMed] [Google Scholar]
- 34.Free A, et al. Retapamulin ointment twice daily for 5 days vs oral cephalexin twice daily for 10 days for empiric treatment of secondarily infected traumatic lesions of the skin. Skinmed. 2006;5(5):224–232. doi: 10.1111/j.1540-9740.2006.05774.x. [DOI] [PubMed] [Google Scholar]
- 35.Schlünzen F, Pyetan E, Fucini P, Yonath A, Harms JM. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol Microbiol. 2004;54(5):1287–1294. doi: 10.1111/j.1365-2958.2004.04346.x. [DOI] [PubMed] [Google Scholar]
- 36.Davidovich C, et al. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci USA. 2007;104(11):4291–4296. doi: 10.1073/pnas.0700041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biedenbach D, et al. 2009. In vitro antibacterial spectrum of BC-3205, a novel pleuromutilin derivative for oral use in humans. Available at: www.jmilabs.com/data/posters/ICAAC2009/F1-1513.pdf. Accessed June 15, 2015.
- 38.Prince WT, et al. Phase II clinical study of BC-3781, a pleuromutilin antibiotic, in the treatment of patients with acute bacterial skin and skin structure infection. Antimicrob Agents Chemother. 2013;57(5):2087–2094. doi: 10.1128/AAC.02106-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paukner S, et al. Antimicrobial activity of the pleuromutilin antibiotic BC-3781 against bacterial pathogens isolated in the SENTRY antimicrobial surveillance program in 2010. Antimicrob Agents Chemother. 2013;57(9):4489–4495. doi: 10.1128/AAC.00358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30(1):26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Noeske J, et al. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22(4):336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 43.Beinsteiner B, Michalon J, Klaholz BP. 2015. IBiSS, a versatile and interactive tool for integrated sequence and 3D structure analysis of large macromolecular complexes. Bioinformatics pii: btv347. [DOI] [PubMed]
- 44.Wall EA, et al. Specific N-terminal cleavage of ribosomal protein L27 in Staphylococcus aureus and related bacteria. Mol Microbiol. 2014;95(2):258–269. doi: 10.1111/mmi.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. A protein component at the heart of an RNA machine: The importance of protein l27 for the function of the bacterial ribosome. Mol Cell. 2005;20(3):427–435. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Wilson DN, et al. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci USA. 2008;105(36):13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ippolito JA, et al. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem. 2008;51(12):3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- 48.Leach KL, et al. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell. 2007;26(3):393–402. doi: 10.1016/j.molcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Bashan A, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11(1):91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 50.Belousoff MJ, et al. Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit. Proc Natl Acad Sci USA. 2011;108(7):2717–2722. doi: 10.1073/pnas.1019406108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Canu A, et al. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2002;46(1):125–131. doi: 10.1128/AAC.46.1.125-131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cannone JJ, et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marshall SH, Donskey CJ, Hutton-Thomas R, Salata RA, Rice LB. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 2002;46(10):3334–3336. doi: 10.1128/AAC.46.10.3334-3336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long KS, et al. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross-resistance. Antimicrob Agents Chemother. 2010;54(11):4705–4713. doi: 10.1128/AAC.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reichert J, Sühnel J. The IMB Jena Image Library of biological macromolecules: 2002 update. Nucleic Acids Res. 2002;30(1):253–254. doi: 10.1093/nar/30.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davidovich C, Bashan A, Yonath A. Structural basis for cross-resistance to ribosomal PTC antibiotics. Proc Natl Acad Sci USA. 2008;105(52):20665–20670. doi: 10.1073/pnas.0810826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee M, et al. Linezolid for treatment of chronic extensively drug-resistant tuberculosis. N Engl J Med. 2012;367(16):1508–1518. doi: 10.1056/NEJMoa1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong L, et al. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J Bacteriol. 2000;182(19):5325–5331. doi: 10.1128/jb.182.19.5325-5331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sander P, et al. Ribosomal and non-ribosomal resistance to oxazolidinones: Species-specific idiosyncrasy of ribosomal alterations. Mol Microbiol. 2002;46(5):1295–1304. doi: 10.1046/j.1365-2958.2002.03242.x. [DOI] [PubMed] [Google Scholar]
- 60.North SE, et al. (2005) Chloramphenicol-selected mutants of Staphylococcus aureus may show cross-resistance to linezolid (abstract C1-1417). Program and abstracts of the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy (American Society for Microbiology, Washington, DC), Vol 81.
- 61.Klitgaard RN, et al. Mutations in the bacterial ribosomal protein l3 and their association with antibiotic resistance. Antimicrob Agents Chemother. 2015;59(6):3518–3528. doi: 10.1128/AAC.00179-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wittmann HG, et al. Biochemical and genetic studies on two different types of erythromycin-resistant mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet. 1973;127(2):175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- 63.Zaman S, Fitzpatrick M, Lindahl L, Zengel J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol Microbiol. 2007;66(4):1039–1050. doi: 10.1111/j.1365-2958.2007.05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harms JM, Schlünzen F, Fucini P, Bartels H, Yonath A. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2004;2:4. doi: 10.1186/1741-7007-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Román F, et al. Detection of linezolid-resistant Staphylococcus aureus with 23S rRNA and novel L4 riboprotein mutations in a cystic fibrosis patient in Spain. Antimicrob Agents Chemother. 2013;57(5):2428–2429. doi: 10.1128/AAC.00208-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. A new mechanism for chloramphenicol, florfenicol, and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol Microbiol. 2005;57(4):1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 67.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother. 2006;50(7):2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.LaMarre JM, Howden BP, Mankin AS. Inactivation of the indigenous methyltransferase RlmN in Staphylococcus aureus increases linezolid resistance. Antimicrob Agents Chemother. 2011;55(6):2989–2991. doi: 10.1128/AAC.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Häuser R, et al. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8(7):e1002815. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X, et al. Structure of ribosomal silencing factor bound to Mycobacterium tuberculosis ribosome. Structure. 2015 doi: 10.1016/j.str.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 72.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 73.Murray RW, Melchior EP, Hagadorn JC, Marotti KR. Staphylococcus aureus cell extract transcription-translation assay: Firefly luciferase reporter system for evaluating protein translation inhibitors. Antimicrob Agents Chemother. 2001;45(6):1900–1904. doi: 10.1128/AAC.45.6.1900-1904.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Otwinowski Z, Minor W. Processing of X-ray diffraction data. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 75.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 78.Gillaspy AF, et al. The Staphylococcus aureus NCTC 8325 genome. In: Fischetti WA, et al., editors. Gram-Positive Pathogens. 2nd Ed American Society for Microbiology; Washington, DC: 2006. [Google Scholar]
- 79.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 80.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chou F-C, Sripakdeevong P, Dibrov SM, Hermann T, Das R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat Methods. 2013;10(1):74–76. doi: 10.1038/nmeth.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schrodinger, LLC 2010. The PyMOL Molecular Graphics System, version 1.3r1. Available from: www.pymol.org. Accessed June 15, 2015.
- 83.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 84.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 85.Kleywegt GJ, Jones TA. Where freedom is given, liberties are taken. Structure. 1995;3(6):535–540. doi: 10.1016/s0969-2126(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 86.Harms J, et al. High-resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107(5):679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]