Significance
Although full mammalian limbs do not regenerate after amputation, the fingertips of select mammalian species do. Understanding digit tip regeneration at the molecular level can potentially provide insight into designing translational therapies for regrowing greater portions of the limbs and other nonregenerative tissues. The nail is known to be critical for digit tip regeneration, at least in part through a mechanism dependent on Wnt signaling. Here, we identify a cell population expressing a mediator of Wnt signaling, Lgr6 (leucine-rich repeat-containing G protein-coupled receptor 6), as key stem cells for the nail. Moreover, we find that Lgr6 is required for proper digit tip regeneration.
Keywords: epimorphic regeneration, Lgr5, Wnt signaling, nail fold, nail matrix
Abstract
The tips of the digits of some mammals, including human infants and mice, are capable of complete regeneration after injury. This process is reliant on the presence of the overlaying nail organ and is mediated by a proliferative blastema. Epithelial Wnt/β-catenin signaling has been shown to be necessary for mouse digit tip regeneration. Here, we report on Lgr5 and Lgr6 (leucine-rich repeat-containing G protein-coupled receptor 5 and 6), two important agonists of the Wnt pathway that are known to be markers of several epithelial stem cell populations. We find that Lgr5 is expressed in a dermal population of cells adjacent to the specialized epithelia surrounding the keratinized nail plate. Moreover, Lgr5-expressing cells contribute to this dermis, but not the blastema, during digit tip regeneration. In contrast, we find that Lgr6 is expressed within cells of the nail matrix portion of the nail epithelium, as well as in a subset of cells in the bone and eccrine sweat glands. Genetic lineage analysis reveals that Lgr6-expressing cells give rise to the nail during homeostatic growth, demonstrating that Lgr6 is a marker of nail stem cells. Moreover, Lgr6-expressing cells contribute to the blastema, suggesting a potential direct role for Lgr6-expressing cells during digit tip regeneration. This role is confirmed by analysis of Lgr6-deficient mice, which have both a nail and bone regeneration defect.
Appendage regeneration in mammals is extremely limited and is found only in cervid antlers (1) and the digit tips of some rodents and primates (2, 3), including humans (4, 5). Deciphering the cell populations and molecular networks used during this process could potentially lend insight into the elements necessary to induce regeneration more broadly in human tissues. The digit tip regenerates via epimorphic regeneration, a process characterized by the intermediate formation of a blastema, a collection of morphologically undifferentiated mesenchymal cells derived from the underlying tissue. Previous work has shown that the digit tip blastema comprises a heterogeneous population of lineage-restricted progenitor cells (6, 7). Digit tip regeneration is under the constraint of the nail organ, a keratinized ectodermal appendage unique to the digit tip. In humans and in mice, amputations that transect the nail can go on to form a blastema and regenerate, yet amputations past the proximal limit of the nail do not mount a regenerative response. Indeed, the nail is necessary for digit tip regeneration (8), and, furthermore, nails implanted proximally to nonregenerative digital positions are sufficient to induce bony outgrowth (9). At least one interpretation of these findings is that molecular signaling normally responsible for continuous nail growth may create a permissive regenerative environment (10).
The nail organ is a specialized ectodermal appendage comprising a superficial hard keratin plate derived from the proliferative matrix at the base of the nail and supported by the nail bed, which has highly vascularized epithelial ridges adhering the plate to the surface of the digit. The nail is further supported by surrounding epithelia, termed the eponychium, perionychium, and hyponychium, that protect the underlying soft tissue from injury and infection. It has been long-recognized that a slowly cycling population of presumptive stem cells resides within the nail matrix (11), and several reports use pulse–chase experiments to demonstrate that nail progenitor cells reside in the matrix (10, 12). However, a definitive marker of this population has not yet been identified. To this end, we focused on Lgr (leucine-rich repeat-containing G protein-coupled receptor) proteins, which are known to be markers of several adult stem cell populations.
Lgr4, Lgr5, and Lgr6 (leucine-rich repeat-containing G protein-coupled receptor 4/5/6) serve as receptors for R-spondins, and together the Lgr–R-spondin complex prevents the constitutive ubiquitination of Wnt receptors via transmembrane proteins RNF43 and ZNRF3, such that cell populations expressing Lgr4/5/6 proteins are more responsive to Wnt signaling in the presence of R-spondin (13). Although Lgr4 is more broadly expressed in a wide range of tissues (14, 15), Lgr5 and Lgr6 are tightly regulated and mark several adult stem cell populations throughout the body (16), including Lgr5 in the intestinal epithelium and hair follicle (17, 18), as well as Lgr6 in the sweat glands and interfollicular epidermis (19). Canonical Wnt signaling has been shown to be necessary for appendage regeneration in many vertebrate epimorphic regenerative systems, including zebrafish fin, axolotl limb, and Xenopus tail (20–22). In addition, conditional deletion of β-catenin in Keratin-14–positive epithelia impairs mouse digit tip regeneration (10). Taken together, the precedent for Lgr5 and Lgr6 to mark epithelial stem cell populations, in combination with the demonstrated necessity of Wnt signaling for epimorphic regeneration, makes Lgr5 and Lgr6 logical candidates to interrogate for expression and function in nail stem cells as they relate to digit tip regeneration.
In this article, we show that Lgr5 is a marker of neither the nail stem cells nor the nail epithelium, but instead marks a mesenchymal population of cells within the proximal nail fold and the distal groove whose expression is not correlated with a regeneration-specific function. Lgr6, however, marks several cell populations within the digit tip, including a small population of cells within the nail epithelium specific to the matrix. Genetic fate mapping during both nail homeostasis and digit tip regeneration shows that the Lgr6-marked cells are adult stem cells giving rise to the nail. Moreover, during digit tip regeneration, Lgr6-marked cell descendants are found within the blastema, suggesting a possible regeneration-specific function. Finally, we find that Lgr6−/− animals have a digit tip regeneration defect.
Results
Canonical Wnt Signaling Occurs Within the Nail Matrix.
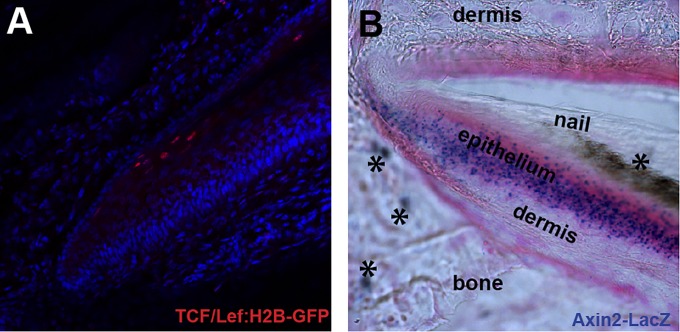
Digit tip regeneration has previously been shown to be dependent upon the epithelially derived nail organ (8). In addition, a recent study demonstrates that this process requires canonical Wnt signaling originating from the epithelium (10). Thus, an attractive hypothesis is that these two dependencies are intertwined and that the nail matrix is a site of important Wnt activity. To directly examine this possibility, we examined mice carrying transgenic reporters for markers of canonical Wnt signaling, TCF/Lef (T-cell factor/lymphoid enhancer factor) and Axin2. In addition, we used immunohistochemistry to probe for nuclear localization of β-catenin, another hallmark of active canonical Wnt signaling. The boundaries of expression identified in TCF/LefH2B-GFP and Axin2LacZ mice and through β-catenin immunohistochemistry varied subtly from one another; however, they collectively showed evidence of canonical Wnt signaling within the nail matrix (Fig. S1) (see Fig. 3C), largely consistent with Takeo et al. (10).
Fig. S1.
Canonical Wnt signaling in the nail matrix. Markers of canonical Wnt signaling are as follows: (A) TCF/LefH2B-GFP digit stained with anti-GFP (red) and counterstained with DAPI (blue) and (B) Axin2LacZ digit stained with X-gal (blue) and counterstained with eosin (pink) showing Axin2 expression within the nail epithelium. Asterisks denote histology artifacts.
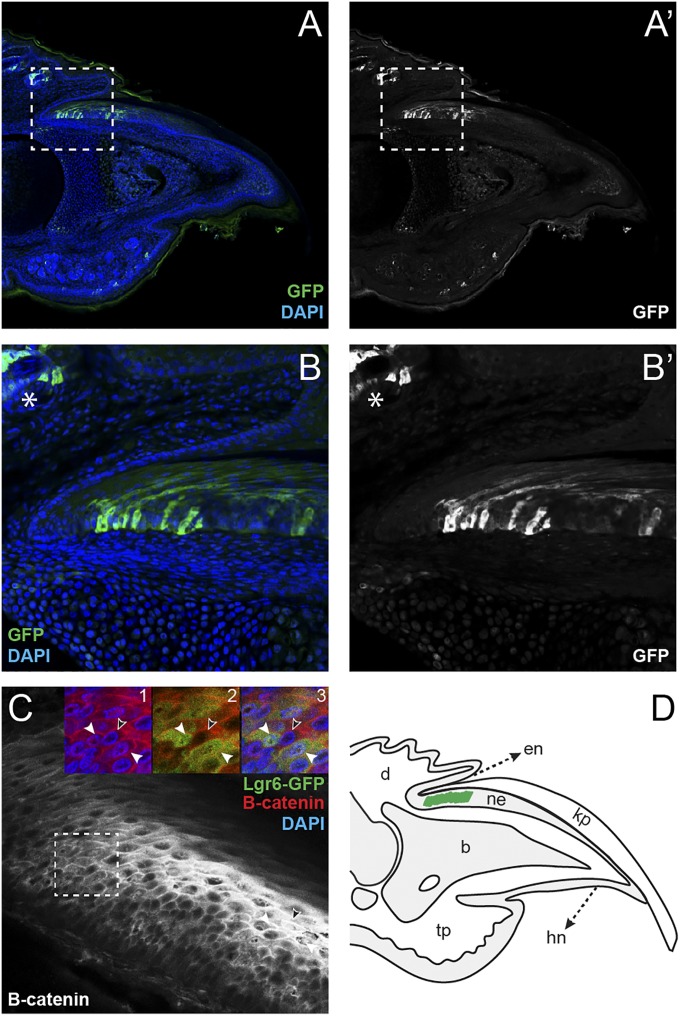
Fig. 3.
Lgr6 is expressed in the nail matrix. Section immunohistochemistry of quiescent Lgr6EGFP-ires-creERT2 mouse digit tips. (A) View of entire digit tip with Lgr6-GFP expression in green (anti-GFP), counterstained with DAPI (blue). (A′) Lgr6-GFP nail matrix expression domain without pseudocolor. (B and B′) Higher magnification regions from A and A′, respectively, with asterisk indicating staining in the hair follicle/sebaceous gland. (C) Anti–β-catenin digit tip immunohistochemistry. (Inset) Close-up of nail matrix cells stained for β-catenin (red), Lgr6-GFP (green), and/or DAPI (blue). Open arrowhead denotes Lgr6-negative cell with no nuclear localization of β-catenin, and filled arrowheads show Lgr6-positive cells with nuclear localization of β-catenin. (D) Schematic of Lgr6 nail epithelium expression domain within the mouse digit tip (green). b, bone; d, dermis; en, eponychium; hn, hyponychium; kp, keratinized plate; ne, nail epithelium; tp, toe pad.
Lgr5 Expression Marks Mesenchymal Progenitors Local to Specialized Nail Epithelia.
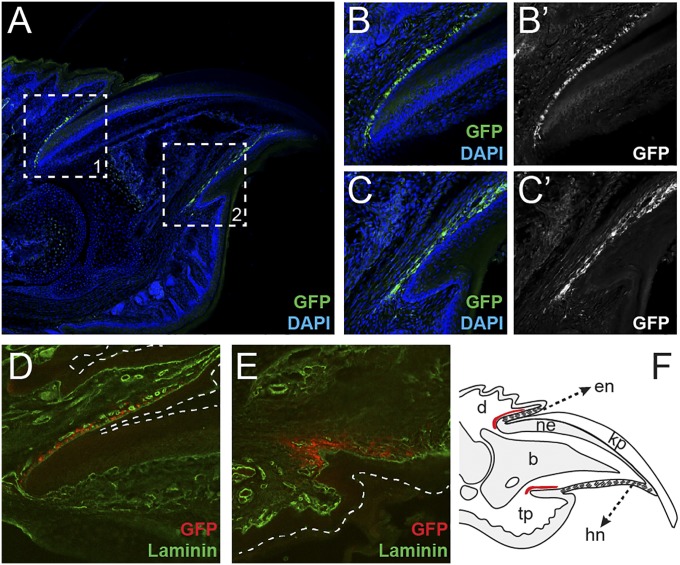
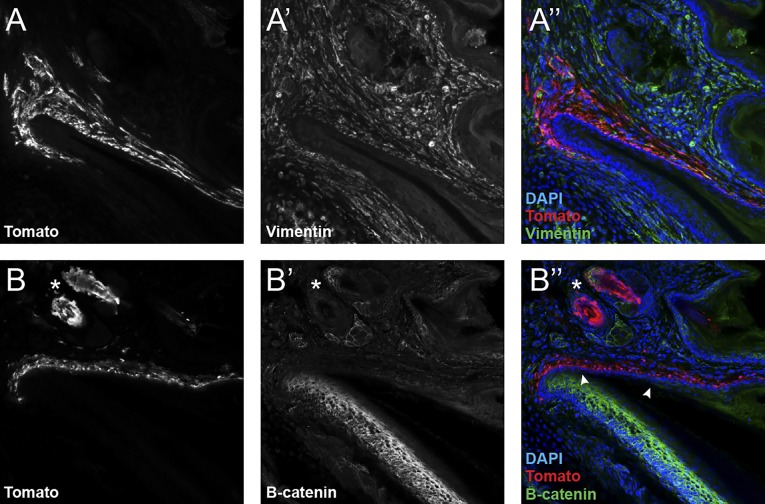
Because canonical Wnt signaling is well-established as a key pathway supporting adult stem cells in a variety of contexts, one possibility was that the Wnt signaling we observed in the nail matrix might include activity in nail stem cells. Extensive work has shown that Lgr5 is a faithful marker of several epithelial adult stem cell populations (16) and has pinpointed Lgr5 as a key modulator of Wnt signaling in adult stem cell biology (23). We therefore set out to determine whether Lgr5 was expressed in the mouse digit tip, specifically in the nail epithelium. We evaluated Lgr5-GFP expression in digit tips of Lgr5EGFP-ires-creERT2 heterozygous mice; however, we found no Lgr5-GFP expression in the nail epithelium (Fig. 1A). Instead, populations of Lgr5-expressing cells were detected dorsal to the nail in the proximal fold (Fig. 1 A–B′ and F) and ventral in the digit within the distal groove (Fig. 1 A, C, C′, and F). Laminin, Vimentin, and B-catenin immunohistochemistry confirmed that the Lgr5-GFP expression was beneath the epithelial basement membrane, within the dermis (Fig. 1 D and E and Fig. S2); however, the cell-type identity and function of this population remains unclear.
Fig. 1.
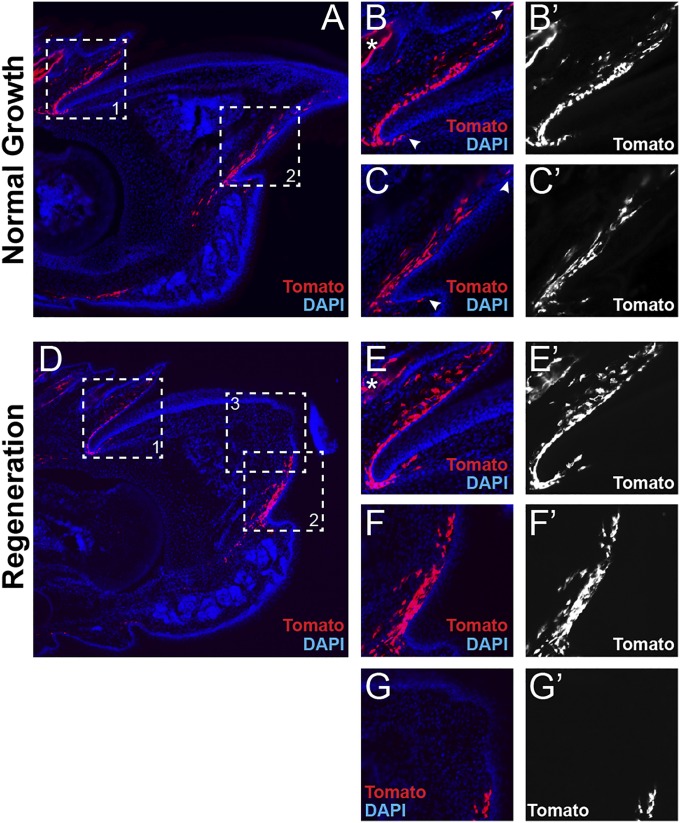
Lgr5 is expressed in a mesenchymal population of cells in the proximal fold and distal groove. Section immunohistochemistry of quiescent Lgr5EGFP-ires-creERT2 mouse digit tips. (A) View of entire digit tip with Lgr5-GFP expression in green (anti-GFP), counterstained with DAPI (blue). (Inset 1) Proximal nail fold. (Inset 2) Distal groove. (B and B′) Higher magnification of dorsal Lgr5-GFP expression domain in proximal nail fold. (C and C’) Ventral Lgr5-GFP expression domain in distal groove. (D and E) Coexpression of Lgr5-GFP (red) and laminin (green) to delineate epithelial boundaries. Dashed lines mark the epidermis. (F) Schematic of Lgr5 expression domains within the mouse digit tip. Red shows region of Lgr5 expression, and hash marks denote specialized nail epithelia eponychium and hyponychium. b, bone; d, dermis; en, eponychium; hn, hyponychium; kp, keratinized plate; ne, nail epithelium; tp, toe pad.
Fig. S2.
Lgr5-expressing cells are localized to the dermis. Lgr5-expressing cell descendants in Lgr5EGFP-ires-creERT2;R26RCAG-LSL-tdTomato 2wkPA digit tips. Imaged region is focused on the dorsal digit tip surrounding the proximal portion of the nail. (A–A′′) Lgr5-expressing cell descendants (Tomato/red) colocalize with the mesenchymal marker Vimentin (green) within the dermis. (B–B′′) Lgr5-expressing cell descendants (Tomato/red) do not colocalize with the epithelial marker B-catenin (green) within the nail epithelium. Note: The left arrowhead depicts the portion of the epithelium that becomes single layer until the stratum corneum is reestablished at the right arrowhead. Asterisks denote hair follicle/sebaceous glands.
Based on the present understanding of Lgr5-expressing cell populations, we hypothesized that the Lgr5-expressing mesenchymal digit tip populations were adult stem cells. In an attempt to assign a potential identity or function, we genetically marked these populations and assessed their cellular contribution to digital tissues during both homeostasis and digit tip regeneration. We treated Lgr5EGFP-ires-creERT2;R26RCAG-LSL-tdTomato heterozygous mice with tamoxifen to permanently genetically mark Lgr5-expressing cells with tdTomato. After 4 wk of normal digit growth, the Lgr5-expressing cell descendants remained dermal and local to the original Lgr5-GFP expression domains in the proximal nail fold and the distal groove (Fig. 2 A–C′), with subtle extensions in the proximal and distal boundaries of the domains (arrowheads in Fig. 2 B and C). To assess whether Lgr5-expressing cells had a specialized function during digit tip regeneration, we evaluated Lgr5 genetically marked digit tips 7 d postamputation (Fig. 2D), and, although there was a qualitative increase in the number of Lgr5-expressing cell descendants in the proximal nail fold and the distal groove (Fig. 2 E–F′), this accumulation was likely associated with increased proliferation during regeneration. Importantly, no Lgr5-expressing cell descendants were found in the blastema (Fig. 2 D, G, and G′), indicating that this unique mesenchymal cell population in the digit does not directly contribute to the regenerating structures. The location and fidelity of these populations allow us to hypothesize that they are dermal fibroblasts that track specifically with the specialized nail epithelia, the eponychium and hyponychium (Fig. 1F). Importantly however, being mesenchymal, they cannot represent the Wnt-responding epithelial cells previously implicated in digit tip regeneration.
Fig. 2.
Lineage analysis of Lgr5-expressing cells during digit tip homeostasis and regeneration. Section immunohistochemistry of tamoxifen-induced Lgr5EGFP-ires-creERT2;R26RCAG-LSL-tdTomato mice. (A) View of entire digit tip after 4 wk of normal growth with Lgr5-expressing cell descendants marked by tdTomato (red) and counterstained with DAPI (blue). (Inset 1) Proximal nail fold. (Inset 2) Distal groove. (B and B′) Higher magnification of dorsal Lgr5 lineage in proximal nail fold and (C and C′) ventral Lgr5 lineage in distal groove. Arrowheads show extension in distal and proximal expression domain boundaries, and asterisks label Lgr5 lineage in the hair follicle. (D) View of entire digit tip 7 d postamputation, with Lgr5-expressing cell descendants marked by tdTomato (red) and counterstained with DAPI (blue). (Inset 1) Proximal nail fold. (Inset 2) Distal groove. (Inset 3) The blastema. (E and E′) Higher magnification of dorsal Lgr5 lineage in proximal nail fold; asterisks label Lgr5 lineage in the hair follicle. (F and F′) Ventral Lgr5 lineage in distal groove. (G and G′) Blastema with no Lgr5-expressing cell descendants found.
Lgr6 Is a Marker of the Nail Matrix and Nail Stem Cells.
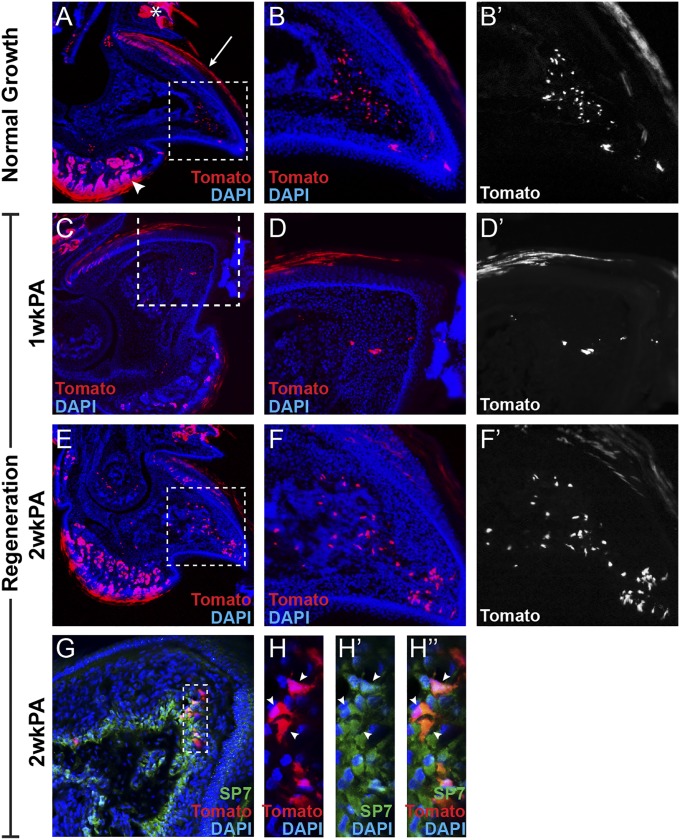
In the absence of Lgr5 expression in the nail epithelium, we turned our attention to Lgr6. Lgr6 expression has also been identified in the context of a number of adult stem cells. Using Lgr6EGFP-ires-creERT2 heterozygous mice, we evaluated the digit tips for Lgr6-GFP expression, and, in this case, we indeed found expression within the nail matrix, the proximal portion of the nail epithelium where the presumptive nail stem cells reside (Fig. 3 A–B′ and D). To assess whether the Lgr6-expressing cells in the nail matrix are in fact nail stem cells, we genetically marked Lgr6-expressing cell populations in Lgr6EGFP-ires-creERT2;R26RCAG-LSL-tdTomato heterozygous mice and followed the contribution of these cells during normal growth and in digit tip regeneration. First, we marked these cells and evaluated their contribution over 4 wk of normal digit/nail growth. Analysis of tdTomato expression in these normally developed digit tips revealed that Lgr6-expressing cells give rise to the nail plate (Fig. 4A, arrow), confirming that Lgr6 is a marker of nail stem cells. Moreover, this analysis revealed and/or confirmed multiple additional sites of Lgr6 expression and cellular contribution within the digit tip. As has been previously characterized, we found that Lgr6 was expressed in the hair follicle/sebaceous gland (Fig. 3B, asterisk) and that this population of Lgr6-expressing cells contributed to the growth of these structures over time (19) (Fig. 4A, asterisk). In addition, we detected previously unidentified Lgr6 expression within the toe pad (Fig. 3A), which gives rise to cells within the eccrine sweat glands, ducts, and toe pad epidermis (Fig. 4A, arrowhead). We also found Lgr6-expressing cell descendants within the bone (Fig. 4 A, box, B, and B′), consistent with the characterized necessity of Wnt signaling in bone development and maintenance (24). Importantly, Lgr5-expressing cells were not found to contribute to the nail epithelium, eccrine sweat glands/ducts, toe pad epidermis, or bone (Fig. 2A), indicating that Lgr5 and Lgr6 have distinct functions in the digit tip.
Fig. 4.
Genetic lineage analysis of Lgr6-expressing cell populations within the digit tip. Section immunohistochemistry of tamoxifen-induced Lgr6EGFP-ires-creERT2;R26RCAG-LSL-tdTomato heterozygous mice with Lgr6-expressing cell descendants expressing tdTomato (red) and counterstained with DAPI (blue). (A) After 4 wk of normal growth, Lgr6-expressing cells contribute to the nail plate (arrow), hair follicle/sebaceous gland (asterisk), eccrine sweat glands/ducts and toe pad epithelium (arrowhead), and the bone (dashed box). Box is magnified in B and B′. (C) One week postamputation (1wkPA), Lgr6-expressing cell descendants are found within the blastema (dashed box). The dashed box is magnified in D and D′. (E) Two weeks postamputation (2wkPA), Lgr6-expressing cell descendants populate the late blastema and the regenerating bone (dashed box). The dashed box is magnified in F and F′. (G) These cells colabel with osteoblast marker SP7. The dashed box is magnified in H, H′, and H′′ where arrowheads show examples of Lgr6-expressing cell descendants that colabel with SP7.
To address whether Lgr6 also has a unique function during digit tip regeneration, we genetically marked Lgr6-expressing cells with tdTomato in Lgr6EGFP-ires-creERT2;R26RCAG-LSL-tdTomato heterozygous mice, subsequently amputated their digit tips, and assessed the digits for Lgr6-expressing cell contribution at different stages during regeneration. By 1 wk postamputation, Lgr6-expressing cell descendants were found in all of the same tissues and domains as were found during normal growth. In addition, we observed that Lgr6-expressing cells also contribute to the blastema (Fig. 4 C–D′). By 2 wk postamputation, Lgr6-expressing cells had accumulated in the “late blastema” as well as the regenerating bone (Fig. 4 E–F′). Previous work has shown that the digit tip bone regenerates via direct ossification, which is facilitated by establishing a new ossification center within the mesenchyme at the distal tip of the digit (25). We hypothesized that the Lgr6-expressing cell descendants within the late blastema are involved in the formation of this new ossification center. Consistent with this hypothesis, Lgr6-expressing cell descendants colabel with the osteoblast marker SP7 by immunohistochemistry (Fig. 4 G and H–H′′) and are likely originating from the resident Lgr6-expressing cells within the bone.
Lgr6 Is Necessary for Digit Tip Regeneration.
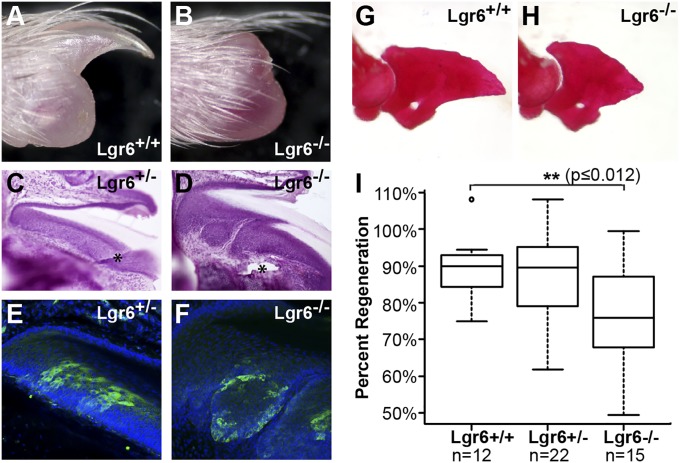
Having found that Lgr6-expressing cells give rise to cells within the blastema and that Lgr6 marks nail stem cells, we next addressed whether Lgr6 is necessary for either nail growth or digit tip regeneration. Different strains of mice possess varying capacities for digit tip regeneration (6). For example, in our hands, mice of the C57BL/6 inbred background, on which the Lgr6 allele is carried, are poor at digit tip regeneration whereas the outbred, CD-1(ICR) strain is much more robust in this respect. We therefore outcrossed the Lgr6−/− allele to CD-1(ICR) mice and assessed their ability to regenerate nails and digit tips relative to heterozygous and WT littermates. The nails developed normally in Lgr6−/− mice; however, after amputation, the nail failed to regenerate in a small subset of cases (3 of 24 digits from eight Lgr6−/− mice) (Fig. 5B). It is of note that we have never observed this phenotype previously on the CD-1(ICR) background (6), thus making a linked genetic mutation, or aberrant amputation, unlikely. Moreover, the contralateral unamputated digits, as well as the preamputation digits, all had normally formed nails (Fig. 5A), implicating the necessity of Lgr6 for nail growth specifically during digit tip regeneration. Histological analysis of the Lgr6−/− dysmorphic nails revealed a defect in the epithelia compared with either Lgr6+/− or Lgr6−/− animals without a phenotype (Fig. 5 C and D). By hematoxylin and eosin staining, it is evident that the Lgr6−/− nails with a regeneration-defective phenotype have a thicker nail epithelium, disorganization of the keratinized plate and nail epithelium, and evidence of dermal cells invading the epithelial layer (Fig. 5 C and D). Interestingly, immunohistochemical analysis of serial sections revealed that the regions of dermal invasion within the epithelium specifically correlate with cells lacking Lgr6 expression (Fig. 5 E and F), suggesting that Lgr6 may function in epithelial organization.
Fig. 5.
The necessity of Lgr6 in nail and bone regeneration. Genetic analysis of digit tip regeneration in Lgr6−/− CD-1(ICR) outbred animals. (A) Representative Lgr6+/+ regenerate digit tip compared with (B) example of nonregenerate Lgr6−/− phenotype. (C and D) Hematoxylin and eosin-stained sections of Lgr6+/− and Lgr6−/− regenerate digit tips. Asterisks denote sectioning artifacts. (E and F) Immunohistochemistry of Lgr6+/− and Lgr6−/− digit tips from serial sections of C and D at higher magnification to focus on the nail matrix: Lgr6-GFP (green) and DAPI (blue). (G) Representative alizarin red-stained digit tip bones from Lgr6+/+ regenerate compared with (H) Lgr6−/− with reduced regeneration. (I) Boxplot of percent regeneration of digit tip bones in Lgr6+/+, Lgr6+/−, and Lgr6−/− cohorts, revealing a significant (P ≤ 0.012) reduction in regeneration of Lgr6−/− animals. The open circle represents outlier.
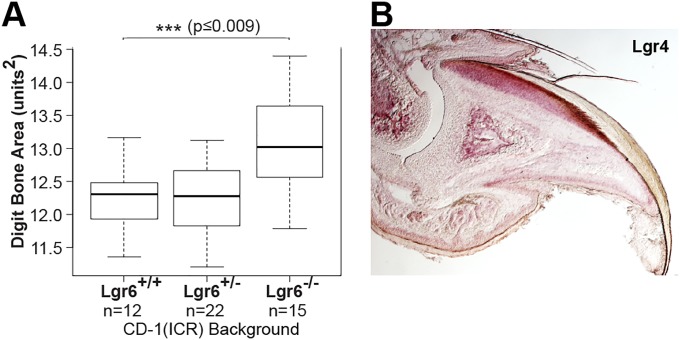
The Lgr6−/− CD-1(ICR) outcross also revealed a bone regeneration phenotype that was more highly penetrant than the dysmorphic nail phenotype. We found that Lgr6−/− regenerate bones were visibly smaller than their WT counterparts (Fig. 5 G and H). Quantitatively, we found that the percent regeneration is significantly less in the Lgr6−/− mice than in Lgr6+/+ littermates (77% vs. 89%, P < 0.012 by Student’s t test) (Fig. 5I). Importantly, as we found in the nail, the effect of Lgr6 is specific to regeneration. Indeed, the digits of Lgr6−/− mice are actually larger than WT and heterozygous controls after normal development (Fig. S3A).
Fig. S3.
Analysis of phenotypic differences of Lgr6 alleles during development. (A) Boxplot of 2D digit bone area measurements from Lgr6+/+, Lgr6+/−, and Lgr6−/− cohorts after normal development in the CD-1(ICR) outbred background. The Lgr6−/− animals have significantly (P ≤ 0.009) larger digits. (B) RNA in situ expression of Lgr4 in the mouse digit tip, with highest levels of expression (dark pink) in the nail epithelium and bone.
Discussion
Lgr6 Marks Nail Stem Cells and Is Required for Digit Tip Regeneration.
Canonical Wnt signaling has been shown to be necessary for epimorphic regeneration, as demonstrated by the conditional deletion of β-catenin in the mouse epidermis during digit tip regeneration, leading to small, dysmorphic regenerate nails/digits (10); this necessity has also been demonstrated in other species (20–22). Because the nail is a continuously growing ectodermal appendage, we hypothesized that Wnt signaling was necessary to maintain the nail stem cell population, which ultimately could induce secondary molecular signaling events facilitating digit tip regeneration. With no identified molecular marker specific to the nail stem cells, we turned to Lgr4/5/6, which have been described as markers of other adult stem cell populations within epithelia, including the hair follicle because it has been hypothesized to be an analogous keratinized ectodermal appendage (26). Based on present research, Lgr5 seemed the most likely candidate of these genes to putatively mark the nail stem cells. Functional experiments and genetic lineage analyses have established that Lgr5 is a stem cell marker of both the intestinal epithelium and the hair follicle (17, 18), and recent experiments have shown it to mark epithelial stem cells in additional tissues, including the stomach, mammary gland, tongue, and ovary (27–30). In most of these cases, Lgr5 expression is driven by canonical Wnt signaling, placing Lgr5 in a feed-forward loop to maintain high Wnt signaling within Lgr5-expressing stem cells. These observations have led to a dogma that Lgr5 is a Wnt target gene, particularly in epithelial stem cell populations (23). However, Lgr5 did not mark nail stem cells and, in fact, did not mark any cells within the nail epithelium, thus setting the growth/maintenance of the nail apart from other epithelial stem cell pools and ectodermal appendages. In contrast, we found that Lgr6 is indeed expressed in the nail matrix, is a marker for nail stem cells, and moreover is necessary for nail regeneration. This population of cells likely represents the key to the necessity of canonical Wnt signaling in the epithelium during digit tip regeneration (10). Moreover, we show that Lgr6 is also expressed in a subset of the osteoblasts in the bone. The role of these cells in normal skeletal homeostasis remains to be determined. Although we found that Lgr6 is also necessary for bone regeneration, it remains unclear whether the Lgr6-positive osteoblasts contribute to this phenotype, or whether the bone regeneration defect is an indirect consequence of the Lgr6 requirement in the nail stem cell population, or both.
Although the requirement of Lgr6 for robust regeneration in the nail and bone is clear, the nail regeneration defect is detected at low penetrance, and the extent of the bone regeneration defect is rather modest. The relatively small magnitude of these phenotypes is likely attributable to redundancy with the third marker of this Lgr subfamily, Lgr4. We find that Lgr4 is broadly expressed in both the nail epithelium and bone, correlating with Lgr6 expression domains (Fig. S3B). Functional redundancy has previously been described between Lgr4 and Lgr5 in the intestinal epithelium where a minimal phenotype is seen in the absence of Lgr5 alone (31). This Lgr4/5 redundancy has been elucidated to act through the canonical Wnt pathway, and, similarly, our data show that Wnt signaling in the nail matrix broadly correlates with the expression domain of Lgr6 and that there is β-catenin localized to the nucleus in Lgr6-expressing nail matrix cells (Fig. 3C). Interestingly, this precedent for Lgr6 to mediate canonical Wnt signaling in vivo stands in contrast to the current thinking that Lgr5, but not Lgr6, is the main mediator of canonical Wnt signaling in epithelial adult stem cell populations (23). This reasoning originated from experiments expressing deltaN-Tcf/Lef in the hair bulge where it prevented proliferation while it promoted sebaceous gland proliferation, supporting canonical Wnt signaling in the hair bulge and noncanonical Wnt signaling in the sebaceous gland (32). Because Lgr6 is expressed in sebaceous gland stem cells, the association has suggested a primary role for Lgr6 in noncanonical Wnt signaling (19) although in vitro experiments have shown that Lgr6 is competent to promote canonical Wnt signaling, albeit at a comparatively weaker level than Lgr4 or Lgr5 (33, 34).
Nail Stem Cells Are Located Within the Nail Matrix.
In 1968, Zaias and Alvarez demonstrated by tritiated glycine incorporation in squirrel monkey that the nail plate originates from the nail matrix (11). They found that, even after 21 d of normal nail/digit growth, the nail plate does not receive cells from the proximal nail fold, nail bed epithelium, or the hyponychium, thus implicating the nail matrix as the single origin of the nail plate. More recently, by similar pulse–chase types of experiments, these findings were corroborated and translated to the mouse model system. Nakamura and Ishikawa identified BrdU label-retaining cells in the basal layer of the nail matrix (12), and Takeo et al. showed that of Keratin14CreER;R26RLSL-LacZ genetically marked epithelial cells, those residing in the nail matrix, give rise to the nail plate (10). Collectively, these experiments demonstrate that a presumptive set of nail stem cells resides within the nail matrix. We confirm that the nail matrix gives rise to the nail plate, and in addition, we show that Lgr6 is a molecular marker specific to the nail stem cells. Interestingly, Leung et al. recently performed a Keratin5TetOff;TreH2BGFP pulse–chase experiment to identify label-retaining cells in the digit tip and did not find a presumptive stem cell population within the nail matrix (35). Instead, they found label-retaining cells within the proximal nail fold that give rise to the eponychium during homeostatic conditions. They also showed that, during long-term growth or wounding/regenerative conditions, these cells have a binary potential and can differentiate into the nail plate, thus leading to the conclusion that the proximal nail fold harbors nail stem cells (35). Although these data are seemingly in conflict with both the preexisting literature as well as the data we present in this article, the differences in experimental markers and timing of pulses/analyses can rectify most of the discrepancies. Importantly, the fact that Leung et al. do not find label-retaining cells in the nail matrix after a 4-wk pulse implies that the notion that discrete populations of stem cells proliferate at an equally “slow” rate should be reconsidered. The cellular properties of quiescence could be entirely context-dependent and likely correlated with the demands of the renewing tissue and the number of resident stem cells.
A Unique Role for Lgr5 in the Digit Tip.
Although Lgr6 is a marker for the stem cells in the nail epithelium, Lgr5 is found in a unique dermal population in the nail fold. Previously, Lgr5 has been associated with epithelial stem cell populations. In this respect, the finding that Lgr5 marked a population of dermal cells associated with the surrounding epithelia of the nail was unprecedented. Furthermore, the lack of expression of canonical Wnt signaling markers (Axin2 and TCF/Lef) in the Lgr5-expressing dermis suggests that Lgr5 is not a target of canonical signaling in these cells (Fig. S1). Our genetic lineage analyses of Lgr5-expressing cells during nail growth and digit tip regeneration show that these cells and their descendants remain dermal, yet closely associated with the eponychium and hyponychium. The onychodermis is a specialized, CD10-positive/CD34-negative dermal tissue, underlying the keratinized plate of the nail, that is thought to play an important role in adhesion and production of hard keratins (36, 37). We hypothesize that Lgr5-expressing cells within the onychodermis in the proximal nail fold and distal groove, are specialized dermal fibroblasts, which are a heterogeneous population of cells involved in the production of connective tissue/extracellular matrix, as well as the facilitation of cellular communication and integrity between the dermis and epidermis (38). Dermal fibroblast properties and function can vary with their embryonic source and/or anatomical location. For example, En1-derived dermal fibroblasts were recently found to be responsible for the bulk of connective tissue deposition and fibrosis during mouse dorsal cutaneous wound healing (39). Importantly, ablation or small molecule inhibition of the En1-derived cell type, in favor of another dermal fibroblast population(s), led to significant reduction in scar formation without compromising the integrity of the skin. In this context, it will be important to evaluate whether dermal Lgr5-expressing cells are dermal fibroblasts involved in the maintenance of the eponychium and hyponychium and, subsequently, to determine whether they can mediate scar-free wound healing as is characteristic of digit tip regeneration.
Do Stem Cells Function Differently in a Regenerative Context?
In this report, we show that loss of Lgr6 in regenerating digit tips results in dysmorphic nails, as well as reduced bone regeneration. Interestingly, unamputated quiescently growing digits from Lgr6−/− animals were morphologically normal. One possible explanation for these observations is that the stem cells marked by Lgr6 could play identical roles in homeostasis and regeneration, but the Lgr6 protein may be more critical during regeneration perhaps due to differential levels of expression of a redundant, compensatory protein during these two processes. Alternatively, however, it is possible that the Lgr6-expressing cells themselves have distinct roles in homeostatic growth versus regeneration. There is already a precedent for a differential function of Lgr6-expressing cells between quiescence and regeneration. Using genetic lineage analyses, Snippert et al. found that, during homeostatic growth of mouse back epidermis, Lgr6-expressing cells give rise to sebaceous glands and interfollicular epidermis, but not the hair follicle. Upon injury however, Lgr6-expressing cells give rise to regenerating hair follicles in addition to the other tissues, much like embryonic Lgr6-expressing cells (19). A similar differential response has been revealed within the digit tip such that the Keratin5/15-marked label-retaining cells within the proximal nail fold serve as the stem cell pool for the eponychium, and only after long term quiescent growth do they give rise to occasional cells in the nail plate. However, upon mechanical ablation of the nail, these stem cells significantly increase their contribution to the nail plate (35), implying an inherent plasticity to this stem cell population during tissue stress. Taken in the context of the Lgr6−/− experiments in this article, this finding suggests that, upon wounding of the nail matrix/plate, the eponychium stem cells may be capable of regenerating the nail matrix or specifically generating Lgr6-expressing nail stem cells; however, if this is the cellular hierarchy, in our experiments, these cells were insufficient to rescue the Lgr6−/− phenotype. This scenario can be likened to that of the intestinal epithelium whereby Lgr5 marks intestinal stem cells within the crypt, but, upon their ablation, Bmi1-expressing cells are competent to repopulate the Lgr5-expressing cells (40). Collectively these examples underscore an inherent plasticity of the adult stem cell populations within epithelia, particularly those expressing Lgr proteins, and perhaps these highly proliferative tissues have a back-up system for times of stress/wounding or depletion of the stem cell pools.
Materials and Methods
All mouse breeding and experimentation was done with approval of the Harvard Medical School Institutional Animal Care and Use Committee. Lgr5 and Lgr6 expression studies were done with Lgr5EGFP-ires-creERT2 and Lgr6EGFP-ires-creERT2 knock-in alleles (JAX 008875 and 016934). These mice can be used for their GFP fluorescent reporters, tamoxifen-inducible cre, and/or genetic null alleles. For clarity, when used as a null allele, Lgr6EGFP-ires-creERT2 is referred to as Lgr6−/−. Genotyping primers for Lgr6 mutant allele were 5′-GCCCACCGACGGCGCAGCCC-3′ and 5′-GCTGAACTTGTGGCCGTTTA-3′ and for Lgr6 WT allele were 5′-CTCGCCCGTCTGAGCG-3′ and 5′-GCAGGCACCACTGAGAGC-3′. Genetic lineage analyses used the R26RCAG-LSL-tdTomato cre reporter allele (JAX 007905). Axin2lacZ and TCF/LefH2B-GFP (JAX 009120 and 013752) were used to analyze canonical Wnt signaling activity. All alleles were maintained as heterozygotes or compound heterozygotes on the C57BL/6J background (JAX 000664), with the exception of the experimental outcross of Lgr6EGFP-ires-creERT2 to CD-1(ICR) (022; Charles River Laboratories), where we performed an intercross of (Lgr6EGFP-ires-creERT2 × CD-1(ICR))F1 animals. Expression and lineage analyses were performed by breeding genetic allele(s) to WT CD-1(ICR) females (022; Charles River Laboratories) to generate large litters for analysis and progeny with robust regeneration (6).
Full methods are provided in SI Materials and Methods.
SI Materials and Methods
Digit Amputations and Genetic Labeling.
For genetic lineage analyses, litters of Lgr5EGFP-ires-creERT2;R26RCAG-LSL-tdTomato or Lgr6EGFP-ires-creERT2;R26RCAG-LSL-tdTomato mice were dosed with 3 mg/40 g body weight i.p. tamoxifen (20 mg/mL in corn oil) on postnatal day 0. These genetically marked animals, as well as all other mice used in regeneration assays, underwent digit tip amputations on postnatal day 3 as previously described (6). In brief, mouse pups were cryoanesthetized, and digits were visualized under a Leica MZ6 stereomicroscope fitted with an eye-piece reticule. Microspring scissors were used to amputate 400 µm of right hindlimb digits 2, 3, and 4, while leaving left hindlimb digits as unamputated controls. Mice were euthanized, and digits were collected for analysis at various time points during regeneration as specifically stated in the text.
Tissue Histology.
Mouse digit tips were collected for section immunohistochemistry and fixed in 4% paraformaldehyde. Digits underwent a 2-d increasing sucrose gradient (5–30%), and were then embedded in OCT (Tissue-Tek). Tissue was cut into 20-µm cryosections, blocked in 5% goat serum in PBST (phosphate-buffered saline with 0.1% Tween 20), and incubated with primary antibody in a humidified chamber at 4 °C overnight. Primary antibodies were as follows: anti-GFP (ab13970, 1:1,000; Abcam), anti-laminin (L9393, 1:100; Sigma-Aldrich), anti-dsRed for tdTomato (632496, 1:1,000; Clontech), anti-β-catenin (ab6302, 1:1,000; Abcam), anti-SP7 (ab22552, 1:2,000; Abcam), and anti-vimentin (NB300-223; Novus Biologicals). Sections were washed with PBST and incubated with a species-appropriate Cy3, A488, or A647 fluorophore-conjugated secondary antibody (1:500) (Jackson Immunoresearch) at room temperature for 1 h [with the exception of anti-SP7 labeling, which was incubated with secondary antibody (1:1,000) at 4 °C overnight]. Slides were washed in PBST, counterstained with DAPI, and mounted with Fluoromount-g aqueous mounting medium (Southern Biotech) for imaging. X-gal staining for β-galactosidase expression in Axin2LacZ digit tips and eosin counterstaining were done as previously reported (6) by standard protocols. Hematoxylin and eosin staining, performed by standard protocol, was done on Lgr6+/− and Lgr6−/− CD-1(ICR) outcross animals, in serial section with anti-GFP immunohistochemistry. Section RNA in situ hybridization was done as previously described (41), with the exception of using 20-µm cryosections and a proteinase K concentration of 3 µg/mL for 10 min at room temperature. Lgr4 cDNA, which was used to generate a DIG-labeled cRNA probe, was amplified by RT-PCR with 7 d postamputation digit tip-derived RNA using 5′-ACAACCAGCTGACCACACT-3′ and 5′-CAGCGGGCTCCATACTCAG-3′.
Skeletal Staining and Bone Regeneration Assay.
Alizarin red-stained digital bones were prepared as previously described (6). In brief, digits were harvested and dehydrated in 95% ethanol, followed by 24 h at 37 °C in alizarin red staining solution (0.005% alizarin red, 5% acetic acid, and 60% ethanol). Tissue was cleared for 48 h with two changes of 2% potassium hydroxide, followed by an increasing glycerol series to 75%. Bones were imaged, and 2D area measurements of the digit tip bone were made using ImageJ (rsbweb.nih.gov/ij/). For each animal, six digits were measured: three regenerates and three contralateral controls. For each digit, three measurements were made, and an average was taken. Percent regeneration was calculated by normalizing the 2D area of the regenerate by the 2D area of the contralateral control unamputated digit. The Lgr6 regeneration assay included 8 Lgr6−/− animals, 11 Lgr6+/− animals, and 6 Lgr6+/+ animals (15, 22, and 12 regenerate digits, respectively). All graphing and statistics were done with the R statistical software package (www.R-project.org).
Acknowledgments
We thank Bryan MacDonald for valuable scientific conversations as well as critical review of this manuscript. We also thank Michael Levin and Jessica Whited for helpful discussion on this work. This work was supported by National Institutes of Health Grant HD045499.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518874112/-/DCSupplemental.
References
- 1.Kierdorf U, Li C, Price JS. Improbable appendages: Deer antler renewal as a unique case of mammalian regeneration. Semin Cell Dev Biol. 2009;20(5):535–542. doi: 10.1016/j.semcdb.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Borgens RB. Mice regrow the tips of their foretoes. Science. 1982;217(4561):747–750. doi: 10.1126/science.7100922. [DOI] [PubMed] [Google Scholar]
- 3.Singer M, Weckesser EC, Géraudie J, Maier CE, Singer J. Open finger tip healing and replacement after distal amputation in rhesus monkey with comparison to limb regeneration in lower vertebrates. Anat Embryol (Berl) 1987;177(1):29–36. doi: 10.1007/BF00325287. [DOI] [PubMed] [Google Scholar]
- 4.Illingworth CM. Trapped fingers and amputated finger tips in children. J Pediatr Surg. 1974;9(6):853–858. doi: 10.1016/s0022-3468(74)80220-4. [DOI] [PubMed] [Google Scholar]
- 5.Douglas BS. Conservative management of guillotine amputation of the finger in children. Aust Paediatr J. 1972;8(2):86–89. doi: 10.1111/j.1440-1754.1972.tb01793.x. [DOI] [PubMed] [Google Scholar]
- 6.Lehoczky JA, Robert B, Tabin CJ. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc Natl Acad Sci USA. 2011;108(51):20609–20614. doi: 10.1073/pnas.1118017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476(7361):409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neufeld DA, Zhao W. Bone regrowth after digit tip amputation in mice is equivalent in adults and neonates. Wound Repair Regen. 1995;3(4):461–466. doi: 10.1046/j.1524-475X.1995.30410.x. [DOI] [PubMed] [Google Scholar]
- 9.Mohammad KS, Day FA, Neufeld DA. Bone growth is induced by nail transplantation in amputated proximal phalanges. Calcif Tissue Int. 1999;65(5):408–410. doi: 10.1007/s002239900722. [DOI] [PubMed] [Google Scholar]
- 10.Takeo M, et al. Wnt activation in nail epithelium couples nail growth to digit regeneration. Nature. 2013;499(7457):228–232. doi: 10.1038/nature12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaias N, Alvarez J. The formation of the primate nail plate: An autoradiographic study in squirrel monkey. J Invest Dermatol. 1968;51(2):120–136. doi: 10.1038/jid.1968.103. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura M, Ishikawa O. The localization of label-retaining cells in mouse nails. J Invest Dermatol. 2008;128(3):728–730. doi: 10.1038/sj.jid.5701062. [DOI] [PubMed] [Google Scholar]
- 13.de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev. 2014;28(4):305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi J, et al. Analysis of LGR4 receptor distribution in human and mouse tissues. PLoS One. 2013;8(10):e78144. doi: 10.1371/journal.pone.0078144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Schoore G, Mendive F, Pochet R, Vassart G. Expression pattern of the orphan receptor LGR4/GPR48 gene in the mouse. Histochem Cell Biol. 2005;124(1):35–50. doi: 10.1007/s00418-005-0002-3. [DOI] [PubMed] [Google Scholar]
- 16.Barker N, Tan S, Clevers H. Lgr proteins in epithelial stem cell biology. Development. 2013;140(12):2484–2494. doi: 10.1242/dev.083113. [DOI] [PubMed] [Google Scholar]
- 17.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 18.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40(11):1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 19.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327(5971):1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 20.Stoick-Cooper CL, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134(3):479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 21.Lin G, Slack JM. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol. 2008;316(2):323–335. doi: 10.1016/j.ydbio.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 22.Kawakami Y, et al. WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell. 2001;104(6):891–900. doi: 10.1016/s0092-8674(01)00285-9. [DOI] [PubMed] [Google Scholar]
- 23.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346(6205):1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 24.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: From human mutations to treatments. Nat Med. 2013;19(2):179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 25.Han M, Yang X, Lee J, Allan CH, Muneoka K. Development and regeneration of the neonatal digit tip in mice. Dev Biol. 2008;315(1):125–135. doi: 10.1016/j.ydbio.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sellheyer K, Nelson P. The ventral proximal nail fold: Stem cell niche of the nail and equivalent to the follicular bulge--a study on developing human skin. J Cutan Pathol. 2012;39(9):835–843. doi: 10.1111/j.1600-0560.2012.01949.x. [DOI] [PubMed] [Google Scholar]
- 27.Yee KK, et al. Lgr5-EGFP marks taste bud stem/progenitor cells in posterior tongue. Stem Cells. 2013;31(5):992–1000. doi: 10.1002/stem.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plaks V, et al. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Reports. 2013;3(1):70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng A, et al. Lgr5 marks stem/progenitor cells in ovary and tubal epithelia. Nat Cell Biol. 2014;16(8):745–757. doi: 10.1038/ncb3000. [DOI] [PubMed] [Google Scholar]
- 30.Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6(1):25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 31.de Lau W, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476(7360):293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs E, Horsley V. More than one way to skin . . . Genes Dev. 2008;22(8):976–985. doi: 10.1101/gad.1645908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong X, et al. LGR6 is a high affinity receptor of R-spondins and potentially functions as a tumor suppressor. PLoS One. 2012;7(5):e37137. doi: 10.1371/journal.pone.0037137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108(28):11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung Y, et al. Bifunctional ectodermal stem cells around the nail display dual fate homeostasis and adaptive wounding response toward nail regeneration. Proc Natl Acad Sci USA. 2014;111(42):15114–15119. doi: 10.1073/pnas.1318848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee DY, et al. The presence and localization of onychodermis (specialized nail mesenchyme) containing onychofibroblasts in the nail unit: A morphological and immunohistochemical study. Histopathology. 2012;61(1):123–130. doi: 10.1111/j.1365-2559.2012.04210.x. [DOI] [PubMed] [Google Scholar]
- 37.Sellheyer K, Nelson P. The concept of the onychodermis (specialized nail mesenchyme): An embryological assessment and a comparative analysis with the hair follicle. J Cutan Pathol. 2013;40(5):463–471. doi: 10.1111/cup.12101. [DOI] [PubMed] [Google Scholar]
- 38.Sorrell JM, Caplan AI. Fibroblast heterogeneity: More than skin deep. J Cell Sci. 2004;117(Pt 5):667–675. doi: 10.1242/jcs.01005. [DOI] [PubMed] [Google Scholar]
- 39.Rinkevich Y, et al. Skin fibrosis: Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science. 2015;348(6232):420–431. doi: 10.1126/science.aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478(7368):255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13(2):225–237. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]