Significance
Chemotherapy has been proven in clinical studies to improve overall survival significantly. Unfortunately, there is a significant degree of heterogeneity in tumor chemosensitivity, often resulting in unnecessary treatment and needless exposure to toxic side-effects. A platform is needed that can identify preemptively which patients will or will not benefit from treatment. Tumor organoids, 3D cultures of cancer cells, present such an individualized platform. In this study we demonstrate that organoid cultures can be established from metastatic biopsy specimens with a high success rate and genetically represent the metastasis they were derived from. These data support the translation of this innovative technology to the clinic as an ex vivo screening platform for tailoring treatment.
Keywords: biopsy-derived tumor organoids, colorectal cancer, DNA sequencing, copy number analysis, personalized medicine
Abstract
Tumor organoids are 3D cultures of cancer cells. They can be derived from the tumor of each individual patient, thereby providing an attractive ex vivo assay to tailor treatment. Using patient-derived tumor organoids for this purpose requires that organoids derived from biopsies maintain the genetic diversity of the in vivo tumor. In this study tumor biopsies were obtained from 14 patients with metastatic colorectal cancer (i) to test the feasibility of organoid culture from metastatic biopsy specimens and (ii) to compare the genetic diversity of patient-derived tumor organoids and the original tumor biopsy. Genetic analysis was performed using SOLiD sequencing for 1,977 cancer-relevant genes. Copy number profiles were generated from sequencing data using CopywriteR. Here we demonstrate that organoid cultures can be established from tumor biopsies of patients with metastatic colorectal cancer with a success rate of 71%. Genetic analysis showed that organoids reflect the metastasis from which they were derived. Ninety percent of somatic mutations were shared between organoids and biopsies from the same patient, and the DNA copy number profiles of organoids and the corresponding original tumor show a correlation of 0.89. Most importantly, none of the mutations that were found exclusively in either the tumor or organoid culture are in driver genes or genes amenable for drug targeting. These findings support further exploration of patient-derived organoids as an ex vivo platform to personalize anticancer treatment.
The considerable variation among patients in sensitivity to antineoplastic treatment emphasizes the need for more accurate treatment selection. Numerous attempts have been made to develop a personalized in vitro platform to predict treatment response in individual patients, but no method has presently gained clinical acceptance (1). Cell lines are not readily established for every individual patient, and although a higher take rate and early signs of predictive value have been reported for patient-derived xenograft models, the 6–8 mo required to gain sufficient material may prohibit its development as a clinical decision-making tool (2).
A recently developed 3D culture system, enriching for the stem cell population, has enabled the indefinite propagation of normal and tumor epithelial cells of a variety of tissues (3–9). These cultures, referred to as “organoids,” are established in a relatively short time frame, are easy to manipulate, and facilitate high-throughput screens (10). However, their most important feature is that cultures can be derived from the tumor tissue of each individual patient. With adequate amounts of starting material, such as tumor resections, the success rate of derivation for colorectal cancer organoids is greater than 90% (10). However, in the metastatic setting there generally is no access to resection material, and acquisition of fresh tumor tissue often is limited to biopsies. To use patient-derived tumor organoids as a personalized screening tool for tailoring treatment, it is imperative that cultures from biopsy specimens have a high success rate and that the genetic landscape of the tumor be preserved in culture. This article addresses both topics.
Materials and Methods
Patients.
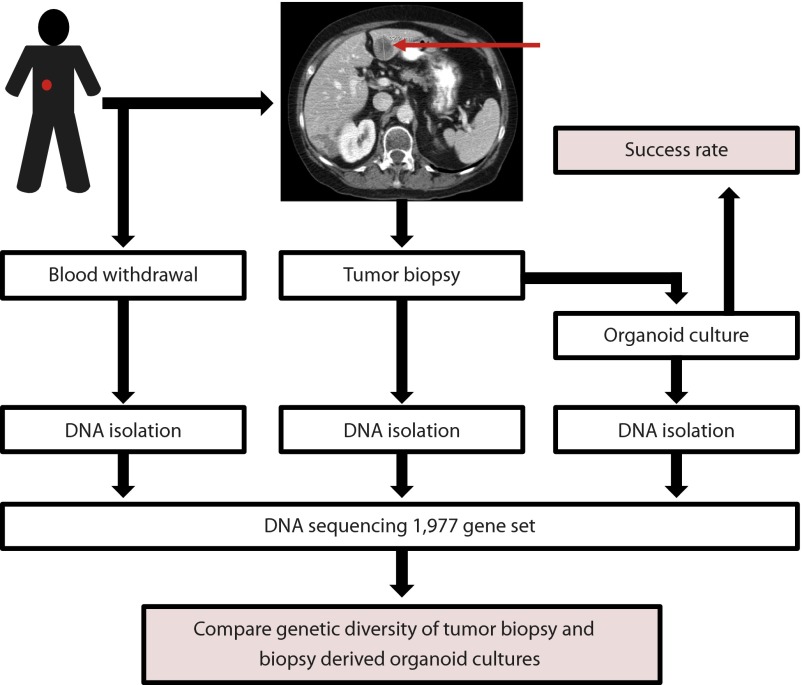
Fourteen patients with metastatic colorectal cancer gave written informed consent to undergo two to four 18-gauge biopsies from accessible metastatic lesions (Fig. 1). This study was approved by the institutional review board of the University Medical Center Utrecht (protocol number NCT01855477).
Fig. 1.
Overview of the study design.
Organoid Cultures.
Organoid culturing was performed according to methods described by Sato et al. (3). Briefly, biopsies were dissected and embedded in Matrigel [Basement Membrane Matrix, growth Factor Reduced, Phenol Red free (MG), (BD; #56231)]. The Matrigel including the embedded cells was overlaid with growth medium optimized for selective outgrowth of tumor cells [Advanced Dulbecco's Modified Eagles Medium with Nutrient Mixture F-12 Hams (Ad-DF) (Invitrogen; #12634), 1% penicillin/streptomycin (Invitrogen; #15140-122), 10 mM Hepes (Invitrogen #15630-056), 1% GlutaMAX (Invitrogen #35050), 20% R-Spondin conditioned medium (3), 10% Noggin conditioned medium (3), 1× B27 supplement (Invitrogen #17504-044), 1.25 mM N-acetyl-cysteine (Sigma-Aldrich; A9165-5G), 10 mM nicotinamide (Sigma-Aldrich; N0636), 50 ng/mL human EGF (Invitrogen Biosource; PMG8043), 10 nM gastrin (Sigma; #G9145), 500 nM TGFb type I Receptor inhibitor A83-01 (Tocris; #2939), 10 μM p38 MAPK inhibitor (p38i) SB202190 (Sigma-Aldrich; S7067), 10 nM prostaglandin E2 (Tocris; #2296), and 1× Primocin (InvivoGen; #ant-pm-1)]. Fresh medium was added every 2 or 3 d. Outgrowing organoids were passaged every 7–10 d after mechanical and enzymatic disruption.
DNA Sequencing.
A separate biopsy was used to compare the mutation and copy number profiles of the tumor biopsy and biopsy-derived cultures. Blood was drawn to obtain control DNA for germline variation. DNA was isolated from organoid cultures after 2–3 mo (depending on organoid growth rate). DNA was isolated using the Tissue and Blood DNA 500 Extraction Kit (Diasorin) according to the manufacturer’s protocol. Matched blood, tumor biopsies, and biopsy-derived cultures were sequenced using the 5500xl SOLiD system (Applied Biosystems) according to the manufacturer’s protocol. The minimum tumor percentage required to enable sequencing was 30%. Tumor cell percentage and percentage of necrosis were determined by a trained pathologist based on H&E staining. Six hundred nanograms of genomic DNA were required per sample to generate barcoded fragment libraries as described by Harakalova et al. (11). Samples were enriched using SureSelect technology (Agilent Technologies) for the actionable cancer genome consisting of 1,977 genes, as previously described by Hoogstraat et al. (12) and Vermaat et al. (13). The actionable cancer genome consists of all exons of genes known to be oncogenes, tumor-suppressor genes, kinases, or functioning in pathways involved in tumorigenesis and comprises ∼16% of the exome. Libraries were sequenced to an average coverage of 150×. Mapping, variant calling, and annotation were done as described by Hoogstraat et al. (12). Somatic mutations were classified as unique for either tumor biopsy or biopsy-derived cultures if the allele frequency in the matching sample was <1%.
DNA Copy Number Analysis.
DNA copy number profiles were generated from the BAM files using CopywriteR as described by Kuilman et al. (14). In short, sequence reads outside the captured genomic regions (off-target reads) were used to generate DNA copy number profiles. A depth-of-coverage method was used for 100-kb bins, and the read count was normalized for GC content and mappability. Log2 ratios were calculated for all tumor or organoid samples versus reference (blood) samples. The normalized and corrected log2 ratios from CopywriteR were analyzed further by circular binary segmentation (CBS) and CGHcall (Bioconductor) (15). CBS allows the detection of segments with nearly identical copy number states. CGHcall is used to classify data points as copy number gain, loss, or neutral.
Results
Organoid culture from metastatic biopsy specimens was successful in 10 of 14 cases (71%). For eight patients DNA from the tumor biopsy and its matched biopsy-derived culture were sequenced to assess whether the original genetic diversity of the tumor was preserved in culture (Fig. 1). The remaining two patients from whom organoid cultures were established were excluded for this analysis because the tumor cell percentage of the biopsy was below 30%.
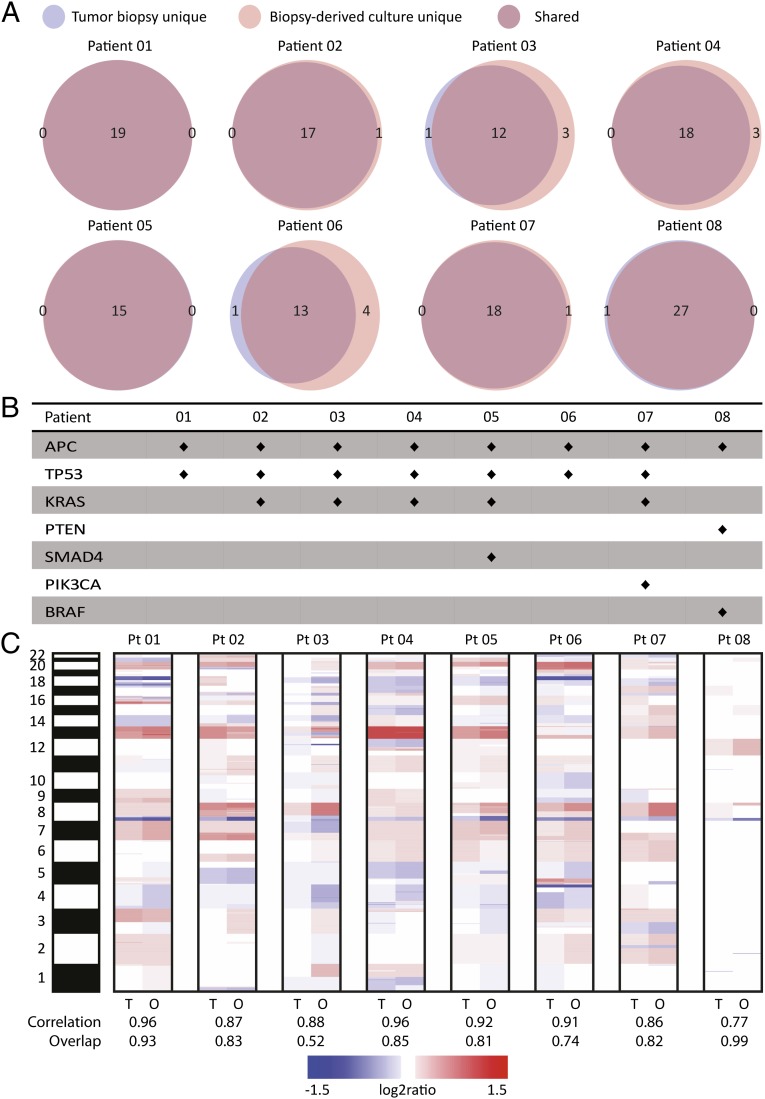
A mean of 19 (±4) somatic mutations were observed per patient (Fig. 2). Ninety percent of somatic mutations were shared between tumor biopsy and organoid cultures. Somatic mutations in driver genes or other genes of interest were invariably detected in both tumor biopsy and biopsy-derived cultures from all patients (Fig. 2). An average correlation of 0.89 (range 0.77–0.96) was detected for DNA copy number profiles between matching tumor biopsy and organoid samples (Fig. 2). The average percentage of data points detected identically as gain, loss, or neutral was 81% (range: 52–99%). Somatic mutations identified are presented in Table S1, sequencing statistics in Table S2, and tumor cell percentage and necrosis percentage of original biopsies in Table S3. Copy number profiles for matched tumor biopsies and biopsy-derived cultures per patient are shown in Fig. S1.
Fig. 2.
Copy number and mutational profiles of biopsy-derived organoid cultures and matched tumor biopsies for patients 01–08. (A) Venn diagrams displaying the number of shared and unique (tumor biopsy or biopsy-derived cultures) mutations per patient as identified by SOLiD sequencing. (B) Mutations in known driver genes or genes of interest for colorectal cancer per patient. Mutations were invariably present in both tumor biopsy and biopsy-derived organoid cultures. (C) Heat map comparing copy number aberrations between matched tumor biopsy and biopsy-derived organoid cultures in a log2 scale. Red colors indicate gains, and blue colors indicate losses. The band on the left represents chromosomes 1–22 and X. For each patient the correlation between the tumor biopsy and biopsy-derived cultures is shown, as well as the percentage of data points detected identically as gain, loss, or neutral (overlap).
Table S1.
SOLiD data somatic mutations
| Subject | Chromosome | Position | Base change | Amino acid change | Mean coverage | Gene | Variant type | Germline coverage | Tumor Coverage | Organoid coverage | Germline allele frequency | Tumor allele frequency | Organoid allele frequency | Shared (S), tumor unique (T), or organoid unique (O) |
| 1 | 1 | 209,797,208 | T/A | K705I | 332 | LAMB3 | SNV | 298 | 459 | 239 | 2 | 85 | 99 | S |
| 2 | 179,407,388 | C/A | NA | 493,67 | TTN | SNV | 489 | 634 | 358 | 0 | 20 | 19 | S | |
| 2 | 179,486,728 | C/A | W6034L | 344,33 | TTN | SNV | 253 | 520 | 260 | 0 | 39 | 43 | S | |
| 3 | 25,665,813 | G/A | P824S | 775,67 | TOP2B | SNV | 694 | 1127 | 506 | 1 | 22 | 24 | S | |
| 3 | 119,582,332 | G/A | R357W | 401 | GSK3B | SNV | 316 | 640 | 247 | 1 | 28 | 39 | S | |
| 3 | 195,605,915 | C/G | NA | 45,33 | TNK2 | SNV | 53 | 71 | 12 | 0 | 20 | 17 | S | |
| 4 | 65,298,248 | A/G | NA | 11,33 | RP11-204H9.2 | SNV | 15 | 17 | 2 | 0 | 76 | 100 | S | |
| 5 | 112,175,021 | C/T | Q1244* | 96,67 | APC | SNV | 96 | 133 | 61 | 1 | 21 | 34 | S | |
| 6 | 4,059,028 | G/A | V514I | 57 | PRPF4B | SNV | 82 | 34 | 55 | 1 | 21 | 7 | S | |
| 8 | 8,999,186 | C/T | NA | 83,33 | PPP1R3B | SNV | 113 | 119 | 18 | 0 | 68 | 100 | S | |
| 11 | 106,810,698 | C/G | E232Q | 693,33 | GUCY1A2 | SNV | 508 | 1145 | 427 | 0 | 27 | 24 | S | |
| 11 | 117,306,398 | C/A | R1673L | 301 | DSCAML1 | SNV | 349 | 390 | 164 | 0 | 31 | 29 | S | |
| 12 | 57,849,972 | C/T | P132S | 180 | INHBE | SNV | 122 | 323 | 95 | 0 | 21 | 32 | S | |
| 13 | 32,972,674 | G/A | E3342K | 846,67 | BRCA2 | SNV | 490 | 1434 | 616 | 0 | 17 | 28 | S | |
| 15 | 99,442,705 | G/A | NA | 351 | IGF1R | SNV | 241 | 603 | 209 | 0 | 26 | 39 | S | |
| 16 | 3,777,892 | C/T | G2348R | 49,67 | CREBBP | SNV | 45 | 76 | 28 | 0 | 53 | 68 | S | |
| 20 | 2,845,936 | C/T | A402V | 187,33 | VPS16 | SNV | 105 | 347 | 110 | 0 | 8 | 19 | S | |
| X | 41,027,389 | C/T | R852* | 164,67 | USP9X | SNV | 251 | 118 | 125 | 0 | 79 | 93 | S | |
| 17 | 75,775,39 | G/A | R248W | 205,33 | TP53 | SNV | 235 | 280 | 101 | 0 | 69 | 79 | S | |
| 2 | 1 | 10,363,653 | G/C | E804Q | 163,67 | KIF1B | SNV | 128 | 184 | 179 | 0 | 8 | 17 | S |
| 1 | 43,814,930 | C/G | NA | 103 | MPL | SNV | 86 | 127 | 96 | 0 | 17 | 21 | S | |
| 2 | 99,172,162 | G/A | NA | 737,33 | INPP4A | SNV | 606 | 996 | 610 | 0 | 24 | 30 | S | |
| 2 | 179,486,342 | A/G | I6005T | 899,67 | TTN | SNV | 1422 | 224 | 1053 | 0 | 16 | 28 | S | |
| 2 | 179,507,003 | G/A | R4567C | 307 | TTN | SNV | 714 | 55 | 152 | 0 | 44 | 45 | S | |
| 3 | 1,414,000 | G/A | V504I | 375,67 | CNTN6 | SNV | 530 | 158 | 439 | 0 | 1 | 16 | S | |
| 3 | 112,991,517 | G/A | A310T | 20,67 | BOC | SNV | 7 | 34 | 21 | 0 | 41 | 14 | S | |
| 4 | 23,825,968 | T/G | K271T | 465 | PPARGC1A | SNV | 793 | 153 | 449 | 0 | 16 | 26 | S | |
| 4 | 146,467,864 | C/T | A262V | 152 | SMAD1 | SNV | 287 | 24 | 145 | 0 | 38 | 42 | S | |
| 5 | 112,155,013 | C/- | NA | 150,67 | APC | Indel | 179 | 155 | 118 | 0 | 60 | 90 | S | |
| 5 | 170,818,803 | G/A | A118T | 240,67 | NPM1 | SNV | 328 | 179 | 215 | 0 | 0 | 18 | O | |
| 6 | 57,075,028 | T/A | I51F | 474 | RAB23 | SNV | 621 | 176 | 625 | 0 | 51 | 43 | S | |
| 11 | 92,533,992 | T/A | Y2605N | 783 | FAT3 | SNV | 877 | 596 | 876 | 0 | 40 | 41 | S | |
| 16 | 77,353,825 | G/A | T818M | 106,33 | ADAMTS18 | SNV | 129 | 122 | 68 | 0 | 53 | 72 | S | |
| 17 | 7,576,927 | C/A | NA | 144 | TP53 | SNV | 175 | 124 | 133 | 0 | 64 | 99 | S | |
| 19 | 39,996,053 | A/C | K352T | 60,33 | DLL3 | SNV | 67 | 70 | 44 | 0 | 13 | 16 | S | |
| 3 | 46,307,365 | G/A | R260Q | 656 | CCR3 | SNV | 693 | 686 | 589 | 0 | 23 | 20 | S | |
| 12 | 25,398,281 | C/T | G13D | 207,33 | KRAS | SNV | 326 | 109 | 187 | 0 | 85 | 91 | S | |
| 3 | 1 | 227,300,460 | C/T | R601Q | 693,67 | CDC42BPA | SNV | 655 | 587 | 839 | 0 | 14 | 35 | S |
| 4 | 48,073,695 | C/G | NA | 237 | TXK | SNV | 247 | 282 | 182 | 0 | 0 | 18 | O | |
| 5 | 112,175,480 | G/T | E1397* | 349 | APC | SNV | 408 | 289 | 350 | 0 | 2 | 36 | S | |
| 6 | 32,184,739 | C/A | C615F | 133,33 | NOTCH4 | SNV | 104 | 172 | 124 | 1 | 5 | 47 | S | |
| 10 | 64,968,097 | G/C | S892C | 285,33 | JMJD1C | SNV | 328 | 204 | 324 | 0 | 24 | 68 | S | |
| 10 | 114,912,204 | G/A | NA | 130,67 | TCF7L2 | SNV | 96 | 187 | 109 | 1 | 7 | 32 | S | |
| 11 | 117,308,068 | C/T | R1287Q | 87 | DSCAML1 | SNV | 61 | 137 | 63 | 2 | 15 | 94 | S | |
| 11 | 118,342,652 | C/G | P293A | 446,33 | MLL | SNV | 390 | 462 | 487 | 0 | 9 | 39 | S | |
| 12 | 120,653,800 | T/G | K4T | 27 | PXN | SNV | 19 | 49 | 13* | 0 | 18 | 0 | T | |
| 12 | 121,416,813 | T/C | F81S | 18,33 | HNF1A | SNV | 8 | 22* | 25 | 0 | 0 | 16 | O | |
| 15 | 44,843,120 | T/C | V65A | 161,33 | EIF3J | SNV | 192 | 145 | 147 | 0 | 0 | 25 | O | |
| 15 | 88,690,618 | G/A | R138W | 494,33 | NTRK3 | SNV | 500 | 490 | 493 | 0 | 26 | 99 | S | |
| 17 | 7,577,095 | G/T | D281E | 192,67 | TP53 | SNV | 160 | 297 | 121 | 0 | 8 | 95 | S | |
| 18 | 45,371,862 | T/C | NA | 276 | SMAD2 | SNV | 297 | 230 | 301 | 0 | 20 | 99 | S | |
| 19 | 7,976,387 | G/T | V377F | 39,67 | MAP2K7 | SNV | 42 | 51 | 26 | 5 | 12 | 77 | S | |
| 12 | 25,398,284 | C/G | G12A | 227,33 | KRAS | SNV | 296 | 265 | 121 | 0 | 4 | 26 | S | |
| 4 | 1 | 160,388,795 | G/T | D66Y | 63,33 | VANGL2 | SNV | 56 | 21 | 113 | 0 | 19 | 46 | S |
| 1 | 226,566,974 | T/A | −538- | 154,67 | PARP1 | SNV | 170 | 53 | 241 | 4 | 38 | 13 | S | |
| 1 | 227,216,861 | C/T | R500H | 284,67 | CDC42BPA | SNV | 357 | 53 | 444 | 0 | 32 | 39 | S | |
| 1 | 228,402,727 | C/A | Q586K | 23,33 | OBSCN | SNV | 26 | 8* | 36 | 0 | 0 | 22 | O | |
| 2 | 179,643,820 | C/T | R1330H | 160,33 | TTN | SNV | 301 | 25 | 155 | 0 | 60 | 43 | S | |
| 3 | 10,070,375 | G/A | D12N | 99,67 | FANCD2 | SNV | 235 | 39 | 25 | 0 | 36 | 20 | S | |
| 5 | 6,743,939 | A/T | I161F | 96,67 | PAPD7 | SNV | 233 | 25 | 32 | 0 | 40 | 72 | S | |
| 5 | 38,960,005 | C/T | E643K | 211,33 | RICTOR | SNV | 537 | 33 | 64 | 0 | 27 | 42 | S | |
| 5 | 112,173,917 | C/T | R876* | 312 | APC | SNV | 591 | 69 | 276 | 0 | 43 | 98 | S | |
| 7 | 19,156,452 | T/A | S33C | 68 | TWIST1 | SNV | 83 | 32 | 89 | 0 | 0 | 17 | O | |
| 8 | 2,887,015 | C/T | E2562K | 81,67 | CSMD1 | SNV | 140 | 6 | 99 | 0 | 50 | 35 | S | |
| 8 | 38,275,833 | C/T | R357Q | 84,33 | FGFR1 | SNV | 108 | 13 | 132 | 1 | 85 | 57 | S | |
| 8 | 103,307,432 | G/A | P1347L | 628 | UBR5 | SNV | 717 | 79 | 1088 | 0 | 68 | 68 | S | |
| 9 | 113,496,548 | C/T | R216W | 318,67 | MUSK | SNV | 431 | 98 | 427 | 1 | 28 | 22 | S | |
| 11 | 11,373,733 | G/A | R312W | 205,67 | CSNK2A3 | SNV | 324 | 46 | 247 | 0 | 13 | 33 | S | |
| 17 | 40,491,345 | C/T | R152Q | 228,33 | STAT3 | SNV | 408 | 47 | 230 | 0 | 70 | 98 | S | |
| 20 | 2,842,501 | G/A | R317H | 45,67 | VPS16 | SNV | 58 | 11 | 68 | 0 | 27 | 21 | S | |
| X | 1,404,820 | C/T | NA | 99,67 | CSF2RA | SNV | 101 | 48 | 150 | 0 | 56 | 40 | S | |
| X | 153,592,746 | G/A | NA | 32,67 | FLNA | SNV | 22 | 9* | 67 | 0 | 0 | 19 | O | |
| 12 | 25,398,285 | C/A | G12C | 140,67 | KRAS | SNV | 240 | 56 | 126 | 1 | 41 | 56 | S | |
| 17 | 7,578,406 | C/T | R175H | 31 | TP53 | SNV | 33 | 5 | 55 | 0 | 80 | 96 | S | |
| 5 | 1 | 242,016,664 | C/T | R96* | 431 | EXO1 | SNV | 532 | 385 | 376 | 0 | 17 | 34 | S |
| 2 | 99,180,106 | C/A | NA | 5,33 | INPP4A | SNV | 1 | 4 | 11 | 0 | 75 | 55 | S | |
| 4 | 103,446,721 | G/A | NA | 74,33 | NFKB1 | SNV | 94 | 58 | 71 | 0 | 14 | 21 | S | |
| 4 | 104,070,038 | T/A | Q1244L | 243,67 | CENPE | SNV | 388 | 226 | 117 | 0 | 22 | 10 | S | |
| 5 | 112,175,621 | C/T | Q1444* | 280 | APC | SNV | 436 | 226 | 178 | 0 | 36 | 88 | S | |
| 6 | 30,856,718 | G/A | R40Q | 200 | DDR1 | SNV | 187 | 206 | 207 | 2 | 33 | 64 | S | |
| 6 | 162,683,713 | C/T | D86N | 210,33 | PARK2 | SNV | 220 | 246 | 165 | 0 | 8 | 19 | S | |
| 7 | 141,424,934 | C/T | R444C | 235 | WEE2 | SNV | 186 | 275 | 244 | 0 | 23 | 40 | S | |
| 10 | 17,142,212 | -/A | NA | 200,67 | CUBN | Indel | 217 | 204 | 181 | 0 | 7 | 18 | S | |
| 15 | 64,275,855 | C/T | R64H | 58,33 | DAPK2 | SNV | 54 | 65 | 56 | 0 | 3 | 20 | S | |
| 17 | 27,208,869 | C/T | E227K | 33 | FLOT2 | SNV | 39 | 26 | 34 | 0 | 50 | 94 | S | |
| 18 | 4,891,888 | G/T | D351Y | 419 | SMAD4 | SNV | 583 | 353 | 321 | 0 | 52 | 100 | S | |
| 12 | 25,398,281 | C/T | G13D | 182 | KRAS | SNV | 202 | 164 | 180 | 0 | 49 | 95 | S | |
| 14 | 21,854,022 | G/T | P2499H | 35,67 | CHD8 | SNV | 31 | 38 | 38 | 0 | 5 | 18 | S | |
| 17 | 7,578,406 | C/T | R175H | 28,67 | TP53 | SNV | 43 | 14 | 29 | 0 | 50 | 97 | S | |
| 6 | 1 | 45,809,000 | G/A | D307N | 125,67 | TOE1 | SNV | 290 | 8 | 79 | 1 | 25 | 47 | S |
| 2 | 170,009,480 | G/A | NA | 96 | LRP2 | SNV | 93 | 32 | 163 | 0 | 38 | 4 | S | |
| 3 | 14,200,245 | T/G | S343R | 127,33 | XPC | SNV | 185 | 98 | 99 | 0 | 33 | 17 | S | |
| 4 | 114,186,052 | G/A | NA | 28,67 | ANK2 | SNV | 55 | 7* | 24 | 0 | 0 | 29 | O | |
| 5 | 112,174,864 | AA/- | NA | 256,33 | APC | Indel | 505 | 100 | 164 | 0 | 36 | 43 | S | |
| 6 | 32,944,234 | T/A | L273H | 316 | BRD2 | SNV | 473 | 124 | 351 | 0 | 81 | 76 | S | |
| 7 | 76,111,907 | G/- | NA | 23,33 | DTX2 | Indel | 31 | 15 | 24 | 0 | 27 | 12 | S | |
| 8 | 41,166,327 | C/T | A118T | 11,67 | SFRP1 | SNV | 10 | 17 | 8 | 0 | 35 | 12 | S | |
| 8 | 103,307,229 | T/G | K1390T | 542 | UBR5 | SNV | 661 | 135 | 830 | 0 | 17 | 0 | T | |
| 9 | 71,504,018 | T/G | F147C | 237,33 | PIP5K1B | SNV | 363 | 51 | 298 | 0 | 0 | 23 | O | |
| 9 | 77,359,089 | T/G | K1690Q | 274,33 | TRPM6 | SNV | 405 | 96 | 322 | 0 | 0 | 21 | O | |
| 18 | 9,522,319 | G/A | E289K | 246,67 | RALBP1 | SNV | 422 | 31 | 287 | 1 | 23 | 3 | S | |
| 19 | 1,455,191 | C/A | L153I | 32,67 | APC2 | SNV | 77 | 10 | 11 | 0 | 50 | 82 | S | |
| 19 | 50,374,886 | T/G | K182T | 104,67 | AKT1S1 | SNV | 91 | 87 | 136 | 0 | 0 | 16 | O | |
| 19 | 56,042,661 | C/T | G102D | 12,33 | SBK2 | SNV | 6 | 19 | 12 | 0 | 21 | 25 | S | |
| 20 | 8,639,221 | G/C | Q244H | 355,33 | PLCB1 | SNV | 440 | 185 | 441 | 1 | 16 | 7 | S | |
| X | 591,711 | G/A | G27R | 54,33 | SHOX | SNV | 45 | 64 | 54 | 0 | 56 | 20 | S | |
| 17 | 7,577,114 | C/T | C275Y | 87,67 | TP53 | SNV | 151 | 11 | 101 | 0 | 82 | 68 | S | |
| 7 | 1 | 41,404,534 | A/- | NA | 13,33 | RP11-348A7.1 | Indel | 8 | 2* | 30 | 0 | 0 | 17 | O |
| 1 | 44,154,629 | G/A | E634K | 125,67 | KDM4A | SNV | 134 | 91 | 152 | 1 | 18 | 2 | S | |
| 1 | 198,225,575 | G/C | E106Q | 232 | NEK7 | SNV | 331 | 156 | 209 | 1 | 19 | 42 | S | |
| 2 | 148,657,319 | C/A | S127* | 442,33 | ACVR2A | SNV | 760 | 303 | 264 | 0 | 50 | 94 | S | |
| 3 | 178,916,725 | C/T | R38C | 603,67 | PIK3CA | SNV | 750 | 459 | 602 | 0 | 54 | 77 | S | |
| 3 | 184,293,701 | G/A | A314T | 60,33 | EPHB3 | SNV | 59 | 49 | 73 | 2 | 16 | 1 | S | |
| 5 | 112,175,423 | C/T | Q1378* | 330 | APC | SNV | 403 | 228 | 359 | 0 | 68 | 91 | S | |
| 7 | 98,565,256 | T/A | Y2476N | 169,67 | TRRAP | SNV | 260 | 64 | 185 | 0 | 75 | 88 | S | |
| 8 | 3,432,515 | T/G | E433D | 148 | CSMD1 | SNV | 256 | 67 | 121 | 0 | 25 | 86 | S | |
| 10 | 412,306 | C/A | R726I | 548,33 | DIP2C | SNV | 616 | 414 | 615 | 0 | 21 | 55 | S | |
| 10 | 8,115,818 | C/G | N388K | 353,67 | GATA3 | SNV | 446 | 314 | 301 | 0 | 6 | 54 | S | |
| 10 | 64,967,678 | G/C | P1032A | 454,67 | JMJD1C | SNV | 659 | 275 | 430 | 0 | 21 | 47 | S | |
| 11 | 116,719,906 | G/A | S983F | 45 | SIK3 | SNV | 41 | 37 | 57 | 0 | 22 | 2 | S | |
| 11 | 117,392,003 | G/A | T412M | 107,33 | DSCAML1 | SNV | 127 | 117 | 78 | 0 | 11 | 26 | S | |
| 13 | 110,804,709 | C/T | A1634T | 141 | COL4A1 | SNV | 122 | 123 | 178 | 1 | 8 | 42 | S | |
| 16 | 68,842,430 | C/A | P87Q | 207,67 | CDH1 | SNV | 275 | 126 | 222 | 0 | 6 | 17 | S | |
| 4 | 153,247,288 | C/A | R505L | 583,33 | FBXW7 | SNV | 783 | 382 | 585 | 0 | 25 | 46 | S | |
| 12 | 25,398,284 | C/A | G12V | 176,33 | KRAS | SNV | 294 | 111 | 124 | 1 | 13 | 28 | S | |
| 17 | 7,578,406 | C/T | R175H | 25 | TP53 | SNV | 39 | 27 | 9 | 0 | 26 | 67 | S | |
| 8 | 1 | 9,793,472 | T/C | E795G | 153,67 | CLSTN1 | SNV | 186 | 161 | 114 | 0 | 14 | 46 | S |
| 1 | 12,164,486 | G/A | E107K | 430,67 | TNFRSF8 | SNV | 453 | 474 | 365 | 1 | 9 | 38 | S | |
| 1 | 165,876,941 | C/T | Q223* | 128 | UCK2 | SNV | 157 | 113 | 114 | 0 | 13 | 57 | S | |
| 2 | 33,482,484 | A/T | K441N | 344,67 | LTBP1 | SNV | 389 | 341 | 304 | 0 | 18 | 34 | S | |
| 2 | 121,708,928 | G/A | V122M | 48,33 | GLI2 | SNV | 57 | 33 | 55 | 0 | 6 | 29 | S | |
| 2 | 171,884,902 | C/T | E347K | 267,67 | TLK1 | SNV | 345 | 323 | 135 | 1 | 6 | 22 | S | |
| 2 | 179,547,627 | A/G | V10647A | 110,67 | TTN | SNV | 168 | 77 | 87 | 0 | 14 | 34 | S | |
| 2 | 179,622,344 | C/A | G3489W | 305 | TTN | SNV | 344 | 299 | 272 | 4 | 12 | 48 | S | |
| 2 | 215,593,455 | G/A | S131L | 597 | BARD1 | SNV | 639 | 555 | 597 | 1 | 12 | 38 | S | |
| 3 | 1,444,098 | A/C | I900L | 305,67 | CNTN6 | SNV | 429 | 231 | 257 | 0 | 38 | 94 | S | |
| 3 | 25,659,978 | C/A | L1089F | 140,67 | TOP2B | SNV | 191 | 136 | 95 | 1 | 3 | 16 | S | |
| 3 | 30,732,969 | C/T | R528C | 49 | TGFBR2 | SNV | 77 | 34 | 36 | 0 | 29 | 89 | S | |
| 3 | 171,455,712 | C/T | V44M | 134,33 | PLD1 | SNV | 208 | 77 | 118 | 0 | 10 | 26 | S | |
| 5 | 112,175,675 | AG/- | NA | 288,33 | APC | Indel | 382 | 257 | 226 | 0 | 11 | 54 | S | |
| 7 | 6,038,819 | C/T | G209R | 590,67 | PMS2 | SNV | 611 | 612 | 549 | 1 | 12 | 38 | S | |
| 8 | 48,715,930 | G/A | R3286W | 173 | PRKDC | SNV | 224 | 173 | 122 | 1 | 6 | 39 | S | |
| 8 | 121,282,371 | C/A | S1057R | 190,67 | COL14A1 | SNV | 303 | 120 | 149 | 1 | 8 | 31 | S | |
| 9 | 94,486,903 | C/T | D485N | 156,67 | ROR2 | SNV | 196 | 139 | 135 | 1 | 24 | 47 | S | |
| 10 | 81,006,67 | C/T | S214L | 38,67 | GATA3 | SNV | 53 | 30 | 33 | 2 | 7 | 39 | S | |
| 10 | 89,692,905 | G/T | R130L | 234 | PTEN | SNV | 281 | 212 | 209 | 0 | 14 | 46 | S | |
| 11 | 108,128,322 | A/C | N789H | 147 | ATM | SNV | 149 | 125 | 167 | 1 | 28 | 54 | S | |
| 12 | 52,374,912 | A/G | E195G | 352 | ACVR1B | SNV | 352 | 298 | 406 | 1 | 35 | 69 | S | |
| 12 | 53,686,729 | G/A | V2046M | 52 | ESPL1 | SNV | 46 | 47 | 63 | 0 | 15 | 38 | S | |
| 19 | 15,276,227 | C/T | G110E | 125,67 | NOTCH3 | SNV | 164 | 81 | 132 | 1 | 26 | 42 | S | |
| X | 44,920,568 | G/A | NA | 55 | KDM6A | SNV | 63 | 47 | 55 | 3 | 19 | 0 | T | |
| X | 71,813,093 | C/A | G1052V | 264,67 | PHKA1 | SNV | 324 | 265 | 205 | 1 | 15 | 72 | S | |
| 7 | 140,453,136 | A/T | V28E | 422 | BRAF | SNV | 474 | 395 | 397 | 0 | 6 | 35 | S | |
| 10 | 89,692,904 | C/T | R130* | 233 | PTEN | SNV | 278 | 212 | 209 | 0 | 16 | 45 | S |
Overview of all somatic mutations identified by SOLiD sequencing. Unique mutations are highlighted in boldface. The allele frequency is the percentage of reads in which the mutant allele has been detected. SNV, single-nucleotide variant.
Low coverage for samples with a unique mutation (might have resulted in the inability to identify a mutation).
Table S2.
Sequencing statistics depicted separately for all samples from each patient
| Sample | Total reads | % mapped | % uniquely mapped | % in target bases | Average coverage | Median coverage | % bases covered | % bases covered >20× |
| Subject01 - blood | 55,725,994 | 87.65 | 82.13 | 55.57 | 231.69 | 174.00 | 94.55 | 80.71 |
| Subject01 - organoid | 40,839,236 | 87.72 | 81.78 | 58.99 | 180.51 | 117.00 | 93.65 | 76.53 |
| Subject01 - tumor | 89,556,002 | 88.03 | 82.11 | 51.25 | 344.62 | 230.00 | 95.82 | 85.74 |
| Subject02 - blood | 78,031,553 | 88.48 | 82.46 | 55.59 | 327.50 | 248.00 | 95.41 | 84.21 |
| Subject02 - organoid | 57,602,210 | 87.38 | 81.41 | 55.31 | 237.68 | 155.00 | 94.80 | 81.20 |
| Subject02 - tumor | 58,474,524 | 85.93 | 80.16 | 54.31 | 232.86 | 157.00 | 96.19 | 86.96 |
| Subject03 - blood | 60,117,496 | 87.51 | 82.05 | 55.64 | 249.91 | 190.00 | 94.68 | 81.40 |
| Subject03 - organoid | 55,752,800 | 88.87 | 83.60 | 55.66 | 235.65 | 160.00 | 94.75 | 80.97 |
| Subject03 - tumor | 72,388,430 | 86.26 | 80.84 | 55.52 | 296.03 | 248.00 | 96.07 | 87.57 |
| Subject04 - blood | 59,027,598 | 86.57 | 80.78 | 55.33 | 241.00 | 189.00 | 95.30 | 83.18 |
| Subject04 - organoid | 42,977,631 | 89.45 | 84.03 | 56.05 | 184.11 | 106.00 | 94.36 | 78.99 |
| Subject04 - tumor | 20,034,396 | 79.87 | 73.20 | 41.30 | 56.46 | 31.00 | 89.01 | 59.71 |
| Subject05 - blood | 58,658,789 | 87.12 | 81.22 | 53.58 | 233.41 | 181.00 | 94.89 | 81.71 |
| Subject05 - organoid | 45,661,474 | 88.40 | 82.37 | 54.46 | 187.51 | 144.00 | 94.87 | 80.89 |
| Subject05 - tumor | 51,527,869 | 88.07 | 82.24 | 56.14 | 217.18 | 157.00 | 94.57 | 81.14 |
| Subject06 - blood | 62,923,615 | 79.67 | 74.27 | 54.87 | 234.91 | 195.00 | 94.68 | 83.73 |
| Subject06 - organoid | 24,908,884 | 68.34 | 62.27 | 45.03 | 65.48 | 42.00 | 94.45 | 70.95 |
| Subject06 - tumor | 61,725,207 | 63.98 | 59.68 | 56.61 | 190.95 | 132.00 | 93.63 | 79.90 |
| Subject07 - blood | 66,097,552 | 82.22 | 76.95 | 58.13 | 269.70 | 226.00 | 95.15 | 85.49 |
| Subject07 - organoid | 65,525,831 | 79.34 | 74.36 | 59.96 | 266.39 | 201.00 | 94.99 | 84.86 |
| Subject07 - tumor | 41,859,372 | 76.29 | 71.42 | 60.34 | 164.69 | 132.00 | 94.54 | 82.40 |
| Subject08 - blood | 57,698,769 | 78.87 | 73.78 | 59.47 | 231.04 | 188.00 | 95.19 | 84.77 |
| Subject08 - organoid | 55,896,268 | 75.59 | 70.73 | 60.17 | 217.12 | 162.00 | 94.67 | 82.34 |
| Subject08 - tumor | 49,018,897 | 76.00 | 71.18 | 60.87 | 193.64 | 156.00 | 94.68 | 82.79 |
Table S3.
Percentage of tumor cells and necrosis of original tumor biopsies per patient
| Sample | Tumor cells, % | Necrosis, % |
| 01 | 80 | 10 |
| 02 | 90 | 10 |
| 03 | 80 | 40 |
| 04 | 90 | 10 |
| 05 | 70 | 10 |
| 06 | 90 | 50 |
| 07 | 80 | 50 |
| 08 | 90 | Not available |
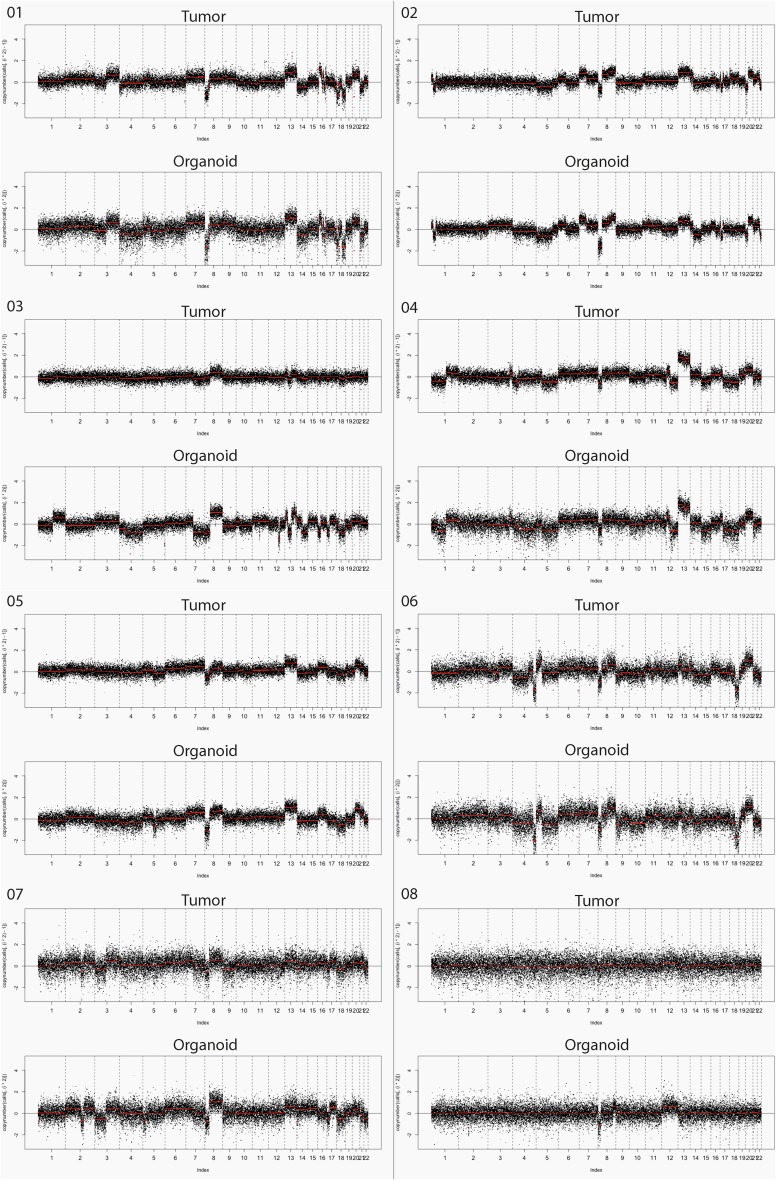
Fig. S1.
Copy number plots for tumor biopsy and biopsy-derived organoid cultures of patients 01–08.
Discussion
These data show that organoid cultures from colorectal cancer can be established readily from biopsies of metastases. The culture success rate of 71% for colorectal cancer compares favorably with the take rate of 15–20% for organoid cultures of biopsies from prostate cancer described by Gao et al. (6). Sequencing data and copy number analysis were largely concordant between organoid cultures and tumor biopsies. The most important observation from our study, however, is that none of the 15 mutations that were found exclusively in either the tumor or organoid culture are in driver genes or genes amenable for drug targeting. Discordance, when present, could be attributed to biological factors such as sampling variation from a heterogeneous tumor or ongoing normal tumor evolution or to selection resulting from culture conditions. With current knowledge it is not possible to distinguish between these factors. Aside from biological processes, technical limitations can result in a misrepresentation of the actual concordance by impeding factors such as low coverage of a particular gene or sample. For instance, 6 of 15 unique mutations have a coverage of <25× of the gene in the matched sample (where the mutation was not identified), and for patient 04 and patient 06 overall coverage of the tumor sample and biopsy-derived culture, respectively, was very low (median coverage <50×) (Tables S1 and S2). Furthermore, the presence of stromal cells and tumor necrosis in biopsies (resulting in a lower tumor percentage) may have resulted in less accurate detection of copy number aberrations and somatic mutations in the tumor biopsy as compared with the cultures. A lower tumor percentage can result in a misrepresentation of allele frequency in a negative way, so that the frequency can fall under the limit of detection. The actual allele frequency of a mutation in tumor biopsies thus is sometimes higher than depicted in Table S1. A lower tumor percentage also affects copy number profiles by reducing the signal toward the diploid state, resulting in discrepancies in the intensity of red and blue shading between tumor biopsies and corresponding biopsy-derived cultures in Fig. 2. This effect could have contributed to the discordance in patient 3 (Fig. S1 and Table S3).
To conclude, organoid cultures from colorectal cancer can be established readily from biopsies of metastases with preservation of the mutational and copy number landscape. This result supports the translation of patient-derived tumor organoids to the clinic as an ex vivo test platform, with the potential to meet the dire need for a means to make more rational treatment decisions for the individual patient. Clinical trials to validate the predictive value of organoids have been initiated.
Acknowledgments
We thank Marja van Blokland (Molecular Pathology, University Medical Center Utrecht) for DNA isolation and Nicolle Besselink (Medical Oncology, University Medical Center Utrecht) for library preparation and sequencing. This work was supported by the Center for Personalized Cancer Treatment, the Barcode for Life Foundation, and Cancer GenomiCs.nl through The Netherlands Organization for Scientific Research Gravitation grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516689112/-/DCSupplemental.
References
- 1.Burstein HJ, et al. American Society of Clinical Oncology American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J Clin Oncol. 2011;29(24):3328–3330. doi: 10.1200/JCO.2011.36.0354. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M, et al. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol Cancer Ther. 2011;10(8):1311–1316. doi: 10.1158/1535-7163.MCT-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 4.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6(1):25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Gao D, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32(20):2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494(7436):247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karthaus WR, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harakalova M, et al. Multiplexed array-based and in-solution genomic enrichment for flexible and cost-effective targeted next-generation sequencing. Nat Protoc. 2011;6(12):1870–1886. doi: 10.1038/nprot.2011.396. [DOI] [PubMed] [Google Scholar]
- 12.Hoogstraat M, et al. Genomic and transcriptomic plasticity in treatment-naive ovarian cancer. Genome Res. 2014;24(2):200–211. doi: 10.1101/gr.161026.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vermaat JS, et al. Primary colorectal cancers and their subsequent hepatic metastases are genetically different: Implications for selection of patients for targeted treatment. Clin Cancer Res. 2012;18(3):688–699. doi: 10.1158/1078-0432.CCR-11-1965. [DOI] [PubMed] [Google Scholar]
- 14.Kuilman T, et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Wiel MA, Picard F, van Wieringen WN, Ylstra B. Preprocessing and downstream analysis of microarray DNA copy number profiles. Brief Bioinform. 2011;12(1):10–21. doi: 10.1093/bib/bbq004. [DOI] [PubMed] [Google Scholar]