Significance
The alarmones guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) [collectively named “(p)ppGpp)”] are important for the adaptation of bacteria and plant chloroplasts to a variety of environmental stress conditions. Their synthesis is carried out by (p)ppGpp synthetases. We delineate the catalytic mechanism of (p)ppGpp synthesis by oligomeric and highly cooperative small alarmone synthetase 1 (SAS1) at atomic resolution. Our structural and biochemical analysis shows that only pppGpp—but not ppGpp—positively affects the activity of SAS1. To our knowledge, this is the first molecular description of a biological activity in which pppGpp and ppGpp execute different functional roles.
Keywords: stringent response, (p)ppGpp, hydrogen–deuterium exchange mass spectrometry, alarmone, crystallography
Abstract
Nucleotide-based second messengers serve in the response of living organisms to environmental changes. In bacteria and plant chloroplasts, guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) [collectively named “(p)ppGpp”] act as alarmones that globally reprogram cellular physiology during various stress conditions. Enzymes of the RelA/SpoT homology (RSH) family synthesize (p)ppGpp by transferring pyrophosphate from ATP to GDP or GTP. Little is known about the catalytic mechanism and regulation of alarmone synthesis. It also is unclear whether ppGpp and pppGpp execute different functions. Here, we unravel the mechanism and allosteric regulation of the highly cooperative alarmone synthetase small alarmone synthetase 1 (SAS1) from Bacillus subtilis. We determine that the catalytic pathway of (p)ppGpp synthesis involves a sequentially ordered substrate binding, activation of ATP in a strained conformation, and transfer of pyrophosphate through a nucleophilic substitution (SN2) reaction. We show that pppGpp—but not ppGpp—positively regulates SAS1 at an allosteric site. Although the physiological significance remains to be elucidated, we establish the structural and mechanistic basis for a biological activity in which ppGpp and pppGpp execute different functional roles.
The ability of living organisms to adapt their metabolism to nutrient limitation or environmental changes is of utmost importance to survival. The stringent response is a highly conserved mechanism that enables bacteria (1–3) and plant chloroplasts (4–6) to respond to nutrient (i.e., amino acid) limitations. However, recent work has indicated that guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) [collectively named “(p)ppGpp”] also impact other, nonstringent response processes such as virulence (7–9) as well as persister (10, 11) and biofilm formation (12). Realization of the importance of (p)ppGpp has also opened new avenues for the design of antimicrobial agents (13, 14).
Central to these processes is the synthesis of two alarmones, pppGpp and ppGpp, which globally reprogram the transcription and translation associated with a variety of cellular processes (summarized in refs. 9 and 15) and which also control the elongation of DNA replication (16). Until now, both alarmones have been collectively named “(p)ppGpp,” because knowledge of their individual roles remained mysterious. Only recently has a study indicated that either alarmone might execute disparate biological functions (17).
Alarmone synthesis is carried out by synthetases of RelA/SpoT homology (RSH) (18) that catalyze the transfer of pyrophosphate (β-, γ-phosphates) from ATP to the ribose 3′-OH of GDP or GTP to synthesize ppGpp or pppGpp, respectively. An in-depth analysis of the catalytic mechanism for this reaction is currently not available. Only one structure describes the GDP-bound state of an RSH synthetase domain, bearing remarkable similarity to members of the nucleotidyltransferase superfamily (19). However, no other structural details relevant to the catalytic mechanism of alarmone synthetases, such as the relative location of either substrate in the active site, are available.
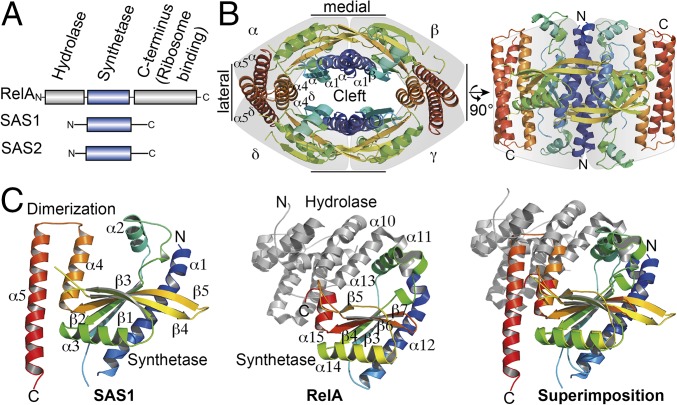
To our knowledge, three types of alarmone synthetases, differing in length and domain composition, have been identified, RelA and the small alarmone synthetases 1 (SAS1) and 2 (SAS2) (Fig. 1A). Best understood is the RelA protein, which senses amino acid starvation during the stringent response by detecting ribosomes blocked by cognate, uncharged tRNAs at the A-site (20, 21). RelA’s synthetase activity is stimulated upon binding to stalled ribosomes, and alarmone synthesis decreases the affinity of this binding. Besides a 40-kDa C-terminal domain that is involved in ribosome binding (21), RelA from Bacillus subtilis also contains an N-terminal hydrolase domain that recycles (p)ppGpp by removing the 3′-diphosphate moiety to generate GDP or GTP (Fig. 1A). Both the synthetase and hydrolase domains of RelA are regulated in a reciprocal manner, ensuring inverse coupling of the opposing activities (19, 22, 23). The recently identified alarmone synthetases SAS1 (synonyms: YjbM and RelQ) and SAS2 (synonyms: YwaC and RelP) (24–27) share high sequence identities of 20–30% with RSH proteins in their synthetase domains (18). However, the absence of the hydrolase and C-terminal domains found in RelA (Fig. 1A) obscures their functional roles in the living cell.
Fig. 1.
Crystal structure of the tetrameric alarmone synthetase SAS1. (A) Domain architecture of the alarmone synthetases RelA, SAS1, and SAS2. (B) Crystal structure of the SAS1 tetramer. Each monomer (α–δ, indicated by a gray shadow) is shown as cartoon in rainbow colors from the N terminus (N) to the C terminus (C). Brackets indicate the lateral and medial interfaces. (C) Cartoon representation of the crystal structures of an SAS1 monomer (Left, this study), RelA [PDB ID code 1VJ7 (19)] (Center), and the superimposition of their synthetase domains (Right). RelA-Syn and SAS1 are shown in rainbow colors from the N terminus (N) to the C terminus (C). Structural elements in RelA-Syn are labeled according to ref. 19. The equivalent elements in SAS1 are described in the text.
Here, we present the biochemical and structural analysis of the small alarmone synthetase SAS1 that forms a homotetramer including the crystal structures of its apo-, α, β-methyleneadenosine 5′-triphosphate (AMPCPP)-, and pppGpp-bound states. Our analysis suggests a plausible model of the catalytic mechanism of alarmone synthesis. Moreover, we show by biochemical and structural means that SAS1 is allosterically activated by pppGpp in a cleft formed by the homotetramer. We discuss the physiological implications of this allosteric regulation.
Results
The Crystal Structure of the Small Alarmone Synthetase SAS1 Reveals a Homotetramer.
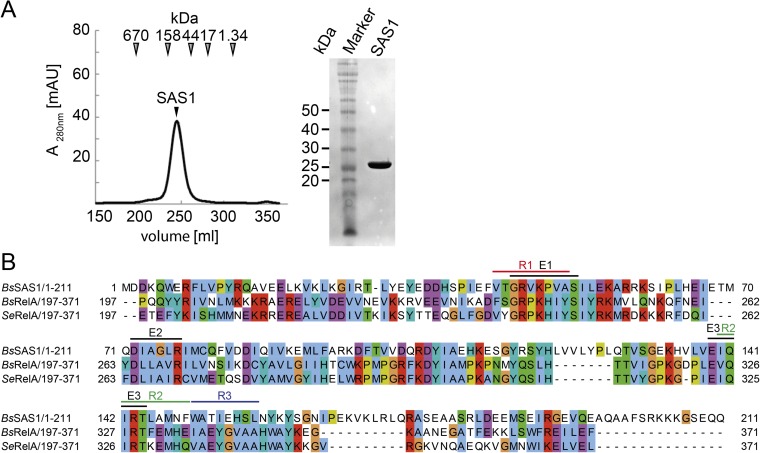
Because SAS1 (and its highly conserved counterpart SAS2) lacks the regulatory domains of RelA (Fig. 1A), we reasoned that it might possess other, thus far unknown, features allowing SAS1 to be regulated. Interestingly, although RelA was shown to function as a monomer, biochemical and biophysical analysis of SAS1 revealed that the purified protein forms homotetramers (Fig. S1A). To understand the architecture of SAS1 and to compare it with RelA, we determined the crystal structure of SAS1 at 1.86-Å resolution (Table S1). The enzyme crystallized as a symmetric, oval-shaped homotetramer (Fig. 1B). Intriguingly, the tetramer contains a central cleft that appears to be of functional relevance (see below). Primarily hydrogen bonds and salt bridges stabilize the medial sides of the tetramer in an interface of ∼1,100 Å2. The lateral sides of the tetramer are formed exclusively by helices α4 and α5 in an interface of ∼1,220 Å2, consisting chiefly of polar contacts.
Fig. S1.
(A) SAS1 forms a homotetramer. (Left) SEC profile of SAS1. Arrowheads indicate the mass of the size standard. (Right) Coomassie-stained SDS/PAGE of the peak fraction containing SAS1. (B) Amino acid sequence alignment of SAS1 and RelA. Colored lines indicate E1–E3 and R1–R3. Bs, B. subtilis; Se, Streptococcus equisimilis.
Table S1.
Data collection and refinement
| Statistics | BsSAS1-apo | BsSAS1-AMPCPP | BsSAS1-pppGpp |
| Data collection | |||
| Space group | P 21 | P 21 | C 2 |
| Cell dimensions | |||
| a, b, c, Å | 78.13, 77.69, 81.61 | 116.67, 103.63, 138.29 | 197.87, 113.49, 139.85 |
| α, β, γ, ° | 90.00, 90.40, 90.00 | 90.00, 104.84, 90.00 | 90.00, 127.17, 90.00 |
| Energy, keV | 12.6616 | ||
| Resolution, Å | 56.63–1.86 (1.96–1.86) | 48.31–2.80 (2.95–2.80) | 50.57–2.94, (3.10–2.94) |
| Rmerge | 0.083 (0.46) | 0.090 (0.48) | 0.103 (0.273) |
| I/σI | 7.4 (2.5) | 10.6 (2.8) | 12.1 (5.8) |
| Completeness, % | 97.7 (98.1) | 100 (100) | 99.3 (96.1) |
| Redundancy | 3.4 (3.5) | 3.8 (3.9) | 6.8 (6.4) |
| Refinement | |||
| Resolution, Å | 56.6–2.00 | 48.3–2.8 | 50.6–2.94 |
| No. reflections | 64,368 | 78,505 | 51,454 |
| Rwork/ Rfree | 19.4/23.8 | 19.6/25.7 | 18.1/24.5 |
| No. atoms | 6,542 | 18,586 | 13,016 |
| Rmsd | |||
| Bond lengths, Å | 0.010 | 0.013 | 0.014 |
| Bond angles, ° | 1.25 | 1.242 | 1.441 |
Highest resolution shell is shown in parentheses.
SAS1 and the synthetase domain of RelA [RelA-Syn; Protein Data Bank (PDB) ID code 1VJ7, residues 197–371 (19)] share sequence identities of 23.5%, and their structures superimpose well with an rmsd of 1.6 Å over 78 Cα atoms, revealing a conserved domain core (Fig. 1C and Fig. S1B). SAS1 and RelA-Syn share the antiparallel β-strands β1–β5 as well as the surrounding helices α1, α2, α3, and a fraction of α4 [named β3–β7 and α12–α15, respectively, in RelA (19)]. Residues involved in substrate binding and catalysis are conserved between the synthetase domains of SAS1 and RelA (Fig. S1B). In RelA, α12 (α1 in SAS1) is preceded by another helix (α11) that establishes the connection to the hydrolase domain (Fig. 1C). In SAS1, helix α1 (α12 in RelA) is important for the formation of the medial side of the homotetramer and marks the beginning of the protein (Fig. 1B). The orientation of α2 [α13 in RelA-Syn (19)] differs in the structures of SAS1 and RelA-Syn by ∼30°. Notably, α13 of RelA-Syn contacts α11 and α10, which mark the end of the hydrolase domain. In SAS1, the α2 helices appear at the surface of homotetramer. For RelA, these elements have been indicated as important for switching the opposing RelA hydrolase and synthetase activities (19).
Another major difference between SAS1 and RelA-Syn comprises the C-terminal helices α4 and α5 of SAS1. Although the N-terminal fraction of α4 (α15 in RelA-Syn) is conserved between SAS1 and RelA-Syn, α4 of SAS1 is elongated by two turns and forms a helical hairpin with α5 that is absent in RelA-Syn. Helices α4 and α5 of SAS1 form the lateral side of the homotetramer interface (Fig. 1B). The structure of RelA-Syn ends with α16 (which has no equivalent in SAS1) and proceeds into the as yet structurally unresolved C-terminal part of RelA that is involved in ribosome binding (21).
SAS1 Binds Its Substrates in an Ordered Sequence During Alarmone Synthesis.
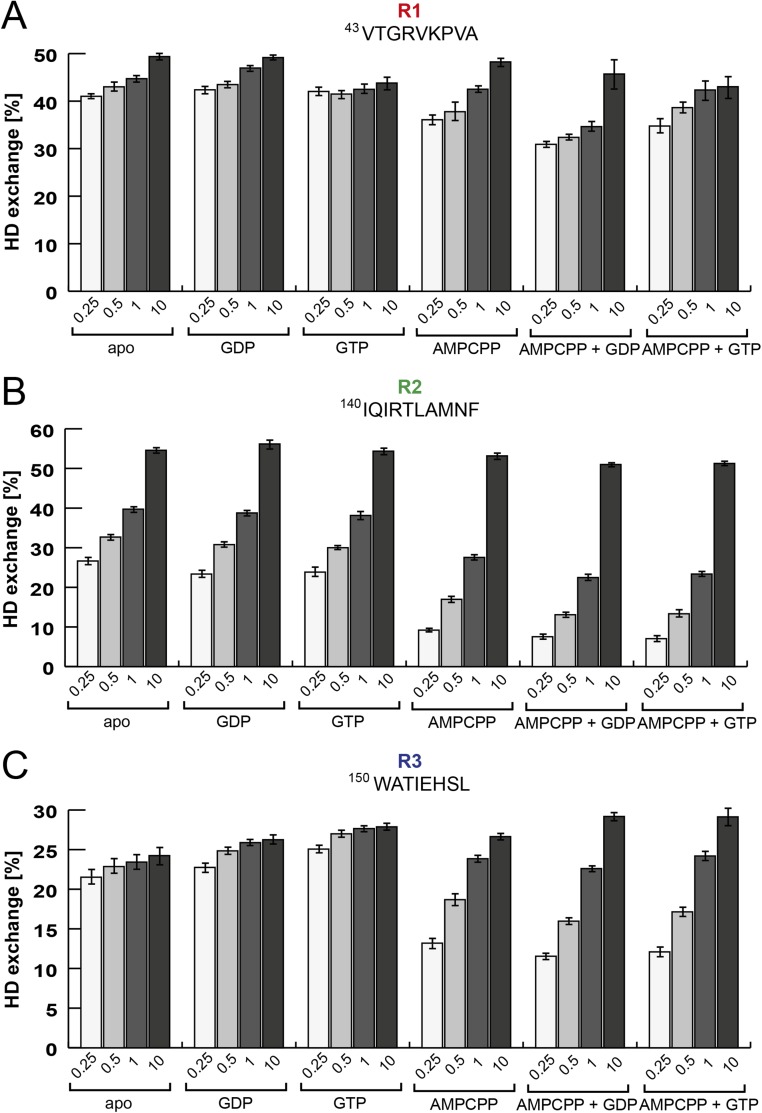
Despite intensive research on alarmone synthetases, the catalytic mechanism and structural basis of alarmone synthesis has not been widely addressed. To examine these properties, we first aimed at defining the binding sequence of the substrates ATP and GDP or GTP. We used hydrogen–deuterium exchange (HDX) mass spectrometry, which allows rapid detection of changes within SAS1 upon binding of substrates. Specifically, SAS1 was incubated with GDP, GTP, or the nonhydrolysable ATP analog AMPCPP. After completion of the HDX-labeling reaction, SAS1 was digested with pepsin, and the peptic peptides were analyzed by electrospray ionization-mass spectrometry. Although no effects could be detected in the presence of either GDP or GTP, incubation with AMPCPP led to a significantly decreased HDX of ∼15% for the R2 region and ∼4% for the R3 region of SAS1, both located within its putative active site (Figs. 1A and 2 A and B and Figs. S1B and S2). This result suggests that ATP should bind to SAS1 before GDP or GTP. To support this finding, we next incubated SAS1 with either GDP or GTP and AMPCPP. The presence of both substrates (i.e., AMPCPP and GDP or GTP) induced a decrease in HDX of 4% (R2) and 2–3% (R3) compared with AMPCPP alone (Fig. 2 A and B and Fig. S2). Taken together, our data suggest that SAS1 binds its substrates in an ordered sequence: first ATP and then GDP or GTP (Fig. 2C).
Fig. 2.
Mechanism of alarmone synthesis. (A) Three regions of SAS1 (R1–R3) show different dynamic responses to the presence of different nucleotides in HDX experiments after 30 s of deuteration. The graph shows the percentage of HDX of R1 (red), R2 (green), and R3 (blue) in the presence or absence of different nucleotides. Error bars represent the SD of three independent measurements. (B) Location of R1 (red), R2 (green), and R3 (blue) on the crystal structure of an SAS1-AMPCPP monomer. An arrow indicates AMPCPP within the active site. (C) The ATP (pale green ball) and GDP or GTP (dark green triangle) substrates bind to the active site of SAS1 (blue) in sequential order. The transition state of the reaction is indicated by a double dagger (‡). The products (p)ppGpp and AMP are shown as orange and gray balls, respectively. (D) ATP (mimicked by AMPCPP, deep teal) binds in a tense, U-shaped conformation in the active site of SAS1. Dashed lines indicate interactions between residues of SAS1 and AMPCPP. The magnesium ion is shown as a green sphere. (E) Superimposition of the active sites of SAS1 bound to AMPCPP (deep teal) and RelA bound to GDP (green). The attacking 3′-OH group of the ribose of GDP is encircled in black, and the arrow indicates the direction of nucleophilic attack toward the β-phosphate of AMPCPP. (F) Scheme of the catalytic mechanism of alarmone synthesis. A detailed description is given in the text.
Fig. S2.
HDX of three regions located within the active site of SAS1. The graphs show the percentage of deuterium uptake of three individual peptides (amino acid sequences are listed above the plots) representing three regions within the active site of SAS1 in different nucleotide-bound states. Deuteration times in minutes are indicated below the bars. Error bars represent the SD of three independent measurements. (A) Peptide R1 (VTGRVKPVA). (B) Peptide R2 (IQIRTLAMNF). (C) Peptide R3 (WATIEHSL).
The Catalytic Mechanism of Alarmone Synthesis.
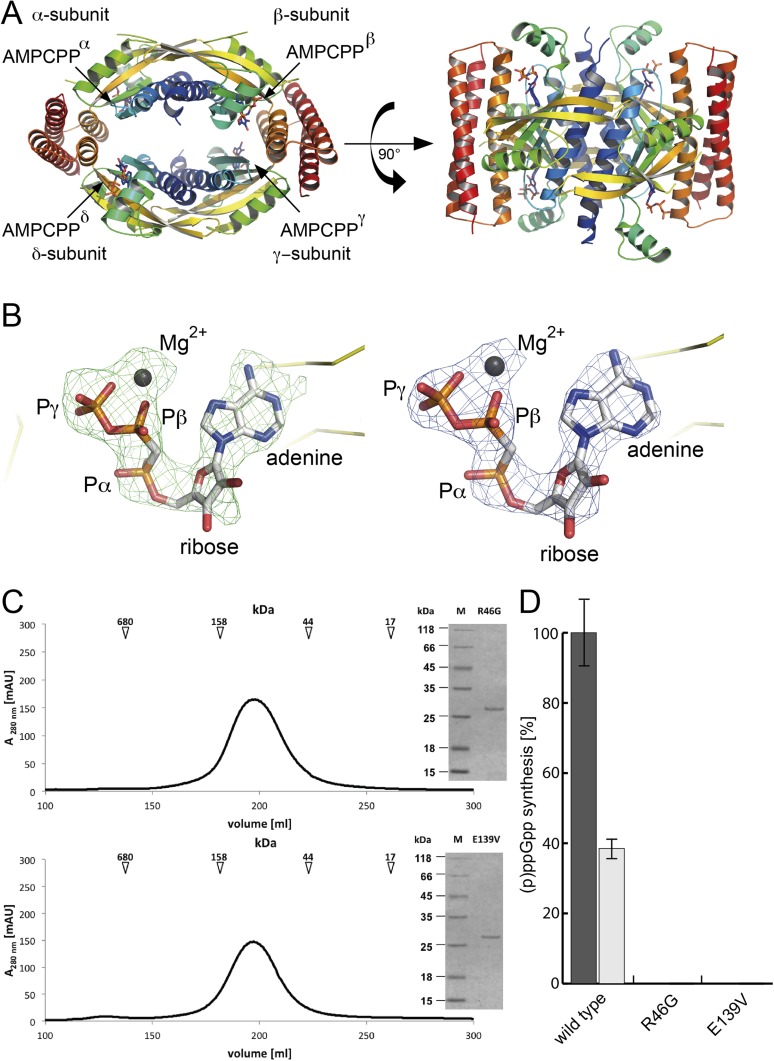
Next, we wanted to determine the structural basis for substrate binding. We obtained crystals of the ATP-bound state of SAS1 (mimicked by AMPCPP) that diffracted to 2.8-Å resolution. After phasing by molecular replacement (MR) with SAS1 (this study) as a search model (Table S1), we observed density in the active site that could be assigned unambiguously to AMPCPP (Fig. S3 A and B). AMPCPP is tightly bound to SAS1 through its interaction with residues from two highly conserved elements, E1 and E2. E1 can be defined as GR(V/P)KxxxS, and E2 can be defined as DIA(G/A)LR (Fig. 2D and Fig. S1B). Arg46 and Arg77 from E1 and E2, respectively, sandwich the adenine base of AMPCPP through π-stacking interactions (Fig. 2D). The phosphate moieties of AMPCPP are found in an unusually tense conformation, bent toward the adenine base. Arg46 (E1) contributes not only by caging the adenine ring but also by interacting with the ribose oxygen and the α-phosphate of AMPCPP. The conformation of the β- and γ-phosphates is stabilized primarily by a magnesium ion that also is coordinated by Asp72 of E2. Furthermore, the negative charge of the γ-phosphate is neutralized in a positively charged pocket formed by Lys48 (E1), Lys56, and Arg59 (Fig. 2D). AMPCPP is in a tense, U-shaped conformation that prepares the nucleotide to donate its β- and γ-phosphates (as pyrophosphate) to the 3′-OH group of the ribose of GDP or GTP. However, crystallization of SAS1 bound to nonreactive AMPCPP and GDP or GTP appeared to be critical, most likely because of the sequentially ordered substrate binding mechanism that prevented stable attachment of the second substrate (see above).
Fig. S3.
(A) The crystal structure of tetrameric SAS1 bound to AMPCPP is shown in cartoon representation. Each monomer appears in rainbow colors from the N terminus to the C terminus. The AMPCPP molecules are shown as sticks and are indicated by arrows. (B) AMPCPP could be assigned unambiguously within the active site of SAS1. (Left) The unbiased Fobs–Fcalc difference electron density of AMPCPP contoured at 2.5 σ is shown as green mesh. Note: The AMPCPP molecule (sticks) and magnesium (gray sphere) were not present during refinement and are placed only for reasons of illustration. (Right) The 2Fobs–Fcalc electron density of AMPCPP/magnesium (sticks and gray sphere, respectively) after final refinement is contoured at 2.5 σ and shown as blue mesh. Yellow ribbons indicate the backbone of SAS1. Figures were generated with PyMOL. (C) SEC profile and Coomassie-stained SDS/PAGE (Inset) of the SAS1 variants R46G (Upper) and E139V (Lower). The arrows indicate the mass of the SEC size standard. (D) Synthesis of ppGpp (black bar) and pppGpp (gray bar) by wild-type SAS1 and the variants R46G and E139V. Two micromoles of SAS1 were incubated in the presence of 5 mM ATP and 5 mM GDP or GTP for 10 min at 37 °C and were subjected to HPLC measurement. Synthesis of ppGpp by wild-type SAS1 is set at 100%. Error bars represent the SD of three independent measurements.
To model the catalytic mechanism of alarmone synthesis, we superimposed the structure of the AMPCPP-bound state of SAS1 (this study) and the proposed hydrolase-OFF/synthetase-ON, GDP-bound state of RelA-Syn [PDB ID code: 1VJ7, chain A (19)]. The synthetase domains of both structures including the catalytically relevant motifs superimpose well, with an rmsd of 1.6 Å over 78 Cα atoms, allowing a convenient estimation of the relative location of GDP and AMPCPP within the active site (Fig. 2E). To our surprise, the GDP and AMPCPP nucleotides were arranged so that pyrophosphate transfer could occur immediately. In our structural superimposition, the distance between the 3′-OH group of GDP and the β-phosphate of AMPCPP is ∼3 Å. The arrangement of both substrates in our model would offer an elegant way for (p)ppGpp synthesis by SAS1 to occur. The close proximity of the 3′-OH group of GDP and the β-phosphate of ATP allow an immediate in-line attack of the β-phosphate moiety ATP by the 3′-OH group of GDP or GTP. Activation of the 3′-OH group is achieved by at least partial deprotonation resulting from its close proximity to the magnesium ion coordinating the β- and γ-phosphates of ATP; furthermore, Glu139, located in the conserved element 3 (E3: EIQIRT; Fig. S1B) may serve as a general base. Thus, the activated 3′-O− group can attack the positively polarized β-phosphate via a second-order nucleophilic substitution (SN2) (Fig. 2 E and F). To solidify our findings, we mutated Arg46 (E1) and Glu139 (E3) to glycine and valine, respectively, and measured the ability of SAS1 to catalyze alarmone synthesis. Although both SAS1 variants formed tetramers as wild-type SAS1, no synthesis of either ppGpp or pppGpp was observed (Fig. S3 C and D).
Next, we performed HDX with the catalytically inactive variants of SAS1 to probe their ability to bind ATP (mimicked by AMPCPP). Interestingly, neither variant showed decreased deuterium incorporation upon incubation with AMPCPP as we observed for the wild-type protein (see above and Fig. S4), indicating that the catalytic deficiency of both SAS1 variants arises from an impaired ability to bind ATP. Taken together, our structural and biochemical analysis delineates the catalytic mechanism of (p)ppGpp synthesis (Fig. 2F). We conclude that alarmone synthetases arrange their substrates in a near-attack (or “pretransition state”) conformation to foster synthesis and stabilize transition state geometry during catalysis.
Fig. S4.
HDX of the active site of SAS1 variants R46G and E139V. The graphs show the percentages of HDX of four individual peptides (amino acid sequences are listed above the plots) located within the active site of SAS1 in the apo- and AMPCPP-bound state. Deuteration times are indicated in minutes below the bars. Error bars represent the SD of three independent measurements. (A) SAS1 R46G. (B) SAS1 E139V.
Alarmone Synthesis in SAS1 Proceeds in a Highly Cooperative Manner.
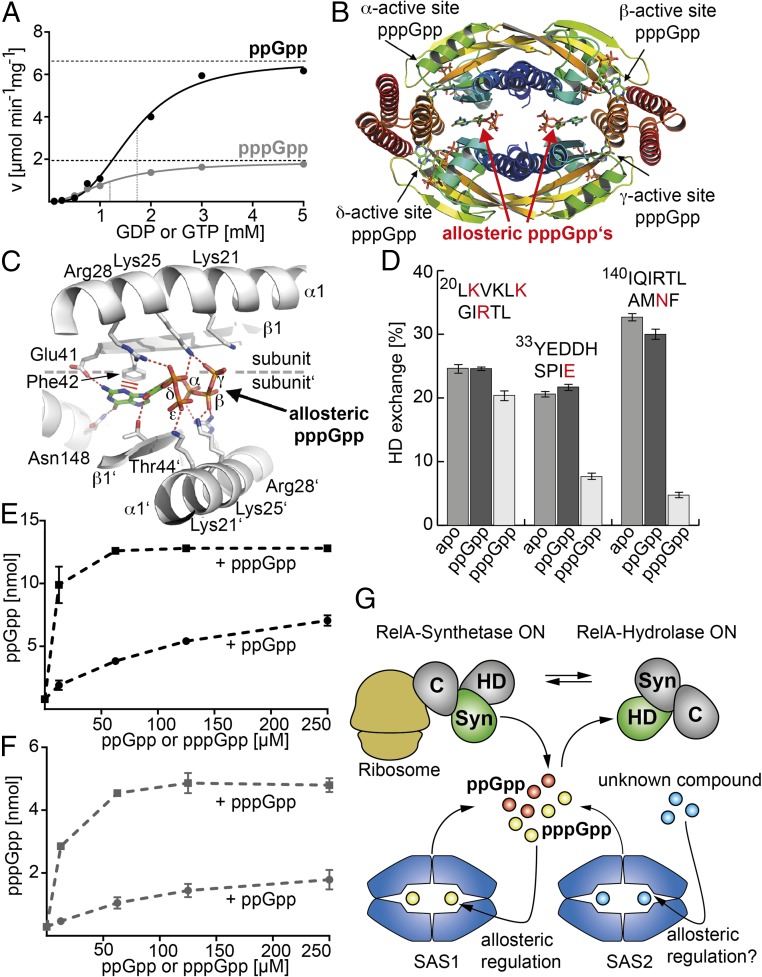
Previous experiments suggest that SAS1 synthesizes ppGpp from GDP more efficiently than pppGpp from GTP (Fig. S3D) (25, 28). To understand this phenomenon in greater detail, we performed an in-depth kinetic analysis of SAS1 (Fig. 3A). We determined the velocity (v) of ppGpp and pppGpp synthesis at varying concentrations of GDP or GTP, respectively, and a constant concentration of ATP (i.e., 5 mM). In both cases, we observed a sigmoidal v/S characteristic of positive cooperativity among the four monomers of the SAS1 tetramer (Fig. 3A and Fig. S5), also reflected by the Hill coefficients of 3.0 ± 0.3 and 2.0 ± 0.1 for ppGpp and pppGpp, respectively. Although the Km values differ only slightly (i.e., 1.7 ± 0.1 mM and 1.2 ± 0.1 mM for GDP and GTP, respectively), the maximal velocity (Vmax) values show an ∼3.5-fold difference, reflecting earlier observations (see above).
Fig. 3.
Allosteric regulation of the tetrameric alarmone synthetase SAS1. (A) v/S characteristic of ppGpp (black) and pppGpp (gray) synthesis by SAS1. Dashed lines indicate Vmax and Km. The velocity is given in micromoles per minute per milligram of SAS1. (B) Crystal structure of the SAS1 tetramer with the pppGpp products (indicated by arrows) in its active sites and two allosteric pppGpp effectors in the central cleft. Each monomer (α–δ) is shown as a cartoon in rainbow colors from N- to C-terminus. (C) Residues from two opposing monomers (indicated as “subunit” and “subunit′”) of the SAS1 tetramer tightly coordinate each of the allosteric pppGpp molecules. (D) The allosteric site of SAS1 shows a different dynamic response to the presence of ppGpp (dark gray bars) or pppGpp (light gray bars) compared with the apo-state (medium gray bars) in HDX experiments after 30 s of deuteration. The graph shows the percentage of HDX of three individual peptides (amino acid sequences are listed above the bars). Amino acid residues also appearing in C are shown in red. Error bars represent the SD of three independent measurements. (E) The ppGpp synthetase activity of SAS1 is stimulated efficiently by pppGpp (black squares) but is stimulated only moderately by ppGpp (black circles). Error bars represent the SD of three independent measurements. (F) The pppGpp synthetase activity of SAS1 is stimulated efficiently by pppGpp (gray squares) but is stimulated only moderately by ppGpp (gray circles). Error bars represent the SD of three independent measurements. (G) Overview of the alarmone synthesis and degradation network. RelA (Syn; green/gray), SAS1 (blue), and SAS2 (blue) represent the three synthetases that establish the ppGpp (orange ball) and pppGpp (yellow ball) pools in B. subtilis. RelA detects amino acid starvation at the ribosome (dark yellow) via its C terminus (C) but also provides the only (p)ppGpp degrading activity known so far via its hydrolase domain (HD). Synthetase and hydrolase activities of RelA are mutually exclusive. SAS1 is positively regulated by pppGpp via an allosteric site in the center of its homotetrameric structure. Whether SAS2 is regulated in a way similar to SAS1 but by an unknown effector (blue balls) remains to be shown.
Fig. S5.
Enzyme kinetic analysis of SAS1. (A) Progress curves of ppGpp production by SAS1 at differing GDP concentrations (0.1, 0.25, 0.5, 0.75, 1, 2, 3, and 5 mM GDP) and 5 mM ATP each. For each GDP concentration, five different time points (1, 3, 5, 7, and 15 min) were measured. For the 2-, 3-, and 5-mM GDP concentrations, the 15-min data point was excluded because substrate limitation was reached. The R2 values for each progress curve are indicated. (B) Progress curves of pppGpp production by SAS1 at differing GTP concentrations (0.1, 0.25, 0.5, 0.75, 1, 2, 3, and 5 mM GDP) and 5 mM ATP each. For each GDP concentration, five different time points (1, 3, 5, 7, and 15 min) were measured. The R2 values for each progress curve are indicated.
pppGpp Allosterically Regulates SAS1.
Because our structural analysis did not provide a plausible explanation as to why SAS1 synthesizes ppGpp more efficiently than pppGpp, we crystallized SAS1 in the presence of ATP and GTP. Crystals were obtained after ∼1 wk and diffracted to 2.94-Å resolution. After phasing by MR with SAS1 (this study) as a search model (Table S1), active-site density could be assigned unambiguously to the pppGpp reaction product (Fig. 3B and Fig. S6A), further supporting the role of SAS1 as an alarmone synthetase.
Fig. S6.
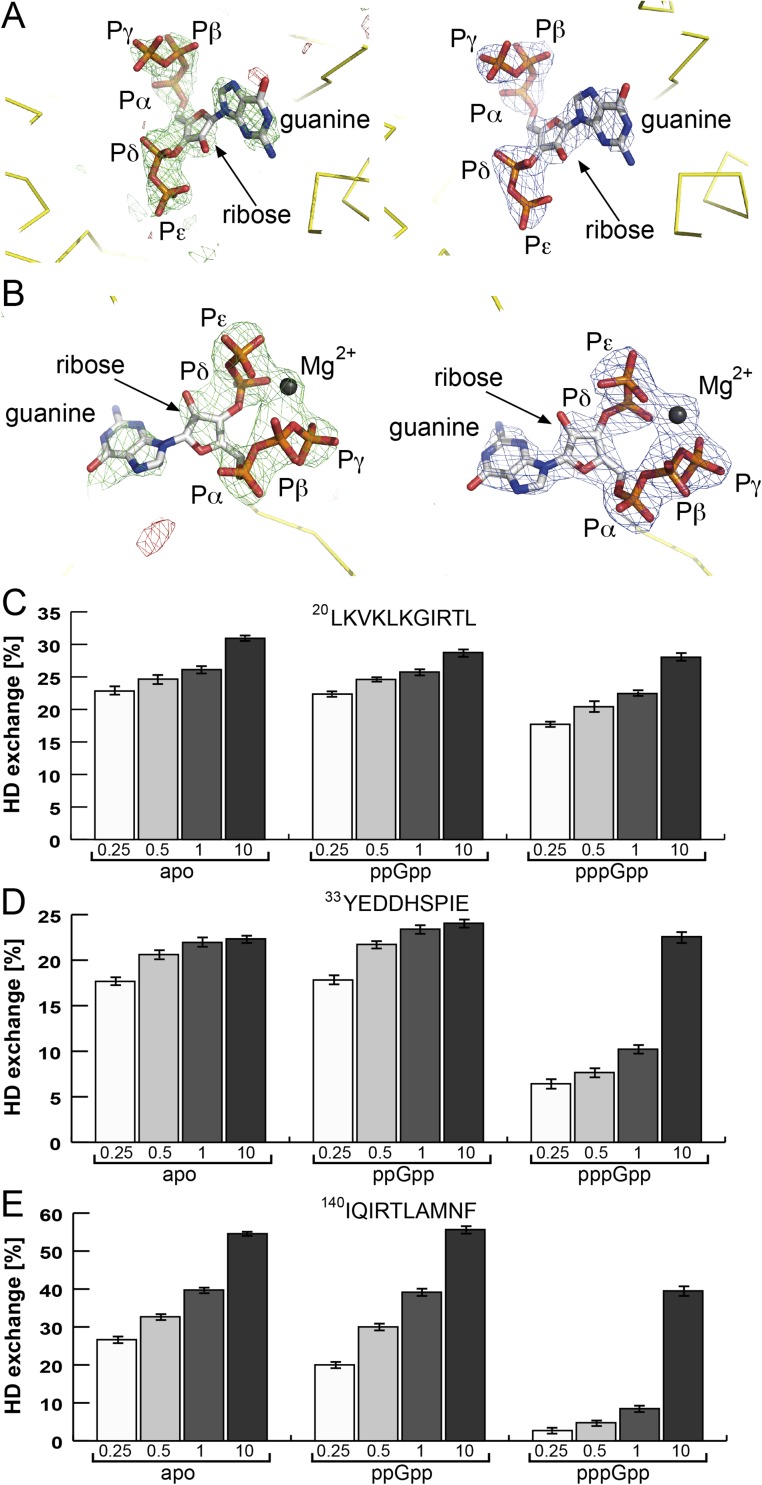
Allosteric binding of pppGpp to SAS1. (A) pppGpp could be assigned unambiguously within the active site of SAS1. (Left) The unbiased Fobs–Fcalc difference electron density of pppGpp contoured at 2.5 σ is shown as green and red mesh for positive and negative differences, respectively. Note: The pppGpp molecule (represented as sticks) was not present during refinement and is placed only for reasons of illustration. (Right) The 2Fobs–Fcalc electron density of pppGpp (represented as sticks) after final refinement is contoured at 2.5 σ and shown as blue mesh. Yellow ribbons indicate the backbone of SAS1. Figures were generated with PyMOL. (B) pppGpp could be assigned unambiguously within the allosteric site of SAS1 that is present in a cleft formed by the subunits of the homotetramer. (Left) The unbiased Fobs–Fcalc difference electron density of pppGpp/magnesium contoured at 2.5 σ is shown as green and red mesh for positive and negative differences, respectively. Note: The pppGpp molecule (represented as sticks) and the magnesium (gray sphere) were not present during refinement and are placed only for reasons of illustration. (Right) The 2Fobs–Fcalc electron density of pppGpp (represented as sticks) and magnesium (gray sphere) after final refinement is contoured at 2.5 σ and shown as blue mesh. Yellow ribbons indicate the backbone of SAS1. Figures were generated with PyMOL. (C–E) The graphs show the percentages of HDX of three individual peptides (amino acid sequences are listed above the plot) comprising the allosteric site of SAS1 in the apo-, ppGpp-, and pppGpp-bound state. Deuteration times are indicated in minutes below the bars. Error bars represent the SD of three independent measurements. (C) Peptide LKVKLKGIRTL. (D) Peptide YEDDHSPIE. (E) Peptide IQIRTLAMNF.
To our surprise, we also observed significant, unassigned density in the cleft formed by the four SAS1 monomers. Two additional pppGpp molecules, tightly coordinated within the cleft, could be unambiguously modeled into this density (Fig. 3B and Fig. S6B). Specificity for the guanine nucleotide is established by the interaction of the N2 amino- and O6 keto-groups with Glu41 and Asn148, respectively (Fig. 3C). Because these residues are located on different monomers, specificity for the allosteric pppGpp is possible only in the context of a tetramer. The guanine base is stabilized further by π-stacking interactions with Phe42. The 2′-OH group of the ribose interacts with Thr44. The α−ε phosphates of the allosteric pppGpp are sequestered in a positively charged cage comprising Lys21, Lys25, and Arg28, which are provided by two monomers with pseudorotational symmetry. In conclusion, the SAS1 tetramer provides a cleft into which two pppGpp molecules can be tightly coordinated.
Our observation raises the question whether the pppGpp bound into the cleft of the SAS1 homotetramer might affect the activity of SAS1 (e.g., as an allosteric regulator). Therefore, we measured ppGpp and pppGpp synthetase activities in the presence of increasing amounts of either ppGpp or pppGpp. For this purpose, ppGpp and pppGpp were enzymatically produced by SAS1 and prepared to purities of 98% and 95%, respectively. The alarmone pppGpp stimulated the catalytic activity of SAS1 more efficiently than ppGpp (∼10-fold) at the lowest concentration tested (12.5 μM) (Fig. 3 E and F).
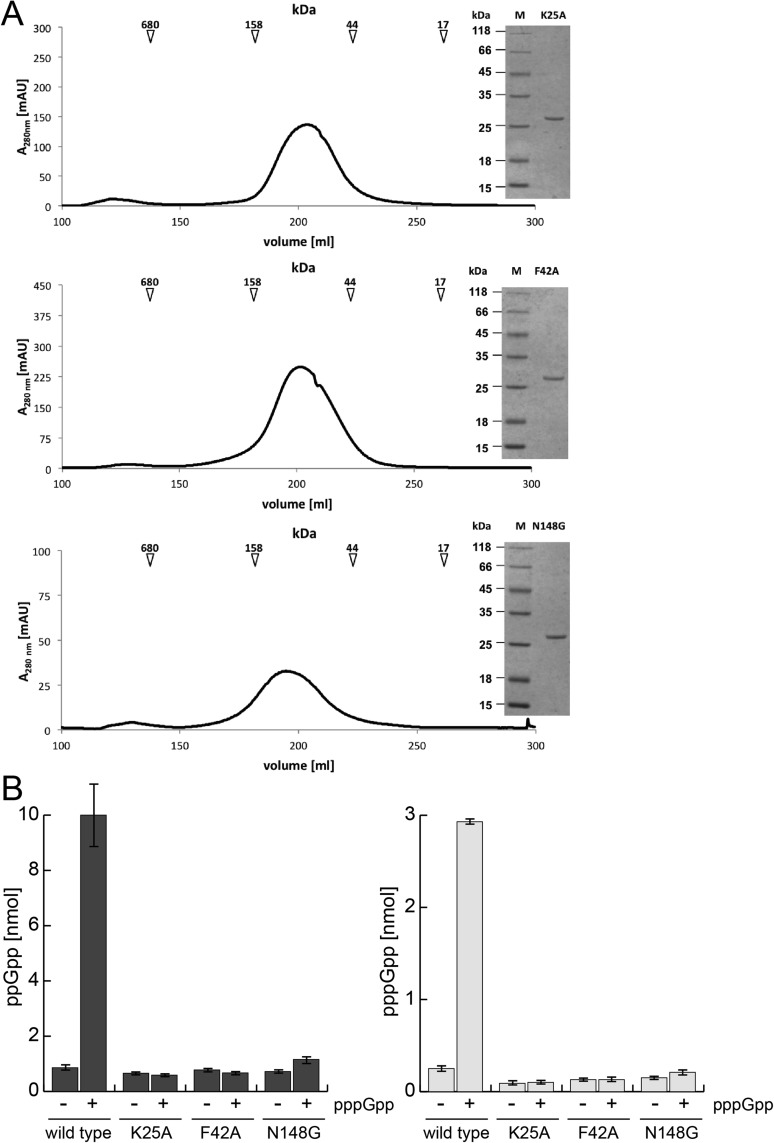
If binding of pppGpp stimulates synthesis, it is not clear whether the functional binding is between the subunits or if binding occurs in one active site that then stimulates synthesis at the other active sites. To discriminate between these scenarios, we measured the (p)ppGpp synthesis of SAS1 variants that should ablate binding of pppGpp between the SAS1 subunits (i.e., K25A, F42A, and N148G, Fig. 3C). Although these variants form homotetramers (Fig. S7A), they did not respond to pppGpp at a concentration of 12.5 μM, at which wild-type SAS1 is stimulated 10-fold (Fig. S7B). Therefore, pppGpp bound into the cleft of the SAS1 homotetramer acts as an allosteric regulator.
Fig. S7.
(A) SEC profile and Coomassie-stained SDS/PAGE (Inset) of the SAS1 variants K25A (Top), F42A (Middle), and N148G (Bottom). The arrows indicate the mass of the SEC size standard. (B) Activation of ppGpp (Left) and pppGpp (Right) synthesis of wild-type SAS1 and its variants by pppGpp. SAS1 (2 µM) was incubated in the presence of 5 mM ATP and 0.25 mM GDP or GTP without (−) or with (+) the addition of 12.5 µM pppGpp for 5 min at 37 °C and was subjected to HPLC measurement. Error bars represent the SD of three independent measurements.
Our structural analysis also shows that the γ-phosphate moiety of pppGpp (which is not present in ppGpp) establishes further contact with the amino acid residue Lys25 of SAS1 (Fig. 3C). This additional contact might allow much stronger binding of pppGpp than of ppGpp. Our HDX experiments support this notion, because we observe significant stabilization of three regions of SAS1 that contain residues involved in its interaction with the allosteric pppGpp (Fig. 3D) Stabilization of these peptides in the regulatory cleft of SAS1 with pppGpp but not ppGpp occurs at concentrations as low as 12.5 µM (Fig. 3D and Fig. S6 C–E). Therefore, it is not surprising that ppGpp also can stimulate SAS1 activity to a certain degree (Fig. 3 E and F). However, the minor stimulation of SAS1 by ppGpp becomes visible only at concentrations that are nonphysiological under nonstringent response conditions (29, 30). These experiments demonstrate that pppGpp—but not ppGpp—acts as a positive effector of the SAS1 enzyme through its ability to bind the regulatory cleft of the SAS1 tetramer.
Discussion
The Catalytic Mechanism, Cooperativity, and Allostery of Alarmone Synthesis.
Synthesis of (p)ppGpp is carried out by synthetases of the RSH type (18), which catalyze the transfer of pyrophosphate from ATP to the ribose 3′-OH of GDP or GTP. A detailed understanding of the catalytic mechanism of this reaction was missing until now. We define the catalytically relevant motifs (E1–E3) required for substrate binding and activation during alarmone synthesis. In particular, our biochemical analysis shows that SAS1 binds its substrates in a sequential and ordered manner initiated by ATP and followed by GDP or GTP (Fig. 2C). Binding of ATP to SAS1 forces the nucleotide into a tense, U-shaped conformation that apparently is necessary to donate the β- and γ-phosphates to the ribose 3′-OH group of GDP (or GTP). Interestingly, such an unusual conformation also has been observed in aminoacyl-tRNA synthetases that use ATP to activate amino acids for subsequent transfer to tRNAs (31, 32).
When loaded with ATP, SAS1 can bind GDP or GTP to assume a near-attack conformation immediately with both substrates in a pretransition state geometry. In this configuration, the β-phosphate of the tense ATP is positioned in ideal proximity to the 3′-OH group of GDP or GTP. However, the 3′-OH group of GDP or GTP is by itself a rather unreactive species that requires activation via deprotonation, enforced by Glu139 and a magnesium ion. Similar activation of hydroxyl groups has been observed in hydroxymethylpterin pyrophosphokinases (HPPKs) (33). Taken together, these observations show that alarmone synthetases position their substrates in a geometry ideal for catalysis. In this configuration, the tense conformation of ATP resembles a loaded spring that is released upon nucleophilic attack of the deprotonated 3′-OH group of GDP or GTP.
Although RelA is catalytically active as a monomer, the small alarmone synthetase SAS1 assembles into a homotetramer. The four subunits of SAS1 display highly cooperative behavior (indicated by the sigmoidal v/S and the Hill coefficient), underlining the functional significance of the homotetramer (see below). The oval-shaped SAS1 homotetramer also contains a central cleft. Our biochemical and structural analyses demonstrate that this cleft provides binding sites for two pppGpp (but not ppGpp) molecules. These molecules serve as allosteric activators that facilitate the synthesis of (p)ppGpp by SAS1. In summary, the action of SAS1 relies on two interconnected but distinct features of the homotetramer: (i) cooperativity of the active sites and (ii) allosteric regulation by pppGpp but not by ppGpp. These intricate features provide the basis for the synthesis of and differential regulation by the two similar (but not identical) alarmones, ppGpp and pppGpp.
Functional Meaning of pppGpp-Dependent Activation of SAS1.
Our data raise two questions: What is the physiological significance of alarmone synthesis at SAS1 being stimulated efficiently by pppGpp but not by ppGpp, and what is the source of the stimulating pppGpp? A simple view is that production of pppGpp by SAS1 leads to an autostimulation of its own activity. In this case, SAS1 would be dependent only on the availability of GTP and GDP substrates characterized by a cellular GTP/GDP ratio of ∼90%/10% (or 5/0.5 mM) under healthy conditions (34). Therefore, SAS1 will produce primarily pppGpp rather than ppGpp simply because GTP is in large excess. This basal alarmone synthesis should be cleared efficiently by the RelA hydrolase activity that exceeds SAS1 activity (23). If so, the SAS1 stimulating pppGpp should originate from another alarmone synthetase (i.e., RelA and SAS2). Amino acid starvation is detected by RelA, which senses the availability of amino acids through the presence of uncharged tRNAs at the ribosome (20). Detection of stalled ribosomes by RelA leads to the synthesis of alarmones but also inhibits their hydrolysis, because RelA activities are mutually exclusive (Fig. 3G) (19, 23). If a stringent response occurs when cellular GTP is in great excess of GDP, RelA will produce primarily pppGpp. This pppGpp can allosterically stimulate SAS1 activity, which in turn would amplify the alarmone signal of RelA during amino acid starvation. However, RelA cannot detect an increase in cellular GDP levels that might occur independently of amino acid starvation. In contrast, SAS1 could use the enlarged pool of GDP as a substrate for ppGpp synthesis. Our data show that SAS1 is much more efficient in producing ppGpp even in the absence of stimulating pppGpp. Under these conditions, SAS1 should be able to produce more ppGpp than could be cleared efficiently by the RelA hydrolase (23). In this way, cells would be able to integrate different stress types (i.e., amino acid starvation by RelA/SAS1 and energy imbalance by SAS1) at the level of ppGpp and pppGpp synthesis. This idea also is supported by the observation that the stringent response seems to be linked to the cellular energy state, because decreased GTP levels render B. subtilis more capable of surviving amino acid starvation, albeit at lower growth rates (35, 36). Both events are closely connected to ppGpp and pppGpp, which also inhibit multiple enzymes for GTP biosynthesis (35, 37, 38). Furthermore, these data strongly indicate that pppGpp and ppGpp execute different biological functions. Cashel and coworkers (17) came to similar conclusions in Escherichia coli, where pppGpp is less potent than ppGpp with respect to growth rate regulation or ribosomal P1 promoter transcription. Nevertheless, further studies are required to dissect the individual physiological roles of ppGpp and pppGpp.
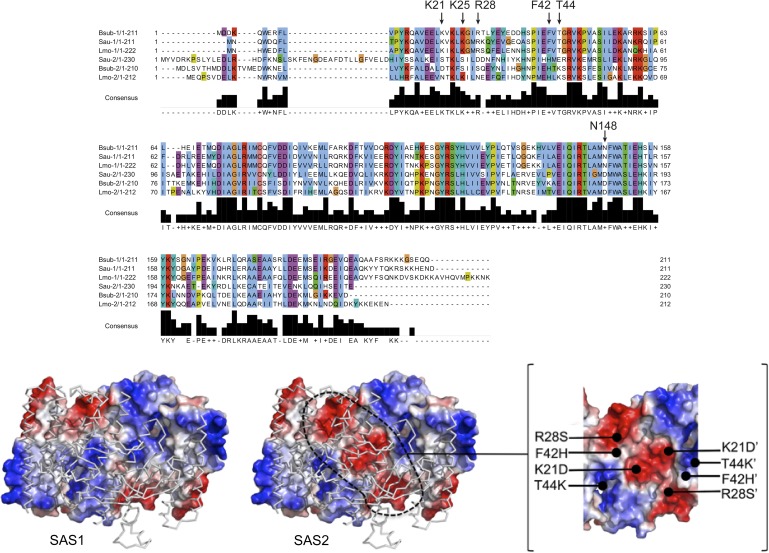
Interestingly, another small alarmone synthetase, SAS2, has been implicated in mediating tolerance against cell envelope stress and blocking translation via ribosome hibernation in Staphylococcus aureus and B. subtilis, respectively (24, 39). Our sequence alignments suggest that SAS2 forms a tetramer with architecture similar to that of SAS1; however, the amino acid residues required for allosteric binding of pppGpp to SAS1 are replaced by a different set of conserved residues in SAS2 (Fig. S8), indicating that alarmone synthesis by SAS2 might be regulated by an allosteric effector other than pppGpp. In this case, another signal could be integrated into the global alarmone pool through differential regulation of SAS2. Future studies should investigate the chemical nature of that putative allosteric ligand. In conclusion, our study provides an in-depth molecular understanding of the complex framework of alarmone synthesis and its potential role in communicating fundamentally different stress signals through two similar but not identical alarmones.
Fig. S8.
SAS1 and SAS2 differ significantly in residues that constitute the allosteric pppGpp-binding site of SAS1. (Upper) Alignment of SAS1 (−1) and SAS2 (−2) from B. subtilis (Bsub), S. aureus (Sau), and Listeria monocytogenes (Lmo). Arrows indicate residues critical for the coordination of allosteric pppGpp in SAS1. Amino acid numbers above the arrows relate to B. subtilis SAS1. (Lower) Crystal structure of SAS1 (Left) and a model of SAS2 (Center) are shown with two monomers in electrostatic surface representation and two as ribbons. Dashed lines indicate differences in the electrostatic properties of the clefts present in SAS1 and SAS2. Amino acids and their locations contributing to these differences are indicated in the Inset (Right).
Materials and Methods
All experiments are described in detail in SI Materials and Methods.
Protein Purification.
Proteins were produced in E. coli BL21 (DE3) and were purified in a two-step protocol consisting of Ni-ion affinity and size-exclusion chromatography (SEC). The SEC buffer was composed of 20 mM Hepes⋅Na (pH 7.5), 200 mM NaCl, 20 mM KCl, and 20 mM MgCl2.
Crystallization and Structure Determination.
Crystallization was performed as detailed in SI Materials and Methods. Data were collected at the European Synchrotron Radiation Facility (ESRF) and were processed with XDS (40) and CCP4-implemented SCALA (41). Structures were determined by MR using ccp4-implemented Phaser (41). The search models were a truncated version of RelA [PDB ID code: 1VJ7 (19)] for apo SAS1 and the structure of apo-SAS1 (this study) for all nucleotide-bound states of SAS1. Structures were built on Coot (42) and refined with Phenix (43). Figures were prepared in PyMOL.
HDX Mass Spectrometry.
Purified SAS1 was preincubated with or without nucleotides for 5 min at 37 °C and diluted 10-fold in deuterated buffer. HDX was stopped after different incubation times (i.e., 15/30/60/600 s) by the addition of ice-cold quench buffer (pH 2.2). Peptic peptides were generated by an online pepsin column and separated by reversed-phase HPLC. Data were analyzed using the ProteinLynx Global Server (PLGS) and DynamX 3.0 software (Waters). HDX raw data are available on request.
Activity Assays.
Kinetic analysis of SAS1 and activation of SAS1 by pppGpp was carried out at 37 °C and stopped by flash-freezing in liquid nitrogen. Samples were subjected to HPLC measurement using an Agilent 1100 Series system (Agilent Technologies) and a C18 column (EC 250/4.6 Nucleodur HTec 3 μm; Macherey-Nagel). Running buffers contained 50 mM KH2PO4 and K2HPO4 each and 10 mM tetrapentylammonium bromide (TPAB). Nucleotides were separated using a gradient up to 90% (vol/vol) acetonitrile at a flow rate of 0.8 mL/min. The reaction products were detected at a wavelength of 260.8 nm in agreement with standards. Allosteric effects of (p)ppGpp on SAS1 activity were measured by HPLC. To discriminate between (p)ppGpp that was added before the reaction and (p)ppGpp synthesized by SAS1, the amount of AMP product released equimolar to the (p)ppGpp product was quantified.
Preparation of ppGpp and pppGpp.
(p)ppGpp was produced enzymatically using SAS1, ATP, and GDP or GTP. After chloroform precipitation of SAS1, nucleotides were separated by anion-exchange chromatography. (p)ppGpp was precipitated by lithium chloride and washed with ethanol. Purity was controlled by analytical HPLC.
SI Materials and Methods
Cloning and Mutagenesis of SAS1.
The yjbM gene encoding SAS1 was amplified from B. subtilis PY79 genomic DNA by PCR using Q5 High-Fidelity DNA Polymerase (New England Biolabs) according to the manufacturer’s manual. The forward primer encoded a hexahistidine tag in-frame with the DNA sequence of yjbM. The PCR fragment was cloned into the pET24d(+) vector (Novagen) at NcoI/XhoI restriction sites. Mutations within SAS1 were generated by overlapping PCRs.
Protein Production and Purification.
E. coli BL21(DE3) cells (New England Biolabs) carrying the expression plasmid were grown in lysogeny broth (LB) medium supplemented with kanamycin (50 μg/mL) and D(+)-lactose-monohydrate (12.5 g/L) for 16 h at 30 °C under rigorous shaking (150 rpm). Cells were harvested (3,500 × g, 20 min, 4 °C) and were resuspended in lysis buffer [20 mM Hepes⋅Na (pH 8.0), 250 mM NaCl, 40 mM imidazole, 20 mM MgCl2, 20 mM KCl]. Cells were lysed by the M-110L Microfluidizer (Microfluidics). After centrifugation (47,850 × g, 20 min, 4 °C), the clear supernatant was loaded on a 1-mL HisTrap column (GE Healthcare) equilibrated with 10 column volumes (CV) of lysis buffer. After washing with 10 CV lysis buffer, the protein was eluted with 15 mL elution buffer (lysis buffer containing 500 mM imidazole). The protein was concentrated to ∼35 mg/mL using an Amicon Ultracel-10K (Millipore) and was applied to SEC (HiLoad 26/600 Superdex 200 pg, GE Healthcare) equilibrated in SEC buffer [20 mM Hepes⋅Na (pH 7.5), 200 mM NaCl, 20 mM MgCl2, 20 mM KCl]. Protein-containing fractions were pooled and concentrated according to the experimental requirements. Protein concentration was determined by a spectrophotometer (NanoDrop Lite; Thermo Scientific).
Crystallization of SAS1 in the Apo- and Nucleotide-Bound States.
All crystallization experiments were carried out with the sitting-drop method in SWISSCI MRC two-well crystallization plates (Jena Bioscience). The reservoir volume was 50 μL, and the drop size was 1 μL with a 1:1 mixture of protein and crystallization solution. All crystallization experiments were performed at room temperature. Crystals of the apo-state of SAS1 were obtained at 20 mg/mL protein concentration after 1 d in 0.1 M MES (pH 5.0), 20% (wt/vol) PEG 6000. For crystallization of the different nucleotide-bound states, SAS1 was incubated for 1 h on ice with 1 mM of the respective nucleotide. Crystals of the AMPCPP-bound state of SAS1 were obtained at a 11.5 mg/mL protein concentration after 1 wk in 0.2 M KSCN, 20% (wt/vol) PEG3350. Crystals of the pppGpp-bound state of SAS1 were obtained at 11.5 mg/mL protein concentration after 1 wk in 0.1 M Citric acid (pH 4.0), 5% (wt/vol) PEG 6000.
Data Collection, Structure Determination, and Analysis.
Before data collection, crystals were flash-frozen in liquid nitrogen after the transfer through a cryoprotection solution consisting of mother liquor supplemented with 20–30% (vol/vol) glycerol. Data collection was performed at the ESRF in Grenoble, France at beamlines ID23-1 (nucleotide-bound states of SAS1) and ID29 (apo-state of SAS1). Data collection was performed under laminar nitrogen flow at 100 K (Oxford Cryosystems 700 Series). Data were recorded with a DECTRIS PILATUS 6M detector and processed with XDS (40) and the CCP4-implemented program SCALA (41). The structure of apo-SAS1 was determined by MR using a truncated structure of RelA [PDB ID code 1VJ7 (19)], which contained only the minimal synthetase fragment. MR was performed using the CCP4-implemented PHASER (44). The structure of apo-SAS1 (this study) was used as search model for MR to determine the nucleotide-bound structures of SAS1. Structures were built in COOT (42) and refined with PHENIX refine (43). Figures were prepared in PyMOL (www.pymol.org).
HDX Mass Spectrometry.
Purified and concentrated SAS1 [200 pmol (50 µM)] was incubated without or in the presence of nucleotides for 5 min at 37 °C before HDX. Nucleotides were used in concentrations of 125 µM (ppGpp and pppGpp) or 1 mM (GDP, GTP, and AMPCPP). The mixtures were diluted 10-fold into D2O-containing SEC buffer [20 mM Hepes⋅Na (pH 7.5), 200 mM NaCl, 20 mM KCl, 20 mM MgCl2] and were incubated at 37 °C during HDX. The reactions subsequently were quenched after different incubation times (i.e., 15/30/60/600 s) by adding one equivalent of ice-cold quench buffer (400 mM KH2PO4/H3PO4, pH 2.2) and were injected directly into a nanoACQUITY system with HDX technology for ultra performance liquid chromatography (UPLC) separation (Waters) (45). In-line pepsin digestion was carried out with an Enzymate BEH Pepsin column 2.1 × 30 mm (Waters) at a flow rate of 100 µL/min in H2O + 0.1% formic acid at 11 °C. The resulting peptic peptides were trapped for 3 min on an AQUITY UPLC BEH C18 1.7-µm 2.1 × 5 mm VanGuard Pre-column (Waters) kept at 0.5 °C. Thereafter, the trap column was placed in line with an ACQUITY UPLC BEH C18 1.7-µm 1.0 × 100 mm column (Waters), and the peptides were separated at 0.5 °C using the following gradient of H2O + 0.1% formic (A) and acetonitrile + 0.1% formic acid at a flow rate of 40 µL/min: 0 min at 95% A, 7 min at 65% A, 8 min at 15% A, 10 min at 15% A, 10.1 min at 5% A, 11 min at 5% A, 11.1 min at 95% A, and 16 min at 95% A. Mass spectra were acquired in positive ion mode by a G2-Si high-density mass spectrometer equipped with an electrospray ionization source (Waters). Lock-mass correction was performed continuously using Leu-enkephalin peptide standard (Waters). The mass spectra of undeuterated SAS1 were obtained similarly, except that samples were diluted 10-fold into H2O-containing SEC buffer. All measurements were performed in triplicate.
Peptide identification and data analysis were carried out basically as described previously (46). Undeuterated peptides were identified in data-independent acquisition mode (MSE) (47) aided by the PLGS software (Waters). Peptides identified in at least two replicates were included in further analysis. Mass spectra were extracted and analyzed using DynamX 3.0 (Waters). The tolerances of retention time and m/z values applied for the identification of the peptic peptides were 0.5 min and 25 ppm, respectively. Absolute deuterium incorporation was calculated by subtracting the centroid of the isotope distribution of the undeuterated peptides from the respective deuterated peptides. Relative deuteration was calculated as the quotient of the absolute deuteration and the number of backbone amide protons (number of amino acids minus one) without prolines (48).
Kinetic Analysis of SAS1.
Activity and the kinetic behavior of SAS1 were monitored by HPLC. All reactions were carried out in a total volume of 50 µL in a reaction buffer containing 100 mM Hepes⋅Na (pH 7.5), 200 mM NaCl, 20 mM MgCl2, and 20 mM KCl. For each assay, 2 μM SAS1 and 5 mM ATP, together with varying concentrations of GDP or GTP (as indicated in figures and text), were used. All assays were carried out at 37 °C. Reactions were stopped by flash-freezing in liquid nitrogen and were stored at −20 °C until measurement. HPLC measurements were performed with an Agilent 1100 Series system (Agilent Technologies) and a C18 column (EC 250/4.6 Nucleodur HTec 3 μm; Macherey-Nagel). After running 30 min with a buffer containing 50 mM KH2PO4, 50 mM K2HPO4, 10 mM TPAB, and 15% (vol/vol) acetonitrile, a linear gradient up to 90% acetonitrile over 20 min was applied at a flow rate of 0.8 mL/min. The reaction products ppGpp and pppGpp were detected at a wavelength of 260.8 nm, in agreement with standards.
Kinetic data analysis was performed in GraphPad Prism version 6.04 for Windows, (GraphPad Software). Velocities of (p)ppGpp formation by SAS1 at varying GDP or GTP concentrations were obtained from linear regression of (p)ppGpp measured at different time points. The Km, Vmax, and the Hill coefficient (h) ± SD were obtained from the sigmoidal fit of the v/S characteristic using the equation v = Vmax Sh/(Kmh + Sh).
Activation of SAS1 by (p)ppGpp.
To determine the allosteric effects of ppGpp and pppGpp on SAS1 (p)ppGpp synthetase activity, 2 µM of SAS1 were incubated in a total volume of 50 µL in a reaction buffer containing 100 mM Hepes⋅Na (pH 7.5), 200 mM NaCl, 20 mM MgCl2, and 20 mM KCl in the presence of 5 mM ATP and 0.25 mM GDP or GTP for 5 min at 37 °C. ppGpp or pppGpp was added to the reactions as stated in figures and text. The reactions were stopped by flash-freezing in liquid nitrogen and were subjected to HPLC measurement as described above. To discriminate between (p)ppGpp added to the reaction and newly synthesized (p)ppGpp, the amount of AMP released equimolar to (p)ppGpp during the reaction was quantified in agreement with standards. The amount of contaminating AMP in the ATP substrate was determined by triplicate measurements of 5 mM ATP for each experiment and typically was below 0.5%.
Preparation of (p)ppGpp.
SAS1 (5 μM) was incubated with 5 mM ATP and 5 mM GDP or GTP in the reaction buffer for 30 min at 37 °C. Thereafter, SAS1 was precipitated by the addition of one volume of chloroform. The aqueous phase that contained the nucleotides was diluted with double-distilled water to a total volume of 5 mL and was subjected to anion exchange chromatography (ResourceQ, 6 mL; GE Healthcare). A gradient ranging from 0–2 M NaCl was used for elution of the nucleotides. ppGpp and pppGpp eluted at 200 and 220 mM NaCl, respectively. Nucleotides were precipitated by adding lithium chloride to a final concentration of 1 M, followed by the addition of four volumes of ethanol. The suspension was incubated on ice for 20 min and centrifuged (5,000 × g, 20 min, 4 °C). The resulting pellets were washed twice with absolute ethanol, dried, and stored at −20 °C. The quality of the ppGpp and pppGpp prepared in this manner was controlled by analytical HPLC.
Acknowledgments
We thank Rudolf K. Thauer and Magdalena Rakwalska-Bange (Max Planck Institute Marburg), Kursad Turgay (Leibnitz University Hannover), and Johann Heider (Philipps University Marburg) for fruitful comments on the manuscript and the European Synchrotron Radiation Facility for support during data collection. This work was supported by the LOEWE program of the state of Hesse (G.B.), Sonderforschungsbereich SFB 987 (M.A.M. and G.B.), and the Scientific Instrumentation Grant 160/621-1 FUGG (G.B. and U.L.) of the Deutsche Forschungsgemeinschaft. J.S.S. is a fellow of the Fonds der Chemischen Industrie.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5DEC (SAS1), 5DED (SAS1-pppGpp), and 5DEE (SAS1-AMPCPP)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505271112/-/DCSupplemental.
References
- 1.Boutte CC, Crosson S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 2013;21(4):174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 3.Liu K, Bittner AN, Wang JD. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol. 2015;24:72–79. doi: 10.1016/j.mib.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Kasai K, Ochi K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc Natl Acad Sci USA. 2004;101(12):4320–4324. doi: 10.1073/pnas.0308555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Biezen EA, Sun J, Coleman MJ, Bibb MJ, Jones JD. Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc Natl Acad Sci USA. 2000;97(7):3747–3752. doi: 10.1073/pnas.060392397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuda S, et al. The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. Plant Cell Physiol. 2008;49(2):135–141. doi: 10.1093/pcp/pcm177. [DOI] [PubMed] [Google Scholar]
- 7.Vogt SL, et al. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun. 2011;79(10):4094–4104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss LA, Stallings CL. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J Bacteriol. 2013;195(24):5629–5638. doi: 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalebroux ZD, Swanson MS. ppGpp: Magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10(3):203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 10.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 11.Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol Cell. 2013;50(4):475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 12.He H, Cooper JN, Mishra A, Raskin DM. Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol. 2012;194(11):2962–2972. doi: 10.1128/JB.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Syal K, Maiti K, Naresh K, Chatterji D, Jayaraman N. Synthetic glycolipids and (p)ppGpp analogs: Development of inhibitors for mycobacterial growth, biofilm and stringent response. Adv Exp Med Biol. 2015;842:309–327. doi: 10.1007/978-3-319-11280-0_20. [DOI] [PubMed] [Google Scholar]
- 14.Wexselblatt E, et al. Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog. 2012;8(9):e1002925. doi: 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanjee U, Ogata K, Houry WA. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol. 2012;85(6):1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128(5):865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013;41(12):6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6(8):e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected] Cell. 2004;117(1):57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 20.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10(4):779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 21.Agirrezabala X, et al. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013;14(9):811–816. doi: 10.1038/embor.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avarbock D, Avarbock A, Rubin H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry. 2000;39(38):11640–11648. doi: 10.1021/bi001256k. [DOI] [PubMed] [Google Scholar]
- 23.Mechold U, Murphy H, Brown L, Cashel M. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol. 2002;184(11):2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geiger T, Kästle B, Gratani FL, Goerke C, Wolz C. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol. 2014;196(4):894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nanamiya H, et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67(2):291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- 26.Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol. 2007;65(6):1568–1581. doi: 10.1111/j.1365-2958.2007.05897.x. [DOI] [PubMed] [Google Scholar]
- 27.Srivatsan A, et al. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4(8):e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaca AO, et al. From (p)ppGpp to (pp)pGpp: Characterization of Regulatory Effects of pGpp Synthesized by the Small Alarmone Synthetase of Enterococcus faecalis. J Bacteriol. 2015;197(18):2908–2919. doi: 10.1128/JB.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryals J, Little R, Bremer H. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol. 1982;151(3):1261–1268. doi: 10.1128/jb.151.3.1261-1268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishino T, Gallant J, Shalit P, Palmer L, Wehr T. Regulatory nucleotides involved in the Rel function of Bacillus subtilis. J Bacteriol. 1979;140(2):671–679. doi: 10.1128/jb.140.2.671-679.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnez JG, Augustine JG, Moras D, Francklyn CS. The first step of aminoacylation at the atomic level in histidyl-tRNA synthetase. Proc Natl Acad Sci USA. 1997;94(14):7144–7149. doi: 10.1073/pnas.94.14.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavarelli J, et al. The active site of yeast aspartyl-tRNA synthetase: Structural and functional aspects of the aminoacylation reaction. EMBO J. 1994;13(2):327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stammers DK, et al. 2.0 A X-ray structure of the ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. FEBS Lett. 1999;456(1):49–53. doi: 10.1016/s0014-5793(99)00860-1. [DOI] [PubMed] [Google Scholar]
- 34.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5(8):593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bittner AN, Kriel A, Wang JD. Lowering GTP level increases survival of amino acid starvation but slows growth rate for Bacillus subtilis cells lacking (p)ppGpp. J Bacteriol. 2014;196(11):2067–2076. doi: 10.1128/JB.01471-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kriel A, et al. GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J Bacteriol. 2014;196(1):189–201. doi: 10.1128/JB.00918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kriel A, et al. Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol Cell. 2012;48(2):231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu K, et al. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell. 2015;57(4):735–749. doi: 10.1016/j.molcel.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tagami K, et al. Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. MicrobiologyOpen. 2012;1(2):115–134. doi: 10.1002/mbo3.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80(17):6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nevin P, Lu X, Zhang K, Engen JR, Beuning PJ. Noncognate DNA damage prevents the formation of the active conformation of the Y-family DNA polymerases DinB and DNA polymerase κ. FEBS J. 2015;282(14):2646–2660. doi: 10.1111/febs.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geromanos SJ, et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9(6):1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 1993;2(4):522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]