Significance
The river sharks of the genus Glyphis, widely feared as man-eaters throughout India, remain very poorly known to science. The group constitutes five described species, all of which are considered highly endangered and restricted to freshwater systems in Australasia and Southeast Asia. DNA sequence data derived from 19th-century dried museum material augmented with contemporary samples indicates that only three of the five currently described species are valid; that there is a genetically distinct, but as-yet-undescribed, species recorded in Bangladesh and Sarawak in Malaysian Borneo; and that these iconic and mysterious sharks are not restricted to freshwater at all but rather appear to be adapted to both marine and freshwater habitats.
Keywords: freshwater sharks, DNA, museum specimens
Abstract
For over a hundred years, the “river sharks” of the genus Glyphis were only known from the type specimens of species that had been collected in the 19th century. They were widely considered extinct until populations of Glyphis-like sharks were rediscovered in remote regions of Borneo and Northern Australia at the end of the 20th century. However, the genetic affinities between the newly discovered Glyphis-like populations and the poorly preserved, original museum-type specimens have never been established. Here, we present the first (to our knowledge) fully resolved, complete phylogeny of Glyphis that includes both archival-type specimens and modern material. We used a sensitive DNA hybridization capture method to obtain complete mitochondrial genomes from all of our samples and show that three of the five described river shark species are probably conspecific and widely distributed in Southeast Asia. Furthermore we show that there has been recent gene flow between locations that are separated by large oceanic expanses. Our data strongly suggest marine dispersal in these species, overturning the widely held notion that river sharks are restricted to freshwater. It seems that species in the genus Glyphis are euryhaline with an ecology similar to the bull shark, in which adult individuals live in the ocean while the young grow up in river habitats with reduced predation pressure. Finally, we discovered a previously unidentified species within the genus Glyphis that is deeply divergent from all other lineages, underscoring the current lack of knowledge about the biodiversity and ecology of these mysterious sharks.
Although little is known about many shark species, the river sharks of the genus Glyphis are an especially enigmatic group. Three different species of river sharks were recognized throughout most of the 20th century: the notorious Ganges man-eater Glyphis gangeticus (Müller and Henle, 1839) (1), represented by one dried skin lectotype and one alcohol-preserved paralectotype; the speartooth shark Glyphis glyphis (Müller and Henle, 1839) (1) of unknown geographic origin represented by a single dried skin holotype, two small, poorly preserved specimens, and a handful of dried jaws; and the Irrawaddy shark Glyphis siamensis (Steindachner, 1896) (2), which originated from the mouth of the Irrawaddy River in Myanmar and is known only from its alcohol-preserved holotype.
All three Glyphis species were believed to be extremely rare or extinct. In 1984, Compagno reported that additional, as-yet-undescribed, species of Glyphis likely existed in Borneo, northern Australia, and Papua New Guinea. In 1996, researchers encountered and collected several juvenile “Glyphis-like” sharks at Kampong Abai in the lower reaches of the Kinabatangan River in northeastern Malaysian Borneo. The Kinabatangan river shark was subsequently described as a new species, Glyphis fowlerae Compagno, White, and Cavanagh, 2010 (3). The discovery fueled optimism that other undiscovered populations of Glyphis might exist in remote regions of the world, prompting a series of expeditions from the late 1990s to 2010 that resulted in the collection of river shark specimens from northern Australia and Borneo. These included additional specimens from the Kinabatangan River, two specimens of uncertain affinity from Mukah in Sarawak, a 2-m specimen from Sampit in the southernmost part of Kalimantan, Indonesian Borneo, and specimens from the Alligator, Adelaide, and Wenlock Rivers in northern Australia. The Australian specimens were examined and compared with reference material from Australia and Papua New Guinea. Two species were identified. One was considered to be conspecific with the speartooth shark G. glyphis, whereas the other was considered new to science and was formally named and described as Glyphis garricki Compagno, White, and Last, 2008 (4).
Initial analyses of DNA sequences derived from several of the specimens sampled in Borneo showed the two specimens from Mukah to be genetically similar to each other but highly divergent from the G. fowlerae specimens from the Kinabatangan River (5). Unfortunately, it was not possible to assess the genetic affinity of the Sampit specimen as no tissue was taken. Morphological measurements, however, suggest that it too is distinct from G. fowlerae. It is not known whether the Sampit specimen differs from the specimens taken in Mukah, as the Mukah specimens are no longer available for comparison. Thus, it is entirely possible that three different species of Glyphis exist in Borneo: G. fowlerae, the “Sampit” Glyphis, and the “Mukah” Glyphis. This would potentially bring the total number of species of Glyphis to seven: three from Borneo; G. garricki and G. glyphis from Australia and Papua New Guinea; and G. gangeticus and G. siamensis from the Indian subcontinent and Myanmar.
Although there has been progress in characterizing the taxonomic diversity within Glyphis, we understand neither the relationships among the various newly discovered Glyphis populations in Australia and Borneo, nor the affinities of these populations to the type specimens of G. glyphis, G. gangeticus, and G. siamensis, nor the processes that have influenced present day species distributions. Resolving these questions with certainty requires obtaining reference DNA sequence data from the museum type material. Although it is increasingly recognized that genetic comparisons of contemporary and historical reference specimens are critically important for understanding global biodiversity, obtaining reliable DNA sequence data from museum specimens is challenging because the quality of DNA derived from historical (>100-y-old) museum specimens is usually poor (6). DNA hybridization capture (7) is a powerful method that can be used to recover large amounts of sequence data while requiring very small amounts of input DNA. Although this approach has successfully been used to assemble complete mitochondrial genomes from highly degraded ancient DNA (8, 9), only a few studies have so far reported full mitochondrial genomes being obtained from museum specimens, and to our knowledge, none of these investigated archival fish specimens (10). Fishes account for more than one-half of the diversity of the vertebrate tree of life and the relationships among many major groups remain unresolved or are based on morphological interpretations alone. Opening up archival fish collections around the world to these types of molecular techniques has promising implications for future ichthyological research.
We therefore assembled a sample set including both modern samples and most available type specimens for the Glyphis river sharks. We applied DNA hybridization capture technology to obtain both mitochondrial and nuclear DNA sequences from these specimens and have used the data to resolve the phylogenetic relationships among these elusive species.
Results
Consistent with the expectation of poor DNA preservation, our attempts using PCR amplification to obtain sequence data from the museum-type material for Glyphis failed to yield results. We therefore modified a recently developed DNA hybridization capture protocol (11) that is well suited to enriching the small amounts of fragmented DNA typical of museum samples, and used it to capture complete mitochondrial genomes for the dried Glyphis-type specimens (see SI Appendix for details of modifications). We used Illumina next-generation sequencing to sequence the captured products. This strategy allowed us to isolate and compare DNA sequence data from both archival and modern specimens including type specimens of all five described Glyphis species, additional dried jaws collected in Pakistan, Bangladesh, India, and Cirebon (northern Java), freshly collected tissues taken from the two Mukah specimens, freshly collected tissues of G. glyphis and G. garricki from northern Australia, and freshly collected tissues of the closely related genus Lamiopsis (Fig. 1 and SI Appendix, Table S1). Our modified protocol yielded complete mitochondrial genome sequences for all 23 Glyphis samples as well as for one of the Lamiopsis samples (SI Appendix, Table S2). Consistent with previous studies (10), we found high variability in the percentage of reads on target ranging from 1% to 97% for the fresh specimens and from 15% to 91% for the museum specimens. After removal of duplicates, this resulted in a sequence depth of the mitochondrial genome between 15 and 21,000 and coverage of the mitochondrial genomes between 96% and 100% (SI Appendix, Table S2). The average read length for the museum specimens was between 100 and 123 bp, slightly longer than true ancient (i.e., subfossil) DNA reads. Thus, as with previous studies investigating other vertebrate classes such as mammals and birds, dried fish specimens appear to be good sources of DNA for comparative genomic studies.
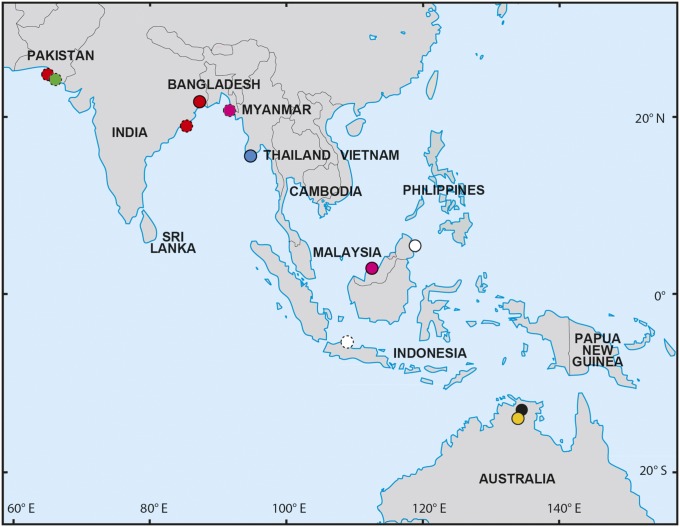
Fig. 1.
Sampling localities used in this study. Colors correspond to taxonomic status assigned at time of collection. Red, Glyphis gangeticus; green, G. sp. 1; blue, G. siamensis; white, G. fowlerae; pink, G. sp. 2; black, G. glyphis; yellow, G. garricki. Solid symbol outlines indicate samples with detailed locality information. Dashed outlines indicate samples for which detailed locality information is not available.
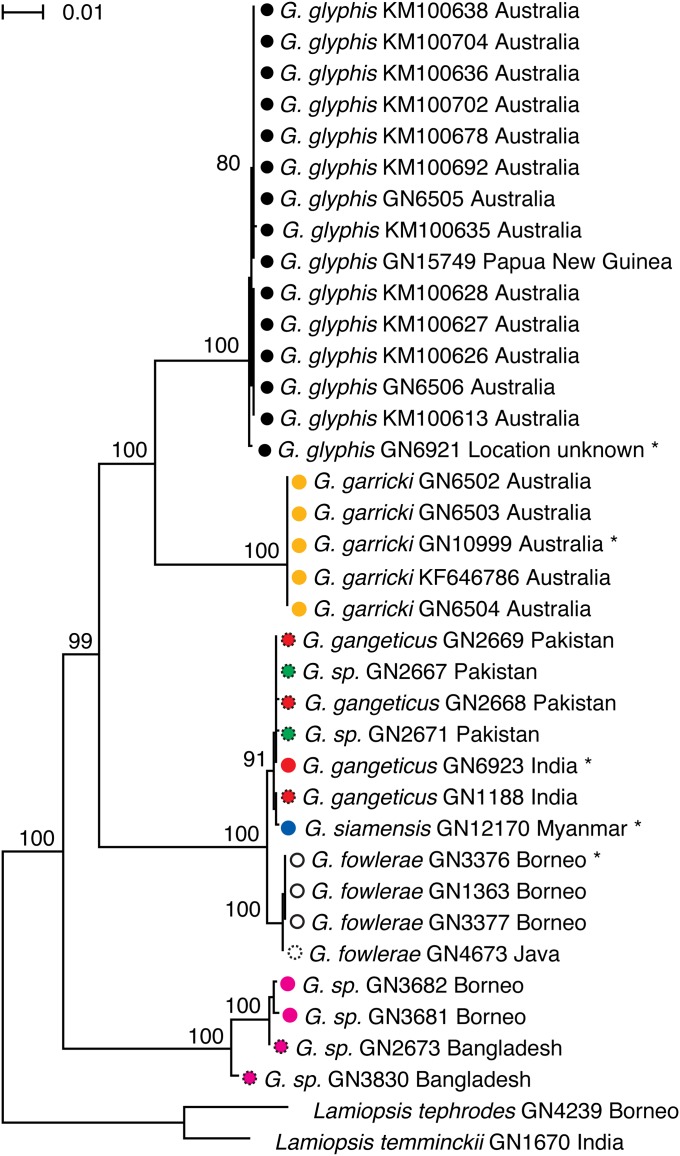
The protein-coding components of the mitochondrial genome sequence were translated to their corresponding amino acid sequences, aligned, and backtranslated to their underlying nucleotide sequences for subsequent analysis. The alignment was augmented with additional protein-coding sequences derived from recently published mitochondrial genome sequences from G. glyphis (12) and subjected to a maximum-likelihood phylogenetic analysis using a GTR + Γ model. The analysis recovered four deeply divergent lineages within Glyphis (Fig. 2). The inferred clade with the highest taxonomic diversity includes sequences from specimens originating from India and Pakistan, the lectotype of G. gangeticus, the holotype of G. siamensis from Myanmar, and four specimens described as G. fowlerae from Borneo and Java. The sequence of the holotype of G. siamensis is nested within the G. gangeticus samples, including the lectotype. The four sequences of G. fowlerae, including the holotype, form a monophyletic sister group to the clade containing the other seven sequences of G. siamensis and G. gangeticus, although all of these sequences are very closely related (p-distance < 0.65%; SI Appendix, Figs. S1 and S2).
Fig. 2.
Maximum-likelihood tree based on a partitioned analysis of the protein-coding portion of the mitochondrial genome using a GTR + Γ (39) model for each codon partition. Type specimens are indicated by an asterisk. Colors are as in Fig. 1. Locality information is available in SI Appendix, Table S1. Bootstrap support values are indicated on the branches.
A second clade consists of 15 Australian sequences from specimens nominally assigned to G. glyphis, including the one obtained from the holotype. The sequence from the holotype of G. garricki, and sequences from four additional G. garricki specimens, form a deeply divergent sister clade to the 15 G. glyphis sequences (Fig. 2).
The last clade consists of four sequences, two from Bangladesh and two from Mukah in Malaysian Borneo. This distinct clade is the first to diverge among the extant Glyphis species examined. The Bangladesh specimens in this clade are paraphyletic with respect to the Borneo specimens.
The statistical parsimony network is generally characterized by relatively small intraspecific divergences, whereas a large number of mutational steps separate all nominal species (SI Appendix, Fig. S1). This is true except in the case of the cluster containing G. gangeticus, G. siamensis, G. sp. Pakistan, and G. fowlerae. Here, the greatest interspecific divergence, that separating G. fowlerae from all others in the cluster, is about one order of magnitude lower than typically observed among nominal species in other areas of the network (SI Appendix, Fig. S1). Likewise, the pairwise p-distance observed among species in this cluster is at the lower end of observed values and is of similar magnitude to those obtained for other comparisons that represent intraspecific diversity for the other nominal species (SI Appendix, Fig. S2).
In addition, a subset of the samples was also successfully subjected to a nuclear gene capture protocol (13). The inferred relationships among the four major clades based on a concatenated maximum-likelihood analysis of 100 single-copy nuclear genes was identical to the inference derived from the mitochondrial data (SI Appendix, Fig. S3).
A species tree analysis using SNAPP (14) of 1,041 independent SNPs (SI Appendix, Table S3 and Fig. S4) yields a result that is consistent with the results of both the mitochondrial analysis (Fig. 2) and that of the concatenated 100 nuclear sequences (SI Appendix, Fig. S3). Interestingly however, a Bayes factor delimitation (*with genomic data) (BFD*) (15) based on the same 1,041 SNPs suggested that G. fowlerae should be considered a separate species (marginal L0 = −7,021 G. fowlerae as distinct species; marginal L1 = −7,082 G. fowlerae grouped as one species with G. gangeticus: Bayesian factor, B01 = 61).
Discussion
To our knowledge, our results represent the first complete mitochondrial genome sequences that have been derived from archival (>100-y-old) museum preserved ichthyological material. Although complete mitochondrial genomes have been recovered from a number of museum specimens (10), such studies are still rare and mostly restricted to mammalian, or more infrequently bird, species. Our results show that DNA hybridization capture approaches can reliably be used to obtain DNA sequence data from museum specimens even in cases where traditional PCR-based approaches have failed because of poor DNA preservation. We also demonstrate that these approaches are particularly useful for recovering large amounts of data from extremely rare specimens of poorly studied vertebrate groups. Ichthyological specimens represent by far the most species-rich, albeit understudied, vertebrate group. Cartilaginous fishes are one of the most threatened vertebrate groups (16), and just like their bony fish counterparts (17, 18), include many species that are known from only single or few specimens (e.g., Mollisquama parini) (19). Obtaining DNA sequence data from such rare preserved ichthyological specimens can have a major impact on evolutionary research.
The phylogenetic tree reconstructed from the protein-coding portion of the mitochondrial genome sequences yields several surprising results. It recovers four deeply divergent lineages within Glyphis, which likely represent different species (Fig. 2). Unexpectedly, the clade with the highest taxonomic diversity includes sequences from specimens originating from India and Pakistan, the lectotype of G. gangeticus, the holotype of G. siamensis, and four specimens described as G. fowlerae, including the holotype. The sequence of the holotype of G. siamensis is nested within the G. gangeticus samples, including the lectotype. Although the four sequences of G. fowlerae from Borneo and Java form a monophyletic sister group to the clade containing the other seven sequences of G. gangeticus and G. siamensis, all of these sequences are very closely related with divergences that range from 0.01% to 0.65%, implying that G. siamensis and G. fowlerae are likely conspecific with G. gangeticus (Fig. 2). This conclusion is further supported by concatenated phylogenetic analysis and a coalescence based SNAPP analysis of SNP loci derived from nuclear data obtained from three G. fowlerae specimens and a single G. gangeticus specimen (SI Appendix, Figs. S3 and S4). However, a SNAPP-BFD* species delimitation analysis of the same nuclear data yielded results that suggest G. gangeticus is distinct from G. fowlerae. That an approach explicitly designed to address species boundaries would yield a conclusion that conflicts with inferences drawn from other types of analysis warrants some scrutiny. Most of the recently developed species delimitation approaches are based on the multispecies coalescent framework (20). The SNAPP-BFD* approach used herein, explicitly bypasses Markov chain Monte Carlo (MCMC) integration over gene trees in an effort to provide a computationally tractable estimate. It is important to recognize that all species delimitation methods, whether explicitly model-based or otherwise, emphasize different aspects of the biological processes that underlie lineage differentiation (21, 22). All make simplifying assumptions that can lead to different conclusions depending on the sampling context and the dynamics of the biological system being examined. Coalescent-based species delimitation approaches are particularly prone to inaccuracies when lineage sampling is sparse (22, 23). In the current study, the rarity of the study organisms and the poor condition of the material that we were able to collect precluded the collection of nuclear data for an extensive sample of lineages (all species are listed as Endangered or Critically Endangered and are thought to have been extirpated from most of their original ranges). Thus, we have based our decision to consider G. fowlerae, G. gangeticus, and G. siamensis as conspecific on multiple independent analyses, including the generally low genetic divergence observed among individuals of each of the three nominal species and the paraphyletic relationships among individuals inferred from mitochondrial (Fig. 2), concatenated nuclear (SI Appendix, Fig. S3), and SNAPP (SI Appendix, Fig. S4) analyses. Although a more robust species delimitation approach would certainly be desirable if more data were available, we doubt that more samples would change the conclusion. For example, the mitochondrial genome of G. siamensis was found to be 99.9% similar to G. gangeticus. This tight clustering within G. gangeticus persists even when multiple representative mitochondrial genomes of G. gangeticus are used in the analysis (Fig. 2).
The network analysis of the mitochondrial data (SI Appendix, Fig. S1) reveals that several hundred steps are required to link haplotypes among nominal species, except for comparisons among G. gangeticus, G. siamensis, G. sp. Pakistan, and G. fowlerae. Although the G. fowlerae specimens are somewhat divergent within this group (separated by 74 substitutions), their divergence is about an order of magnitude lower than the observed divergence between all other nominal species (SI Appendix, Fig. S1) and is considered more in keeping with population variation than it is with being a distinct species. The frequency distribution of pairwise genetic distances clearly shows that the interspecific distances observed within this clade are of a similar magnitude to those representing intraspecific differences for all other nominal species (SI Appendix, Fig. S2). These results imply that G. siamensis was incorrectly described as a new species 117 y ago and that G. fowlerae was incorrectly described more recently.
Given the implicit conflict between the molecular data presented herein that suggest that G. siamensis, G. fowlerae, and G. gangeticus are conspecific, and the morphological data that were used to diagnose the three species as distinct in the original species descriptions, a reexamination of the morphological data is warranted. Compagno et al. (3) distinguishes G. fowlerae from G. gangeticus based on a combination of meristics and morphology. However, although 14 types of G. fowlerae [505- to 778-mm total length (TL)] were examined, comparative material was restricted to only two similar-sized specimens of G. gangeticus (556- and 610-mm TL). The paralectotype of G. gangeticus (MNHN 1141), holotype of G. siamensis (NMW 61379), and several accessible G. fowlerae specimens were remeasured by one of us (W.T.W.) in 2013. This reassessment rendered ambiguous some of the characters that were used by Compagno et al. (3) to separate G. fowlerae from G. gangeticus. These include prenarial length (3.3–5.3% vs. 3.6–5.3% TL), interorbital space (11.5–12.6% vs. 10.9–11.5% TL), head width (11.2–14.2% vs. 11.5–12.6% TL), caudal peduncle width (3.1–4.7% vs. 2.3–4.1% TL), pectoral-fin posterior margin (12.8–17.2% vs. 17.2% TL), and lower postventral caudal margin (4.4–5.5% vs. 3.3–5.9% TL). Upon reexamination, the only characters that were nonoverlapping between the two species were preoral length (7.5–8.3% vs. 6.6–7.4% TL), preorbital length (8.0–10.5% vs. 7.2–7.7% TL), trunk width (11.0–15.3% vs. 9.4–9.5% TL), and pelvic midpoint to second dorsal-fin origin (PDO) (5.8–7.7% vs. 9.4% TL). Limited weight can be placed on the preoral length recorded for G. gangeticus as the two specimens that were measured have been preserved for an extensive period of time and have bent and partially shriveled snouts. Both trunk width and PDO are known to vary greatly in many carcharhinid species and are thus not generally considered to be reliable characters for distinguishing among species. The G. siamensis holotype was also remeasured. There was also ambiguity among comparisons between G. fowlerae and G. siamensis, including prenarial length (4.8–5.3% vs. 4.7% TL), preorbital length (8.3–10.5% vs. 8.3% TL), nostril width (1.9–2.3% vs. 1.8% TL), and pelvic-fin length (8.8–10.3% vs. 8.5% TL). In fact, the second dorsal-fin base length (7.8–9.3% vs. 6.5% TL) is the only character that unambiguously distinguished G. fowlerae and G. siamensis. There is therefore very limited morphological evidence to support the separation of these three species.
Compagno et al. (3) reported slightly higher tooth counts in G. fowlerae (60–63) than G. gangeticus (53–58) and G. siamensis (58). However, counts from an additional 13 jaws of G. gangeticus provided total counts of 57–63, overlapping the distribution of tooth counts reported for G. fowlerae. Examination of the dentition of the additional G. gangeticus jaws and one set of G. fowlerae jaws from Indonesia revealed no differences. Thus, neither tooth counts nor dentition can be used to separate G. fowlerae, G. gangeticus, and G. siamensis.
Vertebral counts are considered an important character for separating Glyphis species. Compagno et al. (3) reported that G. gangeticus has fewer vertebrae (169 total centra) than G. fowlerae (196–209) and G. siamensis (209), making this the only good character distinguishing G. gangeticus from G. fowlerae and G. siamensis. It should be noted, however, that counts are only available for a single specimen of G. gangeticus. It is possible that this specimen represents the lower end of the range in vertebral counts for this species. It is also possible that the observed variation in vertebral counts across G. fowlerae, G. gangeticus and G. siamensis represents population differences in vertebral counts in a single wide-ranging species. A vertebral range of 169–209 is not unrealistic for a carcharhinid shark, as some species are known to have relatively large ranges in vertebral numbers, e.g., Loxodon macrorhinus (148–191 total centra) (24) and Scoliodon macrorhynchos (149–171) (25). Further specimens are required to validate this. Nevertheless, the number of vertebrae alone is not sufficient to separate these species, particularly given the low sample sizes that were examined. Given the morphological and meristic ambiguity discussed above and given that our molecular results show interspecific variation within this group that is similar in magnitude to that representing intraspecific variation in G. glyphis and G. garricki, herein we consider G. siamensis and G. fowlerae to be junior synonyms of G. gangeticus.
The molecular data suggest recent gene flow between the G. siamensis populations in Myanmar and the G. gangeticus populations in India and Pakistan and, more impressively, between the G. gangeticus populations in India and Pakistan and the G. fowlerae populations on Borneo and Java. These locations are currently separated by several thousand kilometers of ocean, which indicates marine dispersal in this group of sharks, which has been presumed to be restricted to freshwater.
The clade derived from 15 Australian mitochondrial genome sequences obtained from specimens nominally assigned to G. glyphis, including the one obtained from the holotype, forms a deeply divergent sister clade to the clade containing five sequences obtained from G. garricki, confirming that Australia is indeed inhabited by two river shark species (4). The last clade consists of four sequences, two from Bangladesh and two from Mukah in Malaysian Borneo. This distinct clade is basal to all other Glyphis species, both in the mitochondrial and nuclear DNA tree (Fig. 2 and SI Appendix, Fig. S3) and almost certainly represents a unique undescribed species. A formal taxonomic treatment of this clade is required to settle its status. Interestingly, rather than being reciprocally monophyletic with respect to location, the Bangladesh specimens in this clade are paraphyletic with regard to the Borneo specimens. Although based on only four specimens, this result suggests marine dispersal in this lineage of river sharks as well—in this case between Bangladesh and Borneo.
That the relationships among the four major clades inferred from the mitochondrial data (Fig. 2) are identical to those derived from the nuclear data (SI Appendix, Fig. S3) strongly suggests that the tree topology presented is the true species tree rather than an inference that is due to incomplete lineage sorting or mitochondrial introgression. The tree topology recovered with both mitochondrial and nuclear DNA suggests that gene flow has occurred between locations that are currently separated by large stretches of marine environment: two recent events in the central Indo-Pacific and a third, older event that resulted in the colonization of Australian waters. Recent fisheries and fisheries-independent surveys of G. glyphis and G. garricki in Australia have provided the first insights (to our knowledge) into the biology, movement, and habitat utilization of Glyphis species. Although rarely encountered, the available data indicate a broad salinity tolerance with animals observed in freshwater, estuarine, and marine environments. In fact, mature individuals have only been encountered in marine environments (26, 27). Further support for regular marine dispersal of Glyphis sharks is provided by a recent phylogeographic study on the speartooth shark G. glyphis, which recovered identical mitochondrial genome sequences in both the Alligator and Adelaide river systems in Australia, and more impressively in both the Wenlock and Alligator river systems, requiring, in the latter case, more than 1,000-km dispersal across marine habitat. These findings are in stark contrast with the description of the genus Glyphis as “river sharks.” Given these euryhaline characteristics and the data presented herein, it seems that the life cycle of Glyphis sharks includes periods of marine dispersal in conjunction with some dependence on rivers and estuaries. In contrast to teleost fishes, 40% of which live in freshwater, only very few (about 5%) of elasmobranch species are able to survive in freshwater (28). Most of these belong to the potamotrygonid stingray family, which is fully adapted to freshwater and no longer capable of surviving in saltwater, as well as a few additional species of rays, some of which are capable of surviving in both freshwater and saltwater (29). Among sharks, only the members of the genus Glyphis and the bull shark (Carcharhinus leucas) are capable of transitioning between saltwater and freshwater environments (30). Although it was assumed that Glyphis represent true freshwater species, our results contribute to a growing body of evidence that suggests that the ecology of the genus Glyphis might be similar to that of the bull shark. Adult bull sharks live in marine environments but use freshwater habitats, where juveniles are frequently encountered, for reproduction (31). It has been speculated that high predation pressure on juveniles in marine environments is promoting the use of freshwater nursery areas in bull sharks (31). It is possible that the same selective pressure is behind the adaptation to euryhaline conditions in the ancestor of the genus Glyphis.
These results underscore how little is known about the biodiversity and ecology of sharks. Our efforts to resolve the phylogenetic relationships among the different lineages of river sharks unexpectedly revealed new information about their taxonomy, evolution, and ecology. We have uncovered a case of taxonomic misdescription that has persisted for more than a century and a previously undocumented lineage that is deeply divergent from all other described river shark lineages and that is likely a species new to science. Our results also uncover a complex evolutionary history for river sharks that encompasses both ancient and recent gene flow across large geographic distances, most likely due to marine dispersal events. Finally, they show that DNA hybridization capture approaches can reliably be used to obtain DNA sequence data from museum specimens even when traditional PCR-based approaches fail because of poor DNA preservation. The approach has potential to yield information that will be useful to future conservation efforts, particularly for critically endangered groups that are represented by museum material but for which specimens are hard to collect in the field.
Materials and Methods
Samples.
Glyphis glyphis (ZMB 5265) and G. gangeticus (ZMB 4474) are the holotype and lectotype specimens, respectively, stored at the Museum für Naturkunde (Berlin, Germany). Glyphis siamensis (NMW 61379) is the holotype specimen, housed at the Naturhistorisches Museum (Vienna, Austria). DNA extraction for the museum samples was performed on dry tissue following an ancient DNA extraction protocol (32, 33). The modern samples, including those derived from the holotypes of G. garricki [Australian National Fish Collection, Commonwealth Scientific and Industrial Research Organisation (CSIRO) Marine and Atmospheric Research H 5262-01] and G. fowlerae (Borneo Marine Research Institute IPMB 38.14.02), were extracted using the E.Z.N.A. Tissue DNA Kit (Omega Bio-Tek) as per the manufacturer’s instructions. Further details regarding the museum and modern samples used in this study, including those derived from the type specimens, are listed in SI Appendix, Table S1. Extracted DNA samples were quantified using a Qubit 2.0 Fluorometer (Life Technologies Corporation). The quality of DNA samples was checked by amplification of one short (131 bp) and one longer (1,048 bp) fragment of the mitochondrial genome. The primers for these amplifications are listed in SI Appendix, Table S4.
Bait Design.
Biotinylated DNA baits were made for capturing the mitochondrial genome of each sample. The entire mitochondrial sequence of a G. garricki sample (GN6502) was amplified using long-range PCR and the primers listed in SI Appendix, Table S4. The amplified products were mixed in equimolar ratios and then sheared to ∼200 bp on a Covaris M220 Focused-ultrasonicator (Covaris). Then, two adapters were added to the sheared product to make a bait template library. Finally, the library was amplified with biotinylated M13 primers and dNTP/dUTP mix to incorporate a biotin label and UTPs into the amplicons to create biotinylated probes for use as baits. For the detailed procedure for making homemade baits, including reaction constituents and PCR cycling conditions, see SI Appendix, Detailed Protocol for Preparing Homemade Baits.
Target Capture and Sequencing.
We prepared the target libraries following the methods of Li et al. (13). For the subsequent target capture, we used “homemade baits,” see SI Appendix, Detailed Protocol for Target Capture. The final captured and indexed libraries were quantified using quantitative PCR on a CFX Connect Real-Time PCR Detection System (Bio-Rad). The libraries were subsequently pooled in equimolar ratios for 100-bp paired-end sequencing using an Illumina MiSeq Benchtop Sequencer (Illumina). The mitochondrial sequence of one G. fowlerae (GN1363) specimen was also determined using PCR with overlapping primers and Sanger sequencing following the method of Aschliman et al. (34) to validate the accuracy of sequence collected by our target capture method. To compare with the mitochondrial data, we also collected sequences of nuclear protein-coding genes for representatives of the major Glyphis lineages following the targeted nuclear gene capture protocol of Li et al. (13).
Reads Assembly.
Sequence reads were sorted into bins based on the 8-bp index that was incorporated with each sample during PCR cycling. Adapter sequences and low-quality sites were trimmed from the reads using Cutadapt-1.1 (35) contained within the wrapper tool “trim_galore_v0” (www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Trimmed reads were assembled de novo using ABySS (36). Assembled contigs were aligned to the reference sequence (GN1363) to obtain the full-length mitochondrial genome sequence. Reads were mapped to the obtained genome sequences using BWA (37). Duplicated reads due to PCR were identified and flagged using Picard (picard.sourceforge.net/). The average read coverage of the museum samples was calculated after PCR duplicates were collapsed.
Phylogenetic Analysis and Network—Mitochondrial Genomes.
DNA sequences of the 13 protein-coding genes of the mitochondrial genomes of all taxa were aligned. The complementary strand sequences were used for ND6, which is encoded on the L-strand. Incomplete stop codons of genes were excluded from the alignment. The final alignment was 11,424 bp in length, including 1,571 parsimony informative sites. Maximum-likelihood analysis was conducted using RAxML, version 7.2.8 (38). The dataset was partitioned by codon position, and the GTR + Γ (39) model was used for each partition. A total of 1,000 distinct runs was performed based on 1,000 random starting trees using the default algorithm of the program. The tree with the best likelihood score was chosen as the final tree. Maximum-likelihood bootstrap analysis (40) was also conducted using RAxML, version 7.2.8 (38). The same partitioning strategy and evolutionary models were used as in the above analyses. The number of nonparametric bootstrap replications was set to 1,000. The resulting trees were imported into PAUP*4.0.b10 (41) to obtain the 50% majority rule consensus tree.
To help visualize intraspecific versus interspecific divergence, the genealogical relationships among complete mitochondrial genome haplotypes were reconstructed using a network analysis based on the statistical parsimony method of Templeton et al. (42) implemented in TCS, version 1.21 (43). Pairwise p-distance, the proportion of sites that differ among unique haplotype sequences, was calculated in MEGA5 (44). All ambiguous positions were removed for each sequence pair. There were a total of 16,714 positions and 34 sequences in the final dataset.
Phylogenetic Analysis—Nuclear Exons.
After removing patchy data, sequences were aligned for 100 nuclear protein-coding loci. The concatenated sequence was 63,492 bp in length, of which 314 sites were parsimony informative. The dataset was partitioned by codon position and analyzed using the GTR + Γ (39) nucleotide substitution model in RAxML, version 7.2.8, with 1,000 bootstrap replicates.
Read Mapping and Nuclear SNP Calling.
Consensus sequences of the best-assembled individuals were obtained for 1,041 nuclear protein-coding loci using a custom Perl script. The trimmed reads of each sample were then mapped to the consensus sequence using BWA-0.7.12 (37). The PCR duplicates in the reads were marked using Picard-1.118 (available from the website picard.sourceforge.net). Base quality score recalibration, local realignment, SNP discovery, and genotyping were performed across all samples simultaneously using standard hard filtering parameters available in GATK-3.2.2 (45). GATK recommendations for best practice were followed (46, 47).
Species Tree Analysis and Bayes Factor Species Delimitation.
A custom Perl script was used to convert the SNP vcf file obtained from GATK to Nexus format for use as input into species tree analyses to be performed using SNAPP (12) available in BEAST 2 (48). Because the SNAPP analyses assume linkage equilibrium among loci, the best SNP site (highest SNP calling score, fewer missing data) was chosen for each target region for subsequent analysis.
SNAPP analyses of the 1,041 SNPs were set up in BEAUTi, and each sample was assigned as a separate taxon. The SNAPP runs were carried out with a chain length of 10 million. The convergence of the MCMC was inspected in Tracer, version 1.6, and the maximum clade credibility tree was calculated using TreeAnnotator, version 2.3.0. BFD* uses a modified version of SNAPP, which is implemented as a plug-in to BEAST 2 (48). Program installation, XML file preparation, and analyses were implemented as detailed on the wiki page for BFD* (beast2.org/bfd/; last accessed June 11, 2015), and as explained in “SNAPP handling missing data and path sampling made easier” (beast2.org/2014/07/21/snapp-handling-missing-data-and-path-sampling-made-easier/; last accessed June 11, 2015). Path sampling with eight steps (100,000 MCMC steps, 10,000 preburnin steps) was conducted to estimate the marginal likelihood for two models: one in which G. fowlerae and G. gangeticus were assumed to be separate species, and another in which they were considered conspecific.
Supplementary Material
Acknowledgments
We thank Gordon Hubbell, Mabel Manjaji, Scott Mycock, Rachel Cavanaugh, Sarah Fowler, Iain Field, Bernard Seret, Janine Caira, Kirsten Jensen, Ernst Mikschi, and Helmut Wellendorf for tissue samples. This research was funded by National Science Foundation Division of Environmental Biology Award 1132229 (to G.J.P.N.). C.L. was supported by “Innovation Program of Shanghai Municipal Education Commission,” “Shanghai Pujiang Program,” and “The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The mitochondrial sequences reported in this paper have been deposited in the GenBank database (accession nos. KT698039–KT698063). The nuclear dataset reported in this paper has been deposited in the TreeBASE database (study ID S18221).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508735112/-/DCSupplemental.
References
- 1.Müller J, Henle FGJ. Systematische Beschreibung der Plagiostomen. Veit; Berlin: 1839. pp. 29–102. [Google Scholar]
- 2. Steindachner F (1896) Bericht über die während der Reise Sr. Maj. Schiff “Aurora” von Dr. C. Ritter v. Microszewski in den Jahren 1895 und 1896, gesammelten Fische. Annalen des Naturhistorischen Museums in Wien 11:197–230.
- 3.Compagno LJV, White WT, Cavanagh RD. 2010. Glyphis fowlerae sp. nov., a new species of river shark (Carcharhiniformes; Carcharhinidae) from northeastern Borneo. Descriptions of New Sharks and Rays from Borneo, eds Last PR, White WT, Pogonoski JJ (CSIRO Marine and Atmospheric Research, Hobart, TAS, Australia), CSIRO Marine and Atmospheric Research Paper 032, pp 29–44.
- 4.Compagno LJV, White WT, Last PR. 2008. Glyphis garricki sp. nov., a new species of river shark (Carcharhiniformes: Carcharhinidae) from northern Australia and Papua New Guinea, with a redescription of Glyphis glyphis (Müller and Henle, 1839). Descriptions of New Australian Chondrichthyans, eds Last PR, White WT, Pogonoski JJ (CSIRO Marine and Atmospheric Research, Hobart, TAS, Australia), CSIRO Marine and Atmospheric Research Paper 022, pp 203–225.
- 5.Naylor GJP, et al. A DNA sequence–based approach to the identification of shark and ray species and its implications for global elasmobranch diversity and parasitology. Bull Am Mus Nat Hist. 2012;367:1–262. [Google Scholar]
- 6.Wandeler P, Hoeck PE, Keller LF. Back to the future: Museum specimens in population genetics. Trends Ecol Evol. 2007;22(12):634–642. doi: 10.1016/j.tree.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39(12):1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 8.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 2013;110(39):15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer M, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2014;505(7483):403–406. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 10.Burrell AS, Disotell TR, Bergey CM. The use of museum specimens with high-throughput DNA sequencers. J Hum Evol. 2015;79:35–44. doi: 10.1016/j.jhevol.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One. 2010;5(11):e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feutry P, et al. Mitogenomics of the Speartooth Shark challenges ten years of control region sequencing. BMC Evol Biol. 2014;14:232. doi: 10.1186/s12862-014-0232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Hofreiter M, Straube N, Corrigan S, Naylor GJ. Capturing protein-coding genes across highly divergent species. Biotechniques. 2013;54(6):321–326. doi: 10.2144/000114039. [DOI] [PubMed] [Google Scholar]
- 14.Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. Inferring species trees directly from biallelic genetic markers: Bypassing gene trees in a full coalescent analysis. Mol Biol Evol. 2012;29(8):1917–1932. doi: 10.1093/molbev/mss086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leaché AD, Fujita MK, Minin VN, Bouckaert RR. Species delimitation using genome-wide SNP data. Syst Biol. 2014;63(4):534–542. doi: 10.1093/sysbio/syu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulvy NK, et al. Extinction risk and conservation of the world’s sharks and rays. eLife. 2014;3:e00590. doi: 10.7554/eLife.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machida Y. A new deep-sea ophidiid fish, Bassozetus levistomatus, from the Izu-Bonin Trench, Japan. Jpn J Ichthyol. 1989;36(2):187–189. [Google Scholar]
- 18.Raj U, Seeto J. A new species of the Anthiine fish genus Plectranthias (Serranidae) from the Fiji Islands. Jpn J Ichthyol. 1983;30(1):15–17. [Google Scholar]
- 19.Grace MA, Doosey MH, Bart HL, Naylor GJP. First record of Mollisquama sp. (Chondrichthyes: Squaliformes: Dalatiidae) from the Gulf of Mexico, with a morphological comparison to the holotype description of Mollisquama parini Dolganov. Zootaxa. 2015;3948(3):587–600. doi: 10.11646/zootaxa.3948.3.10. [DOI] [PubMed] [Google Scholar]
- 20.Rannala B, Yang Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164(4):1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56(6):879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 22.Carstens BC, Pelletier TA, Reid NM, Satler JD. How to fail at species delimitation. Mol Ecol. 2013;22(17):4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- 23.Jones G, Aydin Z, Oxelman B. DISSECT: An assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics. 2015;31(7):991–998. doi: 10.1093/bioinformatics/btu770. [DOI] [PubMed] [Google Scholar]
- 24.Springer V. A revision of the carcharhinid shark genera Scoliodon, Loxodon, and Rhizoprionodon. Proc US Nat Mus. 1964;115:559–632. [Google Scholar]
- 25.White W, Last P, Naylor G. 2010. Scoliodon macrorhynchos (Bleeker, 1852), a second species of spadenose shark from the Western Pacific (Carcharhiniformes: Carcharhinidae). Descriptions of New Sharks and Rays from Borneo, eds Last P, White W, Pogonoski J (CSIRO Marine and Atmospheric Research, Hobart, TAS, Australia), CSIRO Marine and Atmospheric Research Paper 032, pp 61–76.
- 26.Pillans RJ, Stevens JD, Kyne P, Salini J. Observations on the distribution, biology, short-term movements and habitat requirements of river sharks Glyphis spp. in northern Australia. Endanger Species Res. 2010;10:321–332. [Google Scholar]
- 27.Field IC, et al. Distribution, relative abundance and risks from fisheries to threatened Glyphis sharks and sawfishes in northern Australia. Endanger Species Res. 2013;21:171–180. [Google Scholar]
- 28.Ballantyne JS, Robinson JW. Freshwater elasmobranchs: A review of their physiology and biochemistry. J Comp Physiol B. 2010;180(4):475–493. doi: 10.1007/s00360-010-0447-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Yamaguchi A, Zhou Q, Zhang C. Rare occurrences of Dasyatis bennettii (Chondrichthyes: Dasyatidae) in freshwaters of Southern China. J Appl Ichthyology. 2010;26(6):939–941. [Google Scholar]
- 30.Thorson TB. Movement of bull sharks, Carcharhinus leucas, between Caribbean Sea and Lake Nicaragua demonstrated by tagging. Copeia. 1971;1971(2):336–338. [Google Scholar]
- 31.Pillans RD, Good JP, Anderson WG, Hazon N, Franklin CE. Freshwater to seawater acclimation of juvenile bull sharks (Carcharhinus leucas): Plasma osmolytes and Na+/K+-ATPase activity in gill, rectal gland, kidney and intestine. J Comp Physiol B. 2005;175(1):37–44. doi: 10.1007/s00360-004-0460-2. [DOI] [PubMed] [Google Scholar]
- 32.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2(7):1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 33.Rohland N, Siedel H, Hofreiter M. A rapid column-based ancient DNA extraction method for increased sample throughput. Mol Ecol Resour. 2010;10(4):677–683. doi: 10.1111/j.1755-0998.2009.02824.x. [DOI] [PubMed] [Google Scholar]
- 34.Aschliman NC, et al. Body plan convergence in the evolution of skates and rays (Chondrichthyes: Batoidea) Mol Phylogenet Evol. 2012;63(1):28–42. doi: 10.1016/j.ympev.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 36.Simpson JT, et al. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21(4):456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 39.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 40.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 41.Swofford D. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer Associates, Sunderland, MA), 4.0b10.
- 42.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132(2):619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;11(1110):11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.