Significance
We developed antimicrobial polypeptides (AMPs) with unprecedented radial amphiphilicity. Unlike typical AMPs characterized by facial amphiphilicity or biomimetic antimicrobial polymers with randomly distributed charged and hydrophobic groups, this class of AMPs is made up of homo-polypeptides that feature a radially amphiphilic (RA) structure and adopt a stable α-helical conformation with a hydrophobic helical core and a charged exterior shell, which is formed by flexible hydrophobic side chains with terminal charge group. The RA polypeptides appear to offer several advantages over conventional AMPs with regard to stability against proteases and simplicity of design. They also exhibit high antibacterial activity against both gram-negative and gram-positive bacteria and low hemolytic activity. This design may become a general platform for developing AMPs to treat drug-resistant bacteria.
Keywords: antimicrobial peptide, α-helix, polypeptides, radial amphiphilicity, bacteria
Abstract
α-Helical antimicrobial peptides (AMPs) generally have facially amphiphilic structures that may lead to undesired peptide interactions with blood proteins and self-aggregation due to exposed hydrophobic surfaces. Here we report the design of a class of cationic, helical homo-polypeptide antimicrobials with a hydrophobic internal helical core and a charged exterior shell, possessing unprecedented radial amphiphilicity. The radially amphiphilic structure enables the polypeptide to bind effectively to the negatively charged bacterial surface and exhibit high antimicrobial activity against both gram-positive and gram-negative bacteria. Moreover, the shielding of the hydrophobic core by the charged exterior shell decreases nonspecific interactions with eukaryotic cells, as evidenced by low hemolytic activity, and protects the polypeptide backbone from proteolytic degradation. The radially amphiphilic polypeptides can also be used as effective adjuvants, allowing improved permeation of commercial antibiotics in bacteria and enhanced antimicrobial activity by one to two orders of magnitude. Designing AMPs bearing this unprecedented, unique radially amphiphilic structure represents an alternative direction of AMP development; radially amphiphilic polypeptides may become a general platform for developing AMPs to treat drug-resistant bacteria.
Antimicrobial peptides (AMPs) typically contain ∼40–60 amino acids, consisting of both cationic and hydrophobic amino acids. They adopt various secondary structures (e.g., α-helix) and can kill a range of bacteria. As AMPs target generic and necessary lipid components of bacterial membranes (1, 2) and depend less on specific bacterial metabolic status (3–6), development of resistance has been slow. Because of this feature, AMPs have attracted significant attention as potential antimicrobial agents clinically. Among all AMPs developed, the α-helical peptides are the most heavily investigated and generally are facially amphiphilic (FA) in structure with the cationic and hydrophobic amino acids separated to opposite faces of the helix. This structure correlates well to antimicrobial activity (6–10). Recent work has shown how amino acid content of AMPs enables this activity via specific types of membrane curvature generation (11–13).
Despite extensive effort, the commercial development of AMPs has seen limited success, in part due to drawbacks native to peptides. Although much has been learned from fundamental studies on AMP mechanisms (1, 2, 14, 15), precise, quantitative predictions of an AMP’s activity, therapeutic index, and antimicrobial activity compared with off-target eukaryotic cytotoxicity are currently impossible. Designing new AMP-related antibiotics relies on sequence-controlled peptide synthesis and parameter optimization, which is expensive and labor-intensive. Another drawback of most AMP-derived antibiotics is their poor stability in biological systems. LL-37 and magainin, for example, can be degraded by proteases in several minutes in blood circulation and lose their antimicrobial activities (16, 17). Finally, an important drawback of AMPs is related to their FA structure with an exposed hydrophobic helix face (Fig. 1A), which leads to undesired polypeptide interactions with blood proteins and self-aggregation. AMP helix bundles have been reported to cause substantially reduced antimicrobial activity (18, 19).
Fig. 1.
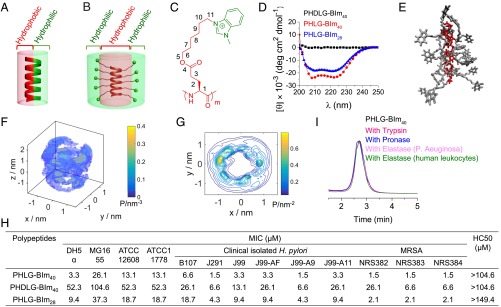
Schematic illustration of (A) FA AMP and (B) RA AMP. (C) The chemical structure of PHLG-BIm. (D) CD spectra of PHLG-BIm40, PHDLG-BIm40, and PHLG-BIm28 in aqueous solution at pH = 7. (E) RA peptide structure predicted from all-atom molecular dynamics simulations. Representative peptide snapshot from our simulation trajectory rendered using VMD. For clarity of viewing, the water molecules have been removed and the backbone and side chains colored in red and gray, respectively. (F) The 3D probability distribution of the side chain N12 atoms around the α-helical backbone. A fly-around of this image is presented in Movie S3. (G) A 2D projection of the 3D probability distribution in F onto a plane perpendicular to the long axis of the peptide. The probability density lies in the range 0–0.83 nm−2 and 20 evenly spaced contours plotted. (H) The antibacterial and hemolytic activity of polypeptides. The antibacterial activity of polypeptides was determined using MIC. The hemolytic activity of polypeptides was determined by HC50 (50% hemolytic concentration) value. (I) The stability of RA polypeptide PHLG-BIm40 when incubated with trypsin, pronase, elastase from P. aeruginosa or elastase from human leukocytes for 8 h.
There has been significant interest in developing AMP analogs, such as β-peptides (20–23), α/β-peptides (24, 25), peptoids (26), and aromatic oligomers (15, 27–30). These compounds have demonstrated improved stability over conventional AMPs. They are in general sequence specific and often require solid-phase peptide synthesis, thereby sharing similar advantages and development drawbacks as AMPs. There has also been growing interest in synthetic polymer-based AMP mimics bearing both cationic and hydrophobic groups, which can be prepared through cost-effective polymerization processes. For example, simplified polymeric AMP analogs have been developed, including poly(methacrylamides) (31), poly(β-lactams) (14, 32–35), polypeptides (36–38), poly(norborenes) (39, 40), and poly(carbonates) (41, 42). These compounds are considerably less expensive than peptides, and much work is being done to optimize them.
Herein, we envision a fundamentally different design of AMPs with radially amphiphilic (RA) structure (Fig. 1B) rather than FA structure (Fig. 1A). This class of antimicrobial homo-polypeptides features a hydrophobic helical core covered with cationic groups in all radial directions of the helix. Because the charged groups form the outer shell of the polypeptides, shielding the hydrophobic helical core, these peptides should have minimal hydrophobic force induced self-aggregation and reduced interaction with blood proteins. With such RA structure, the polypeptide backbone amide bonds should also be well protected and are expected to have improved stability against proteolytic degradation compared with a typical α-helical AMP.
Results
RA Polypeptide Displays High Antibacterial Activity and Selectivity.
We developed a class of helical, charged polypeptides with long hydrophobic side chains and terminal charge groups that demonstrated remarkable helical stability against changes in pH, salts, temperature, and various denaturing conditions (43–45). We adopted this architecture to design RA polypeptides and explored their applications as AMPs. PHLG-BIm (Fig. 1C) was a specific cationic α-helical polypeptide with RA structure. It was synthesized through ring-opening polymerization of amino acid N-carboxyanhydrides (NCAs) (46), followed by amination with 1-methylbenzimidazole (SI Appendix, Scheme S1). With a separation of 11 σ-bonds between the backbone and positive charges, PHLG-BIm, with DP of 40 and 28, adopts α-helical conformation. The helical conformation of polypeptides was evident from the characteristic double minima at 208 and 222 nm in the circular dichroism (CD) spectra at a concentration of 0.40 mg/mL in water (Fig. 1D). The corresponding nonhelical PHDLG-BIm40 was synthesized through the polymerization of dl-NCA to study the effect of helical structure on the antibacterial activity of polypeptide.
Our molecular simulations provide molecular-level theoretical support for the RA structure. A simulation snapshot in Fig. 1E illustrates the cationic side chains forming a hydrophilic shell around the hydrophobic aliphatic side chains and helical core. Simulation movies are presented in Movies S1–S3. Discounting the two terminal residues to eliminate end effects, the peptide backbone is close to the ideal helix, RMSDhelix = (0.04 ± 0.01) nm, with a radius rhelix = (0.232 ± 0.002) nm and twist γhelix = (100 ± 1)° also nearly ideal ( = 0.23 nm, = 100°). The mean per residue molar ellipticity at 222 nm computed from our simulations using Dichro-Calc (comp.chem.nottingham.ac.uk/cgi-bin/dichrocalc/bin/getparams.cgi) (47) of [θ]222 nm = (−21 ± 1) × 103 degrees (deg) cm2/dmol is in excellent agreement with the experimental value of −23 × 103 deg cm2/dmol for PHLG-BIm40 and −19 × 103 deg cm2/dmol for PHLG-BIm28 reported in Fig. 1D. To quantify the RA structure, we computed the probability distribution of the side chain N12 atoms after aligning the peptide backbone to a reference structure and discarding the four side chains at the termini to eliminate end effects (Fig. 1 F and G) (48). We also measured the probability distribution of side chain lengths from the Cα to the N12 atom. The mode of the distribution is 1.24 nm, and the mean value is 1.15 nm with a 95% CI of [0.90, 1.33] nm (SI Appendix, Fig. S3). The result was further confirmed by nuclear overhauser effect (NOE) spectroscopy, with no NOE detected between protons around the nitrogen atoms on the benzimidazole (N12) (d, b, b′, c, c′, e) and protons around Cα (h, i, g) (SI Appendix, Fig. S5). Because the positive charge in the cationic side chains resides primarily in the termini, these distributions are a proxy for the positive charge distribution, providing strong support for an RA structure.
The antibacterial activity of PHLG-BIm was evaluated by the minimal inhibitory concentration (MIC) of the polypeptide against bacteria (49). With an RA structure, PHLG-BIm40 showed strong antibacterial activity against both gram-negative bacteria, DH5α and MG1655, and gram-positive bacteria, ATCC12608 and ATCC11778, with MIC values of 3.3, 26.1, 13.1, and 13.1 µM, respectively (Fig. 1E). The helical PHLG-BIm40 showed higher antibacterial activity than the nonhelical PHDLG-BIm40, with MIC values 16, 4, 4, and 4 times lower against DH5α, MG1655, ATCC12608, and ATCC11778, respectively. Length was also found to influence the antimicrobial activity of the RA polypeptides, as PHLG-BIm28, with DP of 28, showed lower antimicrobial activity compared with PHLG-BIm40. We also tested the antimicrobial activity of RA polypeptides against other bacterial strains, including clinically isolated Helicobacter pylori strains (B107, J291, J99, J99-AF, J99-A9, and J99-A11) (50, 51) and drug-resistant strains (Methicillin-resistant Staphylococcus aureus, NRS382, NRS383, NRS384). Among those H. pylori strains, J99-AF, J99-A9, and J99-A11 are clarithromycin-resistant strains. The RA polypeptides showed high antibacterial activity against these clinically isolated and drug-resistant strains. More interestingly, the antimicrobial activity of the RA polypeptide remained stable against DH5α and ATCC12608 in the presence of different salts (physiological concentrations of 150 mM NaCl, 1 mM MgCl2, and 2.5 mM CaCl2), and mucin, the main component of mucosa (SI Appendix, Table S2). The MIC values of RA polypeptide against DH5α and ATCC12608 decreased in the presence of human serum, fetal bovine serum (FBS), plasma, and artificial tears in comparison with RA polypeptide-only treatment. The results demonstrate that the RA polypeptides are stable in serum and plasma, and polyanionic compounds do not dramatically affect their antimicrobial activity. The decreased MIC in serum and plasma may be attributed to the serum complement system, which provides innate defense against microbial infections. PHLG-BIm40 exhibited low hemolytic activity with an HC50 (50% hemolytic concentration) value higher than 104.6 µM, which indicates a high selectivity of >32 (defined as HC50/MIC), as opposed to a selectivity of >2 for PHDLG-Blm40 against DH5α bacterial cells. It is important to point out that the RA polypeptide is not just bacteriostatic but bacteriocidal. Nearly 100% killing of all four bacterial species was observed at their respective MIC or double MIC within 2 h (SI Appendix, Fig. S6). In addition, the RA polypeptides showed concentration-dependent antimicrobial killing in medium and in conditions with NaCl, human serum, plasma, and artificial tears (SI Appendix, Fig. S7).
Although many AMPs with high antibacterial activity have been developed, the application of AMPs is usually limited by the short durations of activity due to their rapid digestion by endogenous proteases (5, 16, 17, 52, 53). The RA polypeptides, with densely packed hydrophobic side chains forming a hydrophobic cortex that can protect the amide bonds of the polypeptide backbone, in principle should be more stable against proteolysis compared with typical AMPs. We incubated PHLG-BIm40 with trypsin, pronase, and elastase from Pseudomonas aeruginosa or elastase from human leukocytes for 8 h and analyzed polypeptide degradation by HPLC. PHLG-BIm40 exhibited excellent proteolytic stability and experienced almost no degradation by the proteases (Fig. 1F), whereas LL-37, a positive control peptide, was readily degraded under the same conditions (SI Appendix, Fig. S8). Furthermore, after 8 h of protease or trypsin treatment, the antibacterial activity of PHLG-BIm40 remained unchanged, as demonstrated by having the same MIC value against DH5α as the untreated RA polypeptide (3.3 μM).
RA Polypeptide Kills Bacteria by Directly Disrupting the Bacterial Cell Membrane.
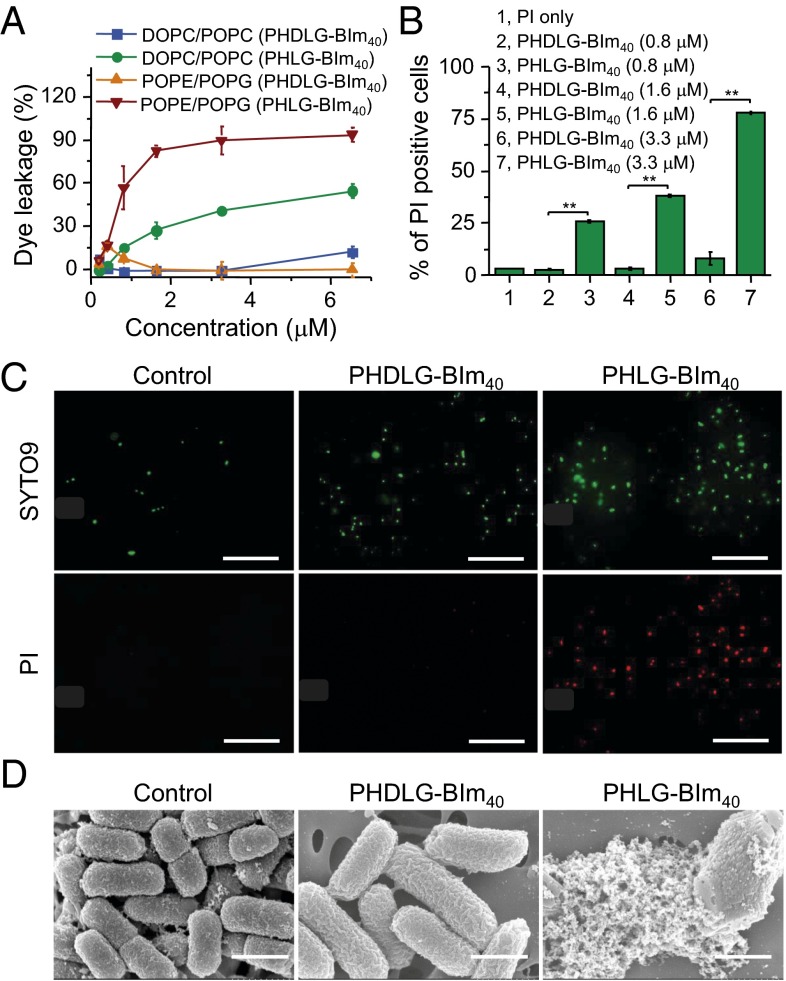
We find that the RA PHLG-BIm kills bacteria by directly disrupting the bacterial cell membrane in a manner similar to typical AMPs, through vesicle leakage, bacterial membrane permeabilization, and bacteria morphology assays. We first investigated the membrane-disruptive activity of helical PHLG-BIm40 and nonhelical PHDLG-BIm40 polypeptides on anionic liposomes 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE)/1-palmitoyl-2-oleoyl- sn-glycero-3-phospho-(1'-rac-glycerol) (POPG) and neutral liposomes 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/1-palmitoyl-2-oleoyl-sn-glycero-3- phosphocholine (POPC), which were used to model phosphatidylethanolamine (PE)-rich bacteria and eukaryotic cell membranes, respectively. At the same concentration, PHLG-BIm40 induced greater dye leakage from both anionic and neutral liposomes than PHDLG-BIm40, suggesting that the helical polypeptide has higher membrane disruption capability (Fig. 2A). PHLG-BIm40 also caused more leakage from the anionic liposomes than the neutral liposomes, which is well-correlated with the observed selectivity against bacterial over mammalian cells. The leakage results also showed the capability of PHLG-BIm to permeabilize model bacteria membranes rich in negative intrinsic curvature-forming lipids. We next used flow cytometry to evaluate permeabilization of PHLG-BIm through bacterial membranes. MG1655 bacteria cells were incubated with PHLG-BIm40 and propidium iodide (PI), a membrane impermeable dye. The total number of PI-containing bacteria cells was greater for those treated with PHLG-Blm40 than PHDLG-BIm40 and increased with higher concentration of RA polypeptide (Fig. 2B). An uptake study analyzed by fluorescence imaging provided additional evidence of enhanced membrane activity of PHLG-BIm40, which permeabilized MG1655 bacterial cell membranes more effectively than PHDLG-BIm40 when the bacterial cells were coincubated with polypeptide and dye (PI and SYTO9) (Fig. 2C). Using scanning electron microscopy, we observed drastic changes and damage of the bacterial membranes after incubation with PHLG-BIm40, whereas PHDLG-BIm40 minimally affected bacteria morphology (Fig. 2D). Taken together, the results indicate that membrane disruption and permeation are an important component of the antimicrobial activity of PHLG-BIm.
Fig. 2.
PHLG-BIm40 kills bacteria by directly disrupting the bacterial cell membrane. (A) Extent of calcein efflux in neutral vesicles (DOPC/POPC) and negatively charged vesicles (DOPE/DOPG) after treatment with PHDLG-BIm40 (nonhelical, lacking radially amphiphilic structure) or PHLG-BIm40 (helical with radially amphiphilic structure) at various concentrations for 1 h. (B) Flow cytometry analysis of propidium iodide (PI) uptake after incubation with free PI, PI with PHDLG-BIm40, or PI with PHLG-BIm40 at various concentrations. All of the data are represented as average ± SD and analyzed by Student t test (**P ≤ 0.01). (C) The fluorescence microscopy of stained Escherichia coli MG1655 in the absence and presence of PHDLG-BIm40 and PHLG-BIm40 (3.3 µM). (Scale bar, 50 μm.) (D) SEM images of MG1655 after treatment with PBS, PHDLG-BIm40, or PHLG-BIm40. (Scale bar, 1 μm.)
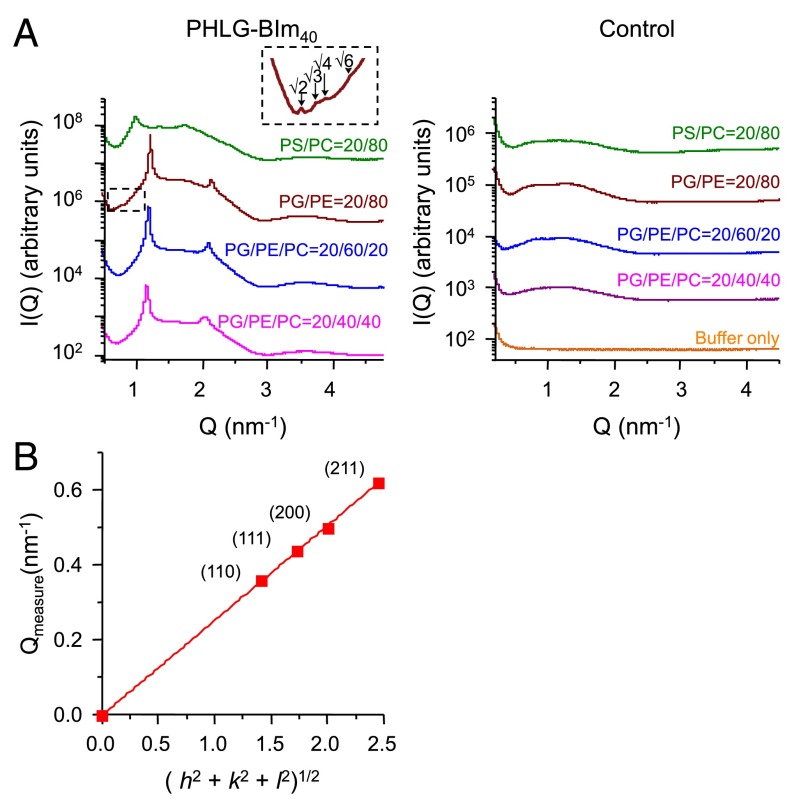
To examine in detail the root causes of selective membrane activity for RA polypeptides, we use synchrotron small angle X-ray scattering (SAXS) to investigate the type and quantity of membrane curvature deformations induced by PHLG-BIm. Small unilamellar vesicles (SUVs) were prepared with lipid compositions representative of bacterial (DOPG/DOPE = 20/80) and eukaryotic (DOPS/DOPC = 20/80) membranes. Compositions DOPG/DOPE/DOPC = 20/60/20 and 20/40/40 were also used as model systems to isolate the role of negative intrinsic curvature lipids such as PE, because eukaryotic membranes are known to have lower PE content relative to bacterial membranes. The SUVs were incubated with PHLG-BIm40 at a peptide/lipid (P/L) molar ratio of 1/400, which is equivalent to a charge ratio of 1/2, and the resulting structures were characterized using SAXS. Synchrotron SAXS spectra from the lipid vesicle solutions showed a broad characteristic feature consistent with a single lipid bilayer form factor of unilamellar vesicles. When exposed to PHLG-BIm40, the lipid vesicles undergo a structural transition, resulting in correlation peaks with specific ratios of Q values in the diffraction data (Fig. 3). For each membrane composition, we found a set of correlation peaks having integral Q-ratios of 1:2 consistent with a lamellar (Lα) phase, characterized by d-spacings of 6.7–8.8 nm. These lamellar phases resulting from exposure of PHLG-BIm to the SUVs indicate intermembrane attraction without the generation of significant curvature. Interestingly, for the model bacterial membrane composition, we identified a second set of correlation peaks with characteristic Q-ratios of √2:√3:√4:√6, which indexed to a cubic (QII) Pn3m “double-diamond” lattice having a lattice parameter of 24.8 nm. Bicontinuous cubic phases, such as the Pn3m, consist of two nonintersecting water channels that are separated by a lipid bilayer. The center of this bilayer traces out a minimum surface characterized by negative Gaussian curvature (NGC), also known as saddle-splay curvature, at every point. Our SAXS data showed that PHLG-BIm promotes saddle-shaped membrane deformations in model bacteria membranes to stabilize a bulk Pn3m cubic phase. NGC is the saddle-shaped curvature that manifests along the inside of pores, around the base of a bleb, and the neck of a bud, the basic membrane permeation mechanisms. Earlier studies have found a strong correlation between the formation of cubic phases and membrane permeation induced by AMPs (1, 14, 54). The ability of PHLG-BIm to generate cubic phases in bacteria-like PE-rich membranes suggests that the RA polypeptide may permeate bacteria membranes via the induction of NGC consistent with that of natural AMPs. By systematically examining a range of membranes, we determined how lipid composition affects the ability of PHLG-BIm to restructure vesicles. We observed the general trend of decreasing PE content resulting in the suppression of nonlamellar phase formation. More specifically, we found that the peptide does not disrupt membranes (no NGC generation) with a PE content of 60% and lower, which includes those representative of eukaryotes. The preference for PHLG-BIm to generate cubic phases at high PE levels of ∼80% suggests a mechanism of selectivity for bacterial over animal membranes based on their specific lipid distributions, again consistent with generic AMP trends.
Fig. 3.
(A) SAXS spectra show generation of NGC by PHLG-Blm40 in membranes rich in PE. SAXS profiles from lipid vesicle solutions after exposure to PHLG-Blm40 at molar ratio P/L = 1/400. SUV-only controls at each composition show a broad characteristic feature consistent with the form factor of the unilamellar vesicles. (B) PHLG-Blm induces a Pn3m cubic phase. The measured Q positions of the diffraction peaks were plotted to show indexing of the Pn3m cubic phase for DOPG/DOPE = 20/80.
RA Polypeptide Enhances the Antibacterial Effects of Traditional Antibiotics.
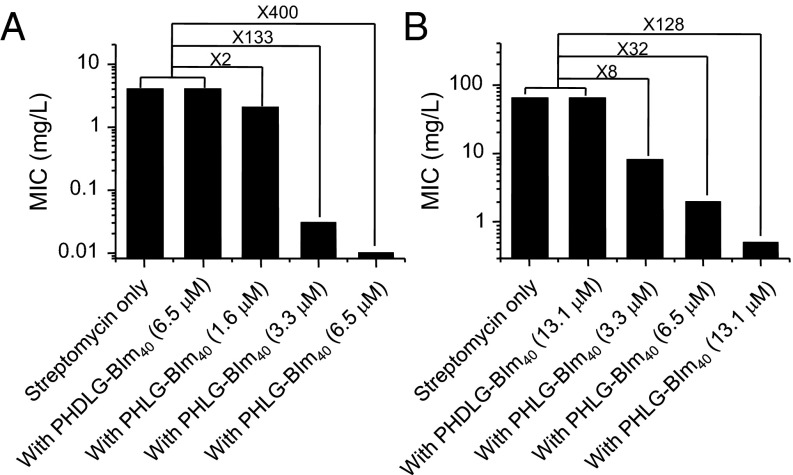
Because PHLG-BIm can cause membrane disruption and permeabilization, we explored its potential to enhance the antibacterial effect of traditional antibiotics. Synergistic-like enhancement of bacteria killing has been previously reported from coadministration of membrane-disruptive agents with commercial antibiotics (41, 55). We selected four antibiotics: streptomycin (aminoglycoside), doxycycline (tetracycline), rifampicin (rifamycin), and gentamicin (aminoglycoside). Streptomycin, doxycycline, and gentamicin are protein synthesis inhibitors, whereas rifampicin inhibits DNA-dependent RNA synthesis. We tested the antibacterial activity of these antibiotics coadministered with helical PHLG-BIm40. Nonhelical PHDLG-BIm40 served as the control. Bacteria were incubated with either antibiotic alone, antibiotic coadministered with PHDLG-BIm40, or antibiotic coadministered with PHLG-BIm40 at varying concentrations. For MG1655 bacteria, coadministration of streptomycin with PHDLG-BIm40 resulted in MIC values identical to those of streptomycin alone (Fig. 4A). However, when streptomycin was coadministered with PHLG-BIm40 at concentrations of 1.6, 3.3, and 6.5 µM, the respective MIC values against MG1655 were 2, 133, and 400 times lower than that of streptomycin alone. Because the MIC against MG1655 with PHLG-BIm40 alone was 26.1 µM, this result suggested a synergistic effect from combining treatments of streptomycin and PHLG-BIm40. We also observed a similar synergistic effect of PHLG-BIm40 and antibiotics against C101 (P. aeruginosa), a bacterium with high resistance to many antibiotics. For C101 bacteria, the MIC of PHLG-BIm alone was 52.3 µM. A synergistic effect when streptomycin was coadministered with PHLG-BIm40 (13.1 µM) was indicated by an MIC that was 128 times lower than that of streptomycin alone (Fig. 4B). The synergistic effect of combination therapy was also detected against three other bacterial strains, DH5α, ATCC11778, and ATCC12608 (SI Appendix, Table S3) and for three other antibiotics: doxycycline, rifampicin, and gentamicin (SI Appendix, Table S4). The synergistic effect of combination therapy may be a result of enhanced antibiotic uptake due to RA polypeptide-induced membrane permeation. To test this hypothesis, bacterial cells were incubated with rifampicin without or with RA polypeptides. After 0.5- or 1-h incubation with RA polypeptide, we observed increased intracellular uptake of rifampicin in MG1655 and C101 cells (SI Appendix, Fig. S9). These findings suggest that the synergy from coadministering antibiotics with RA polypeptides may potentially result from increased antibiotic uptake, such as that caused by peptide-induced defects, or suppressed efflux activities. Our study showed combination therapy to be a promising application for this class of membrane-active RA polypeptides, which can significantly improve the effectiveness of traditional commercial antibiotics by killing bacteria through a distinct mode of action.
Fig. 4.
PHLG-BIm40 enhances the antibacterial activity of commercial antibiotics. Antimicrobial activity of streptomycin codelivered with PHLG-BIm40 or PHDLG-BIm40 at various concentrations against MG1655 (A) (MIC of PHLG-BIm40: 26.1 µM) and C101 (B) (MIC of PHLG-BIm40: 52.3 µM).
Discussion
We developed a class of cationic, helical antimicrobial homo-polypeptides with unprecedented radial amphiphilicity. Unlike typical AMPs characterized by facial amphiphilicity or biomimetic antimicrobial polymers with randomly distributed charged and hydrophobic groups, these AMPs are homo-polypeptides with RA structure. They adopt a stable α-helical conformation with a hydrophobic helical core and a charged exterior shell, formed by long hydrophobic side chains with terminal charge group. The RA polypeptides offer several advantages over typical AMPs. They can be easily synthesized through controllable NCA polymerization followed by side chain modification. The RA structure enables the polypeptides to bind effectively to the negatively charged bacterial surface, and exhibit high antimicrobial activity against both gram-positive and gram-negative bacteria. Moreover, the shielding of the hydrophobic core by the charged exterior shell decreases nonspecific interactions with eukaryotic cells and contributes to low mammalian cytotoxicity. The RA polypeptides also demonstrate excellent stability against enzymatic degradation, potentially due to suppressed protease access to the polypeptide backbone. In addition, the antibacterial and hemolytic activities of the RA polypeptides can be tuned by varying the terminal amine group and the hydrophobicity of helical core, both of which can be easily attainable based on our previous studies (43–45). Thus, the RA polypeptides provide an excellent platform for the design of new AMP analogs.
The antibacterial activity of these RA polypeptides likely results from electrostatic interactions between their cationic groups and anionic bacteria cell membranes, followed by the disruption of the bacteria cell membranes by the membrane-active polypeptide helix. Additional factors are involved in bacterial membrane destabilization, as suggested by our SAXS studies on model bacterial membranes in which helical polypeptide PHLG-BIm promoted saddle-shaped membrane deformations (NGC), a topological requirement for membrane destabilizing events such as pore-formation. All tested model membranes had a fixed anionic charge, yet we observed selective generation of NGC in the PE-rich bacteria model membrane. This result is consistent with previous work, which showed that membranes with greater amounts of negative intrinsic curvature lipids promote destabilization and are more susceptible to pore formation (2). Moreover, both anionic and negative intrinsic curvature lipids are necessary for activity, whereas neither alone is sufficient. Therefore, the preference for the helical RA polypeptides to generate NGC at high PE content points to a mechanism of selectivity that involves membrane curvature effects.
The observed synergistic bactericidal effect may result from increased cellular penetration of antibiotics that is facilitated by the membrane permeabilization activity of the RA polypeptides. Induced NGC is broadly enabling in the context of membrane permeation mechanisms, such as transmembrane pores, blebbing, budding, and scission. It is likely that permeation can involve a hierarchy of mechanisms. We observe NGC in the form of a cubic phase; the size of the defect on a 2D membrane will depend on the type of defect it is. The Pn3m cubic phase induced by the RA polypeptide PHLG-BIm40 has an average Gaussian curvature 〈K〉 value of −0.01065 nm−2. For example, this is the amount of NGC found in a transmembrane pore with a size of ∼40 nm, if we ignore all other effects. If on the other hand, the destabilization mechanism is that of a budding event followed by scission, then we can estimate the size of the defect using a catenoid surface, which is an approximate representation of the surface of a scission pore (56, 57). The Gaussian curvature of a minimal catenoid surface with a neck radius of c along its z axis is defined by K(z) = −[sech4(z∕c)]/c2. The measured 〈K〉 value corresponds to a catenoid neck diameter of ∼19.4 nm. If we account for the approximate bilayer membrane thickness of 4 nm, this diameter translates to a pore size of ∼15.4 nm. In both of the above cases, the size of the defect is significantly larger than the size of a typical antibiotic.
In conclusion, designing AMPs bearing the unique RA structure represents an alternative direction of AMP development; the RA polypeptide may become a general platform for developing AMPs to treat drug-resistant bacteria. Although outside the scope of this investigation, specifics pertaining to the mechanism and selectivity of these polypeptides can aid the design and optimization of future synthetic AMPs based on a RA structure. We suggest further studies on this class of polypeptides using ellipsometry, electrochemistry, NMR, and atomic force microscopes to examine these details (58–61). Future studies to determine the toxicity of the degradation products will also be necessary to assess the potential applicability of the peptide as a clinical therapeutic.
Materials and Methods
PHLG-BIm was synthesized by mixing PCHLG, NaI, and 1-methylbenzimidazole in DMF and acetonitrile in a 25-mL Schlenk tube. The mixture was stirred at 80 °C for 48 h. Molecular dynamics simulations of PHLG-BIm with a DP of 20 follow a procedure similar to that we previously detailed in ref. 62. Details describing preparation and characterization of PHLG-BIm; simulation methods; antibacterial assays and hemolytic assay; and small-angle X-ray scattering experiments can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Paul J. Hergenrother for kindly supplying bacterial strains. J. C. acknowledges support from the National Science Foundation (NSF) (Grant CHE-1153122) for the design and synthesis of polypeptide and the National Institutes of Health (NIH) (Director’s New Innovator Awards 1DP2OD007246 and 1R21EB013379) for the biological evaluation of the polypeptides. L.-F.C. and J.C. acknowledge support from NIH Grant R21AI117080 for antibacterial peptides against H. pylori. G.C.L.W. and M.W.L. acknowledge support from NSF Grant DMR-1411329 for the characterization of the polypeptide-membrane interaction. R.M.P. acknowledges support from NIH Grants R01 DK 58587, R01 CA 77955, P01 CA 116087, and P30 DK 058404.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507893112/-/DCSupplemental.
References
- 1.Schmidt NW, Wong GC. Antimicrobial peptides and induced membrane curvature: Geometry, coordination chemistry, and molecular engineering. Curr Opin Solid State Mater Sci. 2013;17(4):151–163. doi: 10.1016/j.cossms.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, et al. Mechanism of a prototypical synthetic membrane-active antimicrobial: Efficient hole-punching via interaction with negative intrinsic curvature lipids. Proc Natl Acad Sci USA. 2008;105(52):20595–20600. doi: 10.1073/pnas.0806456105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurdle JG, O’Neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat Rev Microbiol. 2011;9(1):62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engler AC, et al. Emerging trends in macromolecular antimicrobials to fight multi-drug-resistant Infections. Nano Today. 2012;7(3):201–222. [Google Scholar]
- 5.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24(12):1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 6.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 7.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 8.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462(1-2):55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 9.Breukink E, de Kruijff B. The lantibiotic nisin, a special case or not? Biochim Biophys Acta. 1999;1462(1-2):223–234. doi: 10.1016/s0005-2736(99)00208-4. [DOI] [PubMed] [Google Scholar]
- 10.Matsuzaki K, et al. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry. 1998;37(34):11856–11863. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
- 11.Dürr UHN, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta. 2006;1758(9):1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA. 1987;84(15):5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt NW, et al. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J Am Chem Soc. 2011;133(17):6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MW, et al. Two interdependent mechanisms of antimicrobial activity allow for efficient killing in nylon-3-based polymeric mimics of innate immunity peptides. Biochim Biophys Acta. 2014;1838(9):2269–2279. doi: 10.1016/j.bbamem.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, et al. Synthetic antimicrobial oligomers induce a composition-dependent topological transition in membranes. J Am Chem Soc. 2007;129(40):12141–12147. doi: 10.1021/ja072310o. [DOI] [PubMed] [Google Scholar]
- 16.Strömstedt AA, Pasupuleti M, Schmidtchen A, Malmsten M. Evaluation of strategies for improving proteolytic resistance of antimicrobial peptides by using variants of EFK17, an internal segment of LL-37. Antimicrob Agents Chemother. 2009;53(2):593–602. doi: 10.1128/AAC.00477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng H, Kumar K. Antimicrobial activity and protease stability of peptides containing fluorinated amino acids. J Am Chem Soc. 2007;129(50):15615–15622. doi: 10.1021/ja075373f. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, et al. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J Biol Chem. 2005;280(13):12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013;6(12):1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamuro Y, Schneider JP, DeGrado WF. De novo design of antibacterial β-peptides. J Am Chem Soc. 1999;121(51):12200–12201. [Google Scholar]
- 21.Porter EA, Weisblum B, Gellman SH. Mimicry of host-defense peptides by unnatural oligomers: Antimicrobial beta-peptides. J Am Chem Soc. 2002;124(25):7324–7330. doi: 10.1021/ja0260871. [DOI] [PubMed] [Google Scholar]
- 22.Porter EA, Wang X, Lee HS, Weisblum B, Gellman SH. Non-haemolytic beta-amino-acid oligomers. Nature. 2000;404(6778):565–565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]
- 23.Liu D, DeGrado WF. De novo design, synthesis, and characterization of antimicrobial beta-peptides. J Am Chem Soc. 2001;123(31):7553–7559. doi: 10.1021/ja0107475. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt MA, Weisblum B, Gellman SH. Interplay among folding, sequence, and lipophilicity in the antibacterial and hemolytic activities of alpha/beta-peptides. J Am Chem Soc. 2007;129(2):417–428. doi: 10.1021/ja0666553. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt MA, Weisblum B, Gellman SH. Unexpected relationships between structure and function in alpha,beta-peptides: Antimicrobial foldamers with heterogeneous backbones. J Am Chem Soc. 2004;126(22):6848–6849. doi: 10.1021/ja048546z. [DOI] [PubMed] [Google Scholar]
- 26.Patch JA, Barron AE. Helical peptoid mimics of magainin-2 amide. J Am Chem Soc. 2003;125(40):12092–12093. doi: 10.1021/ja037320d. [DOI] [PubMed] [Google Scholar]
- 27.Tew GN, et al. De novo design of biomimetic antimicrobial polymers. Proc Natl Acad Sci USA. 2002;99(8):5110–5114. doi: 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tew GN, Clements D, Tang H, Arnt L, Scott RW. Antimicrobial activity of an abiotic host defense peptide mimic. Biochim Biophys Acta. 2006;1758(9):1387–1392. doi: 10.1016/j.bbamem.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Tang H, Doerksen RJ, Jones TV, Klein ML, Tew GN. Biomimetic facially amphiphilic antibacterial oligomers with conformationally stiff backbones. Chem Biol. 2006;13(4):427–435. doi: 10.1016/j.chembiol.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Liu D, et al. Nontoxic membrane-active antimicrobial arylamide oligomers. Angew Chem Int Ed Engl. 2004;43(9):1158–1162. doi: 10.1002/anie.200352791. [DOI] [PubMed] [Google Scholar]
- 31.Palermo EF, Sovadinova I, Kuroda K. Structural determinants of antimicrobial activity and biocompatibility in membrane-disrupting methacrylamide random copolymers. Biomacromolecules. 2009;10(11):3098–3107. doi: 10.1021/bm900784x. [DOI] [PubMed] [Google Scholar]
- 32.Mowery BP, Lindner AH, Weisblum B, Stahl SS, Gellman SH. Structure-activity relationships among random nylon-3 copolymers that mimic antibacterial host-defense peptides. J Am Chem Soc. 2009;131(28):9735–9745. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]
- 33.Mowery BP, et al. Mimicry of antimicrobial host-defense peptides by random copolymers. J Am Chem Soc. 2007;129(50):15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 34.Liu R, et al. Nylon-3 polymers with selective antifungal activity. J Am Chem Soc. 2013;135(14):5270–5273. doi: 10.1021/ja4006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu R, et al. Structure-activity relationships among antifungal nylon-3 polymers: Identification of materials active against drug-resistant strains of Candida albicans. J Am Chem Soc. 2014;136(11):4333–4342. doi: 10.1021/ja500036r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou C, et al. High potency and broad-spectrum antimicrobial peptides synthesized via ring-opening polymerization of alpha-aminoacid-N-carboxyanhydrides. Biomacromolecules. 2010;11(1):60–67. doi: 10.1021/bm900896h. [DOI] [PubMed] [Google Scholar]
- 37.Engler AC, et al. Effects of side group functionality and molecular weight on the activity of synthetic antimicrobial polypeptides. Biomacromolecules. 2011;12(5):1666–1674. doi: 10.1021/bm2000583. [DOI] [PubMed] [Google Scholar]
- 38.Chin W, et al. Biodegradable broad-spectrum antimicrobial polycarbonates: Investigating the role of chemical structure on activity and selectivity. Macromolecules. 2013;46(22):8797–8807. [Google Scholar]
- 39.Gabriel GJ, et al. Comparison of facially amphiphilic versus segregated monomers in the design of antibacterial copolymers. Chemistry. 2009;15(2):433–439. doi: 10.1002/chem.200801233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colak S, Nelson CF, Nüsslein K, Tew GN. Hydrophilic modifications of an amphiphilic polynorbornene and the effects on its hemolytic and antibacterial activity. Biomacromolecules. 2009;10(2):353–359. doi: 10.1021/bm801129y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng VW, Ke X, Lee AL, Hedrick JL, Yang YY. Synergistic co-delivery of membrane-disrupting polymers with commercial antibiotics against highly opportunistic bacteria. Adv Mater. 2013;25(46):6730–6736. doi: 10.1002/adma.201302952. [DOI] [PubMed] [Google Scholar]
- 42.Nederberg F, et al. Biodegradable nanostructures with selective lysis of microbial membranes. Nat Chem. 2011;3(5):409–414. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- 43.Yin L, et al. Supramolecular self-assembled nanoparticles mediate oral delivery of therapeutic TNF-α siRNA against systemic inflammation. Angew Chem Int Ed Engl. 2013;52(22):5757–5761. doi: 10.1002/anie.201209991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu H, et al. Ionic polypeptides with unusual helical stability. Nat Commun. 2011;2:206. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]
- 45.Gabrielson NP, et al. Reactive and bioactive cationic α-helical polypeptide template for nonviral gene delivery. Angew Chem Int Ed Engl. 2012;51(5):1143–1147. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu H, Cheng J. Hexamethyldisilazane-mediated controlled polymerization of alpha-amino acid N-carboxyanhydrides. J Am Chem Soc. 2007;129(46):14114–14115. doi: 10.1021/ja074961q. [DOI] [PubMed] [Google Scholar]
- 47.Bulheller BM, Hirst JD. DichroCalc--circular and linear dichroism online. Bioinformatics. 2009;25(4):539–540. doi: 10.1093/bioinformatics/btp016. [DOI] [PubMed] [Google Scholar]
- 48. (2014) MATLAB (The MathWorks, Natick, MA)
- 49.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2002;49(6):1049–1050. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 50.Peek RM, Jr, et al. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 1999;59(24):6124–6131. [PubMed] [Google Scholar]
- 51.Israel DA, et al. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci USA. 2001;98(25):14625–14630. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Findlay B, Zhanel GG, Schweizer F. Cationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffold. Antimicrob Agents Chemother. 2010;54(10):4049–4058. doi: 10.1128/AAC.00530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brogden NK, Brogden KA. Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int J Antimicrob Agents. 2011;38(3):217–225. doi: 10.1016/j.ijantimicag.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu K, et al. A critical evaluation of random copolymer mimesis of homogeneous antimicrobial peptides. Macromolecules. 2013;46(5):1908–1915. doi: 10.1021/ma302577e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eckert R, et al. Enhancement of antimicrobial activity against pseudomonas aeruginosa by coadministration of G10KHc and tobramycin. Antimicrob Agents Chemother. 2006;50(11):3833–3838. doi: 10.1128/AAC.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boucrot E, et al. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149(1):124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GC. Influenza virus A M2 protein generates negative Gaussian membrane curvature necessary for budding and scission. J Am Chem Soc. 2013;135(37):13710–13719. doi: 10.1021/ja400146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidtchen A, et al. Membrane selectivity by W-tagging of antimicrobial peptides. Biochim Biophys Acta. 2011;1808(4):1081–1091. doi: 10.1016/j.bbamem.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 59.Ringstad L, et al. An electrochemical study into the interaction between complement-derived peptides and DOPC mono- and bilayers. Langmuir. 2008;24(1):208–216. doi: 10.1021/la702538k. [DOI] [PubMed] [Google Scholar]
- 60.Bechinger B. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim Biophys Acta. 1999;1462(1-2):157–183. doi: 10.1016/s0005-2736(99)00205-9. [DOI] [PubMed] [Google Scholar]
- 61.García-Sáez AJ, Chiantia S, Salgado J, Schwille P. Pore formation by a Bax-derived peptide: Effect on the line tension of the membrane probed by AFM. Biophys J. 2007;93(1):103–112. doi: 10.1529/biophysj.106.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mansbach RA, Ferguson AL. Machine learning of single molecule free energy surfaces and the impact of chemistry and environment upon structure and dynamics. J Chem Phys. 2015;142(10):105101. doi: 10.1063/1.4914144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.