Significance
Heart failure occurs when the heart cannot pump enough blood to meet the body’s demands. The heart most often weakens through the loss of muscle cells, called cardiomyocytes. Heart failure is a devastating disease, and one possible cure is to replace the lost cardiomyocytes through regeneration. Unlike humans, zebrafish can efficiently regenerate their hearts after injury. Interestingly, zebrafish heart cells are similar to human heart cells at the molecular level. Understanding how zebrafish can regenerate cardiac tissue can help identify regenerative therapies for humans. Here, we find that NF-κB signaling is a link between the injury response and the regenerative program in zebrafish.
Keywords: heart regeneration, cardiomyocyte, zebrafish, NF-κB, epicardium
Abstract
Heart regeneration offers a novel therapeutic strategy for heart failure. Unlike mammals, lower vertebrates such as zebrafish mount a strong regenerative response following cardiac injury. Heart regeneration in zebrafish occurs by cardiomyocyte proliferation and reactivation of a cardiac developmental program, as evidenced by induction of gata4 regulatory sequences in regenerating cardiomyocytes. Although many of the cellular determinants of heart regeneration have been elucidated, how injury triggers a regenerative program through dedifferentiation and epicardial activation is a critical outstanding question. Here, we show that NF-κB signaling is induced in cardiomyocytes following injury. Myocardial inhibition of NF-κB activity blocks heart regeneration with pleiotropic effects, decreasing both cardiomyocyte proliferation and epicardial responses. Activation of gata4 regulatory sequences is also prevented by NF-κB signaling antagonism, suggesting an underlying defect in cardiomyocyte dedifferentiation. Our results implicate NF-κB signaling as a key node between cardiac injury and tissue regeneration.
Heart failure is an epidemic, with 870,000 new cases diagnosed annually in the United States (1). Failing hearts are characterized by progressive myocyte loss and replacement fibrosis, ultimately leading to susceptibility to fatal arrhythmia and pump dysfunction. Although major strides have been made in the treatment of heart failure, mortality remains high and additional therapies are needed. Recently, low-grade cardiomyocyte turnover has been reported in the mammalian heart (2, 3). Therapeutically augmenting this turnover toward regeneration of lost cardiac tissue is an attractive strategy for the treatment of heart failure. However, a better understanding of the mechanisms for heart regeneration is needed before regenerative therapies can be developed for patients.
In contrast to humans, zebrafish are capable of impressive regeneration following cardiac injury (4). Heart regeneration in zebrafish proceeds by reexpression of developmental factors along with cardiomyocyte proliferation and cytokinesis. Notably, gata4 regulatory sequences are induced at the site of injury, and it is these cardiomyocytes that primarily contribute to the regenerate (5). The induction of a developmental program is seemingly required for regeneration, because functional inhibition of Gata4 impairs both cardiomyocyte proliferation and heart regeneration (6). Although most attention has been paid recently to cardiomyocytes, heart regeneration is a complex process that also involves the activation of embryonic programs in epicardial and endocardial cells (7–9). Remarkably, mechanisms for heart regeneration appear to be conserved across species; thus, studies in zebrafish have the potential to inform strategies for mammalian heart regeneration (10).
Regeneration is broadly defined as the replacement of a lost or damaged body part following injury. As such, tissue growth during regeneration is inextricably linked to the injury response. Earlier studies have identified response pathways such as retinoic acid signaling, JAK/Stat3 signaling, H2O2 signaling, and HIF1 signaling that are required for heart regeneration (7, 11–13). More recently, the inflammatory response itself has been implicated in guiding regeneration. Inflammatory responses are sufficient to stimulate neurogenesis through a regenerative program in the adult zebrafish brain (14). Conversely, loss of macrophages impairs appendage regeneration in zebrafish and salamanders, and ablation of macrophages disrupts heart regeneration in neonatal mice (15–19). However, molecular links between the injury response and the induction of a developmental program in regenerating tissues await discovery.
NF-κB factors were first identified nearly 30 years ago as a family of transcription factors capable of binding κ light chain enhancers in lymphocytes (20). Since then, NF-κB signaling has been shown to have broad effects in a variety of tissues, influencing cell survival, tissue growth and proliferation, and chromatin structure (21). In the heart, NF-κB signaling has been implicated as a hypertrophic influence and has been linked to expression of cardiac response genes like ANF and β-MHC (22–24). Classically, NF-κB factors are sequestered in the cytoplasm through interaction with IκB. Following stimulation, IκB is targeted for proteasomal degradation, and NF-κB factors are released for activation. Not surprisingly, NF-κB signaling is the prototype inducible transcription factor and an important component of the early injury response in a variety of tissues (21). Here, we show that NF-κB activity is induced following cardiac injury and required for heart regeneration. Importantly, NF-κB has broad effects on the regenerative response, with evident contributions to cardiomyocyte dedifferentiation, cardiomyocyte proliferation, and epicardial injury responses. These studies offer a link between the injury response and the induction of a regenerative program.
Results
NF-κB Activity Is Induced During Zebrafish Heart Regeneration.
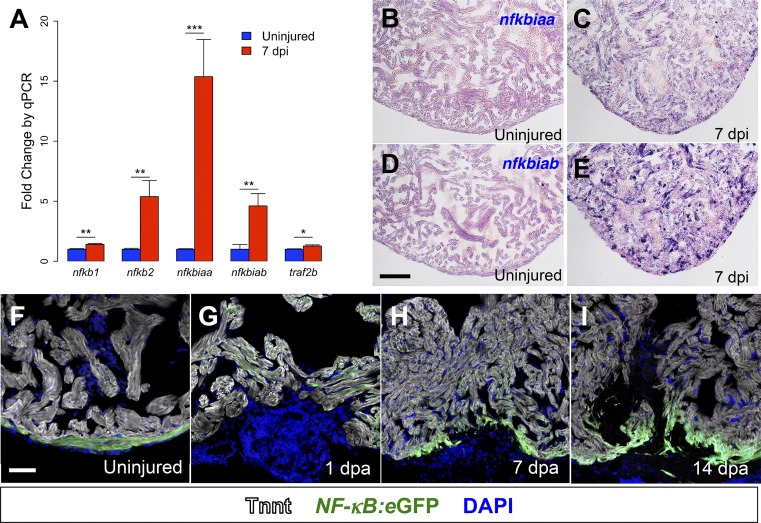
We previously established a model for genetic cardiomyocyte ablation that results in widespread regenerative responses, inducing proliferation in a high proportion of spared cardiomyocytes (25). To identify pathways related to regeneration, we used RNA sequencing to compare transcriptional profiles of ventricles at 7 days after genetic cardiomyocyte ablation (dpi) to profiles of uninjured ventricles. We found large changes in global gene expression during regeneration, with enrichment for targets of NF-κB signaling. To confirm increased NF-κB activity during regeneration, we performed quantitative PCR (qPCR) for NF-κB target genes by using RNA from regenerating ventricles at 7 dpi and uninjured ventricles. Regenerating hearts were significantly enriched for the NF-κB targets genes nfkb1, nfkb2, nfkbiaa, nfkbiab, and traf2b, further suggesting increased NF-κB activity during regeneration (Fig. 1A). We confirmed induction of increased NF-κB activity following injury by in situ hybridization for the NF-κB target genes nfkbiaa and nfkbiab in regenerating hearts (Fig. 1 B–E). As in mammals, zebrafish have five members in the family of NF-κB transcription factors. To identify predominant NF-κB transcription factors during regeneration, we performed in situ hybridization for the annotated NF-κB members on sections from regenerating hearts following cardiomyocyte ablation (Fig. S1). We found NF-κB1 to be induced in the compact muscle of regenerating zebrafish hearts. As additional evidence for the presence of NF-κB1 in cardiomyocytes, we assayed for the presence NF-κB1 in sections of mouse hearts. We found the murine NF-κB1 colocalizes with cardiomyocyte nuclei, further supportive of NF-κB1 activity in cardiomyocytes (Fig. S1). Of note, NFKB1 polymorphisms have been linked to human cardiomyopathy, and Nfkb1−/− mice develop more fulminant heart failure following coronary ligation compared with wild-type controls (26–28). To more clearly identify which cell types displayed increased NF-κB activity during zebrafish heart regeneration, we used a recently described transgenic reporter strain that reports NF-κB activity with eGFP fluorescence (29). In the uninjured heart, we found eGFP fluorescence throughout the compact muscle, indicative of increased NF-κB activity. As early as 1 d after resection injury (dpa), NF-κB activity could be detected by eGFP fluorescence adjacent to the injury, and this activity increased during regeneration (Fig. 1 F–I). Colocalization studies with cardiac troponin revealed strong NF-κB activity, most notably in regenerating muscle.
Fig. 1.
NF-κB activity is induced by injury and during regeneration. (A) qPCR for NF-κB target genes in regenerating ventricles at 7 dpi in cmlc2:CreER; βact2:RS-DTA animals treated with tamoxifen (n = 6) compared with ventricles from cmlc2:CreER; βact2:RS-DTA animals treated with vehicle (n = 6). Error bars indicate SE. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test, two-tailed. Each replicate consists of a pool of 2–3 ventricles. (B–E) In situ hybridization for nfkbiaa or nfkbiab on sections from ventricles of cmlc2:CreER; βact2:RS-DTA animals treated with vehicle (B and D) or tamoxifen (C and E). Violet staining indicates expression. (F–I) Time course of NF-κB:eGFP induction during regeneration in sections from an uninjured ventricle, a 1 dpa ventricle, a 7 dpa ventricle, and a 14 dpa ventricle. Ventricles are counterstained with an antibody against Tnnt (gray) to delineate cardiomyocytes. (Scale bars: 100 μm.)
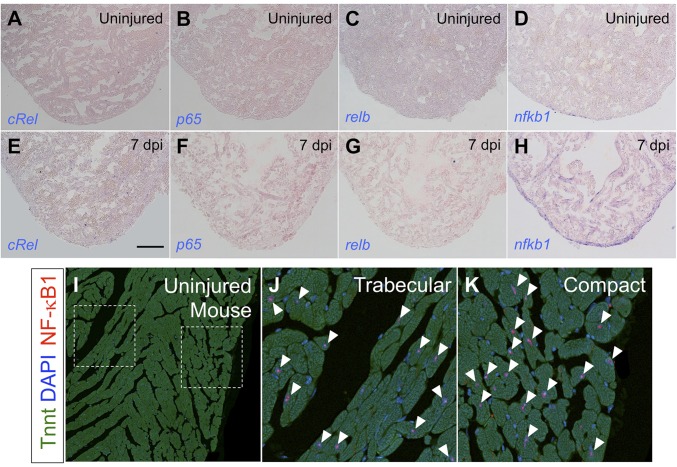
Fig. S1.
NF-κB factors during heart regeneration. (A–H) In situ hybridization for NF-κB transcription factors on sections from ventricles of cmlc2:CreER; βact2:RS-DTA animals treated with vehicle (A–D) or tamoxifen (E–H) to diffusely ablate cardiomyocytes. nfkb1 was the only gene from this group to show a detectable signal in cardiomyocytes. (I–K) Staining of uninjured adult mouse hearts for NF-κB1 and Tnnt, indicating that NF-κB1 is present in murine cardiomyocytes (arrowheads). We did not detect an obvious difference in NF-κB1+ cardiomyocytes in compact versus trabecular muscle. (Scale bar: 100 μm.)
NF-κB Activity Is Required for Zebrafish Heart Regeneration.
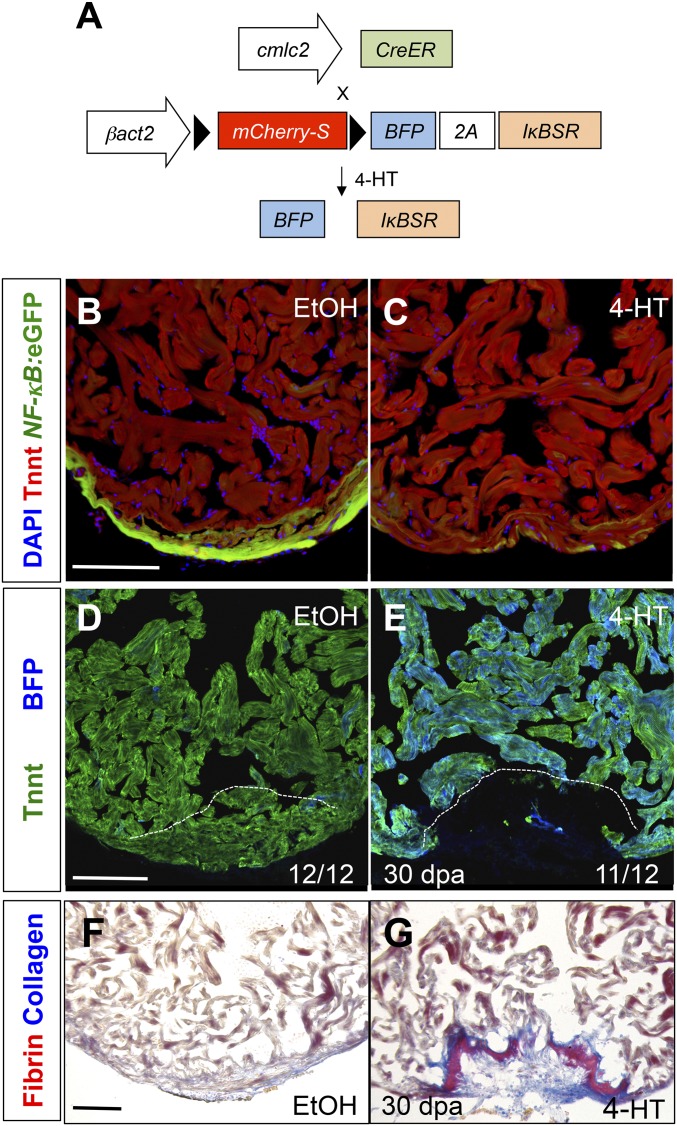
The strong spatiotemporal association of NF-κB activity with regenerating muscle suggests a functional role for NF-κB signaling during heart regeneration. To test this idea, we established a new transgenic zebrafish line for conditional expression of a superrepressor IκB (IκBSR) to inhibit NF-κB signaling [Tg(βactin2:loxP-mCherry-STOP-loxP-mTagBFP-2A-IκBSR)pd114]; hereafter referred to as a βact2:RS-IκBSR) (Fig. 2A). Because IκBSR is resistant to phosphorylation, NF-κB transcription factors remain in the cytoplasm and NF-κB activity is diminished (30). This mutant IκBSR has been well described in mammalian systems and has also been shown to inhibit NF-κB signaling in zebrafish embryos (29, 31). To determine the effect of diminished NF-κB signaling in cardiomyocytes, we crossed βact2:RS-IκBSR with cmlc2:CreER fish (5), enabling Cre-mediated recombination and expression of BFP and IκBSR specifically in cardiomyocytes upon 4-HT treatment. First, we generated cmlc2:CreER ; βact2:RS-IκBSR; NF-κB:eGFP transgenic fish to confirm inhibition of NF-κB activity in cardiomyocytes. As expected, induced transgenic IκBSR suppressed NF-κB reporter activity in adult cardiomyocytes (Fig. 2 B and C). Because we noted marked NF-κB activity in the compact muscle of adult hearts, we assessed whether long-term NF-κB inhibition has adverse effects on cardiac homeostasis. We did not observe any alteration in fish survival or cardiac structure, even at 60 d after recombination (Fig. 2 B and C).
Fig. 2.
NF-κB is required for heart regeneration. (A) Schematic of βact2:RS-IκBSR transgene. (B and C) Section images of cmlc2:CreER; βact2:RS-IκBSR; NF-κB:eGFP hearts 60 d after treatment (dpt) with vehicle (n = 12) or 4-HT (n = 12). (D and E) Section images of cmlc2:CreER; βact2:RS-IκBSR ventricles at 30 d after amputation after treatment with vehicle or 4-HT. Images are stained for troponin to denote cardiac muscle. Dashed line indicates approximate resection plane. (F and G). Section images of cmlc2:CreER; βact2:RS-IκBSR ventricles at 30 d after amputation after treatment with vehicle or 4-HT. Images are stained with acid fuchsin orange G (AFOG) to identify scar (blue) and fibrin (red). (Scale bars: 100 μm.)
To determine the requirement for myocardial NF-κB signaling during heart regeneration, we treated cmlc2:CreER; βact2:RS-IκBSR fish with 4-HT and resected ventricular apices. We noted that animals with impaired NF-κB signaling had defective regeneration at 30 dpa, evidenced by large gaps in muscle (Fig. 2E). By contrast, control animals consistently regenerated a contiguous wall of heart muscle (Fig. 2 D and F). With a scoring system to assess regeneration, our analysis indicated that animals with a block in NF-κB signaling had significant regenerative defects at 30 dpa compared with control animals (P < 0.001, Kruskall–Wallis test, n = 25). Similarly, fibrin/collagen staining revealed scar tissue at the amputation site in IκBSR-expressing animals that was typically not present in control animals (Fig. 2 F and G). These experiments reveal a requirement of NF-κB signaling in zebrafish heart regeneration, but not for homeostatic cardiac maintenance.
NF-κB Activity Is Required for the Cardiomyocyte Regenerative Program.
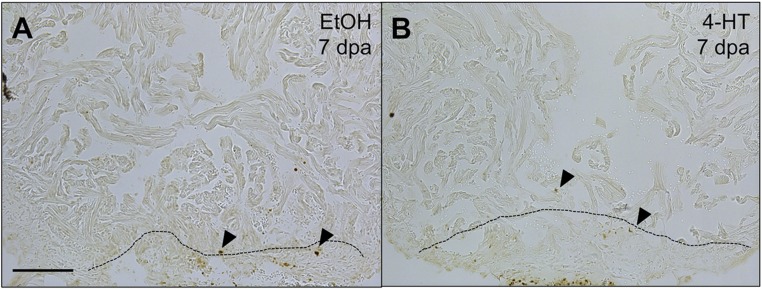
During regeneration, new cardiomyocytes emerge through the division of preexisting cardiomyocytes (5, 32). Based on the localization of NF-κB activity to regenerating cardiomyocytes and the effects on overall muscle regeneration, we tested whether altering NF-κB activity affects cardiomyocyte proliferation. We treated cmlc2:CreER; βact2:RS-IκBSR zebrafish with 4-HT or vehicle and assayed ventricles at 7 dpa, a stage of peak cardiomyocyte proliferation. We observed a nearly 50% decrease in cardiomyocyte proliferation in animals with impaired NF-κB activity (9.1%) versus animals with intact NF-κB (17.3%) (P = 0.005, Mann–Whitney) (Fig. 3 A–C). Because NF-κB is also known to affect cell survival, we also examined the effect of NF-κB inhibition on cardiomyocyte apoptosis following injury. TUNEL staining of hearts with and without intact NF-κB signaling revealed no gross differences in cardiomyocyte apoptosis at 7 dpa (Fig. S2). Together, these data reveal a cell-autonomous role for injury-induced NF-κB activity to promote or enable cardiomyocyte proliferation during heart regeneration.
Fig. 3.
NF-κB modulates cardiomyocyte proliferation and dedifferentiation. (A and B) Section images from cmlc2:CreER; βact2:RS-IκBSR hearts treated with vehicle or 4-HT at 7 dpa. Sections are stained for Mef2 (red) and PCNA (green). Arrowheads indicate Mef2+/PCNA+ nuclei. Dashed line indicates approximate resection plane. (C) Quantification of cardiomyocyte proliferation in cmlc2:CreER; βact2:RS-IκBSR hearts treated with vehicle (n = 15) or 4-HT (n = 17) at 7 dpa. Error bars indicate SEM. *P = 0.004, Mann–Whitney test. (D–I) Sections from NF-κB:eGFP; gata4:DsRed2 hearts at 14 and 30 dpa. (J–Q) Section images from cmlc2:CreER; βact2:RS-IκBSR; gata4:eGFP hearts treated with vehicle (n = 9) or 4-HT (n = 10) at 7 dpa. Inset shows unique high-magnification images for Tnnt, eGFP, and a merge of both channels. (R) Plot for density of putative NFκB1 sites in the gata4 promoter. seqLogo is for the consensus NFκB1 binding site used for analysis. Arrow indicates the region of the promoter with highest density of NFκB1 sites. (S) ChIP-PCR for the region of the gata4 promoter highlighted by arrowhead in R after immunoprecipitation for NFκB1 from cardiac chromatin extracts of cmlc2:CreER; βact2:RS-IκBSR fish treated with vehicle or 4-HT at 7 dpa. (Scale bars: 100 μm.)
Fig. S2.
Cardiomyocyte apoptosis assays during NF-κB inhibition. TUNEL staining on sections of 7 dpa hearts from cmlc2:CreER; βact2:RS-IκBSR fish treated with vehicle (A) or 4-HT (B). No gross differences in TUNEL staining were noted during NF-κB inhibition. Arrowheads indicate TUNEL + nuclei. (Scale bar: 100 μm.)
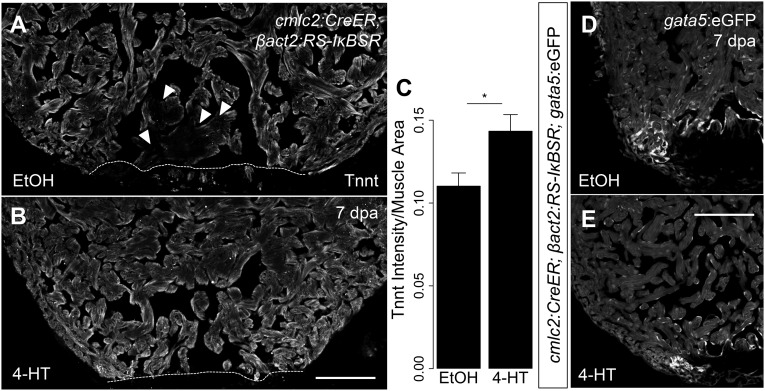
The pattern of NF-κB activation in cardiomyocytes was similar to the induction of GATA binding protein 4 (gata4) regulatory sequences, previously described in compact muscle and cardiomyocytes adjacent to the injury site (5). Moreover, the inhibition of cardiomyocyte proliferation by blockade of NF-κB was similar to the decrease in proliferation that we have observed in transgenic animals expressing a dominant negative version of Gata4 in cardiomyocytes (6). To test for an association of NF-κB with gata4, we crossed NF-κB:eGFP animals with a new gata4:DsRed2 reporter strain that we generated for this study. We noted that gata4 regulatory sequences were activated in cardiomyocytes with NF-κB activity (Fig. 3 D–I), suggesting a functional relationship. Although NF-κB has been linked to a fetal gene program during hypertrophy, we were unaware of a direct interaction of NF-κB with gata4 regulatory sequences. To test the requirement of NF-κB signaling in injury-induced activation of gata4 regulatory sequences, we crossed cmlc2:CreER; βact2:RS-IκBSR with gata4:eGFP transgenic fish. In these experiments, control ventricles displayed typical induction of gata4 regulatory sequences throughout the compact layer and adjacent to the injury site. However, fish with diminished NF-κB activity did not activate detectable reporter fluorescence from gata4 regulatory sequences (Fig. 3 J–Q). This requirement of NF-κB activity seems specific to induction of gata4 regulatory sequences, because suppression of NF-κB was not sufficient to silence gata5 regulatory sequences during regeneration (Fig. S3).
Fig. S3.
NF-κB signaling contributes to cardiomyocyte dedifferentiation. (A and B) Representative images of 7 dpa ventricles from cmlc2:CreER; βact2:RS-IκBSR treated with vehicle (n = 6) or 4-HT (n = 7) stained for Tnnt (gray). Arrowheads show diminished Tnnt staining in cardiomyocytes adjacent to the wound in control animals, suggestive of sarcomere disassembly. (C) Quantification of intensity of Tnnt intensity per muscle area suggests decreased sarcomere disassembly in ventricles with defective NF-κB signaling (n = 13). (D and E) Section images of 7 dpa ventricles from cmlc2:CreER; βact2:RS-IκBSR; gata5:eGFP fish treated with vehicle (n = 5) or 4-HT (n = 5). eGFP fluorescence is shown in grayscale. *P < 0.05, Student’s t test, two-tailed. (Scale bars: 100 μm.)
To further test direct interaction of NF-κB with gata4 regulatory sequences, we performed chromatin immunoprecipitation with an antibody against NF-κB1 on extracts from hearts with and without intact cardiomyocyte NF-κB signaling. We performed a bioinformatics analysis of the gata4 promoter by using the JASPAR database and noted an increased density of NF-κB1 binding sites ∼12 kb upstream of the gata4 transcriptional start site (Fig. 3R) (33). Interestingly, this site is just proximal to an enhancer required to direct gata4 expression in the cardiac ventricle of zebrafish embryos (34). Using primers specific to this region of the gata4 promoter, we observed evidence for direct interaction of NF-κB1 with the gata4 promoter in animals with intact NF-κB signaling. By contrast, animals without intact NF-κB signaling displayed less binding of NF-κB1 with the gata4 promoter (Fig. 3S). These data suggest that NF-κB directly interacts with gata4 regulatory sequences and is required for their induction during regeneration.
The induction of gata4 regulatory sequences during regeneration has been suggested as evidence for cardiomyocyte dedifferentiation along with sarcomere dissociation (5, 32). We noted that cardiomyocytes with intact sarcomeres, such as in uninjured ventricles, display intense immunofluorescence for cardiac Troponin T (Tnnt). By contrast, regenerating ventricles show reduced Tnnt staining intensity in cardiomyocytes at the injury site by 7 dpa. Interestingly, we found that cardiomyocytes with diminished NF-κB signaling stained more brightly for Tnnt during regeneration than controls, suggestive of a defect in sarcomere disassembly (P = 0.028, n = 13 ventricles; Fig. S3). Together, alterations in sarcomere disassembly and in the induction of gata4 regulatory sequences suggest that NF-κB signaling contributes to cardiomyocyte dedifferentiation.
Blocking NF-κB Activity in Cardiomyocytes Has a Pleiotropic Effect on Regeneration.
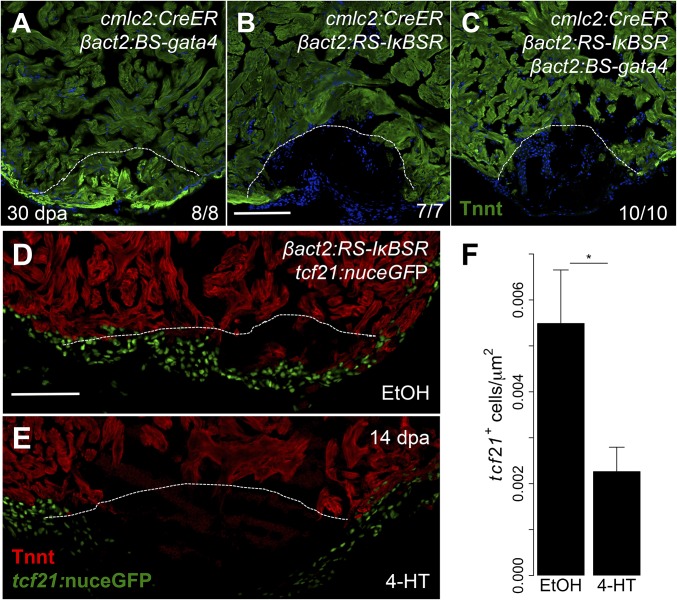
The colocalization of NF-κB activity with activated gata4 regulatory sequences and the requirement for NF-κB to activate these sequences suggest that NF-κB acts upstream of gata4 during regeneration. We hypothesized that restoration of gata4 to cardiomyocytes might be able to rescue the defect in regeneration we observed with NF-κB blockade. To test our hypothesis, we established a new transgenic line Tg(βactin2:loxP-mTag-STOP-loxP-gata4)pd115 (hereafter referred to as βact2:BS-gata4) to overexpress gata4 in cardiomyocytes (Fig. S4). As we noted earlier, blockade of NF-κB in cardiomyocytes resulted in impaired regeneration by 30 dpa (Fig. 4B). By contrast, we found that animals activating gata4 overexpression are able to regenerate normally (Fig. 4A). When we examined cmlc2:CreER; βact2:BS-gata4; βact2:RS-IκBSR transgenic fish for regenerative capacity, we found that regeneration remained impaired. This finding indicates that gata4 is insufficient to restore regenerative capacity in the absence of NF-κB signaling (Fig. 4 B and C). Similarly, gata4 overexpression was insufficient to rescue cardiomyocyte proliferation defects at 7 dpa caused by NF-κB inhibition (Fig. S5).
Fig. S4.
gata4 overexpression in cardiomyocytes. (A) Schematic of βact2:BS-gata4 transgene. (B and C) Section images of cmlc2:CreER; βact2:BS-gata4 ventricles after treatment with vehicle or 4-HT. Images are stained with an anti-FLAG antibody to identify overexpressed Gata4. (Scale bar: 100 μm.)
Fig. 4.
NF-κB has pleiotropic inhibitory effects on heart regeneration. (A–C) Section images of cmlc2:CreER; βact2:BS-gata4, cmlc2:CreER; βact2:RS-IκBSR, and cmlc2:CreER; βact2:BS-gata4; βact2:RS-IκBSR ventricles at 30 dpa after treatment with 4-HT. Images are stained for Tnnt to demarcate cardiac muscle. Dashed line indicates approximate resection plane. (D and E) Tilescan images of cmlc2:CreER; βact2:RS-IκBSR; tcf21:nuceGFP ventricles at 14 dpa after treatment with vehicle or 4-HT. Images are stained for Tnnt to denote cardiac muscle. (F) Quantification of tcf21+ cell responses in cmlc2:CreER; βact2:RS-IκBSR hearts treated with vehicle (n = 10) or 4-HT (n = 12) at 14 dpa. Error bars indicate SE. *P < 0.05, Student’s t test, two-tailed. (Scale bars: 100 μm.)
Fig. S5.
gata4 overexpression does not alter cardiomyocyte proliferation defects caused by NF-κB depletion. (A–C) Section images of 7 dpa ventricles from cmlc2:CreER; βact2:RS-IκBSR; βact2:BS-gata4 fish treated with 4-HT. Sections are stained for Mef2 (red) and PCNA (green). Arrowheads indicate Mef2+/PCNA+ nuclei. Dashed line indicates approximate resection plane. (D) Quantification of cardiomyocyte proliferation from 7 dpa cmlc2:CreER; βact2:BS-gata4 (n = 7), cmlc2:CreER; βact2:RS-IκBSR (n = 5), and cmlc2:CreER; βact2:RS-IκBSR; βact2:BS-gata4 (n = 8) ventricles treated. Error bars indicate SEM. *P < 0.05, Mann–Whitney test.
Upon closer examination of cardiac wounds in transgenic animals with myocardial NF-κB blockade, we noted that regenerates were typically pauci-cellular and displayed a thin layer of cells on the outermost surface. This abnormality was reminiscent of phenotypes that we have observed with defective epicardial integration during regeneration (8). To assess epicardial responses in the context of NF-κB blockade, we crossed cmlc2:CreER; βact2:RS-IκBSR fish with tcf21:nuceGFP fish, which marks epicardial and epicardial-derived cells. In control animals, we noted infiltration of the wound by epicardial cells at 14 dpa; by contrast, animals with defective NF-κB in cardiomyocytes had notably fewer epicardial cells in the wound (Fig. 4 D–F). Together, these data reveal that NF-κB signaling in cardiomyocytes has broad, multicellular effects on the heart regeneration program.
Discussion
In many contexts, injury provides a facilitative milieu for regeneration (35). Inflammatory cells are recruited to the wound and provide an array of paracrine signals, some of which are thought to improve regenerative capacity. Macrophages, in particular, have been implicated in regeneration of limbs, the nervous system, skeletal muscle, fins, and heart muscle (15–19, 36, 37). Tissue resident macrophages are suspected to support heart regeneration, whereas systemic inflammatory macrophages may limit regenerative potential (18). A major goal of regenerative medicine is the replacement of lost or damaged organs ravaged by chronic disease. Manipulation of the systemic inflammatory milieu may be an important adjuvant strategy to increase the efficiency of regenerative therapies. However, altering the inflammatory composition in chronic disease states could prove challenging, and targeting downstream tissue signaling responses may be more amenable to therapeutic modification.
NF-κB signaling is known to have a wide array of effects under a variety of contexts. Although most famously studied in inflammatory cells as a stress response, developmental roles for NF-κB have also been described. NF-κB has roles in mesoderm formation and development of the liver, skin, and skeletal system (38–42). More recently, NF-κB has been demonstrated to be required for transdifferentiation of the dorsal aortic hemogenic endothelium into hematopoietic stem cells in zebrafish (31). By way of its activation in response to a variety of stresses and its role in development, NF-κB signaling is ideally suited to couple the injury response to tissue growth. Indeed, NF-κB modulates developmental signaling pathways during hair follicle regeneration and skeletal muscle regeneration (43). Further, NF-κB signaling can be tumorigenic by promoting dedifferentiation. Excessive NF-κB signaling in intestinal epithelial cells results in reversion to a stem cell-like state and increased tumorigenesis (44). One mechanism by which NF-κB signaling may broadly work is to induce lineage-restricted developmental programs by activation of lineage-restricted enhancers. In macrophages, for instance, NF-κB signaling coordinates rapid activation of tissue-restricted enhancers in response to inflammatory stimuli (45). Here, we add to this paradigm by showing a link between NF-κB and regeneration through cardiomyocyte proliferation and dedifferentiation. Further, our observed defects in epicardial responses during regeneration by suppression of NF-κB signaling in cardiomyocytes suggests that NF-κB contributes to a cross-talk of cardiomyocytes and epicardial cells that is required to coordinate regeneration.
Although NF-κB is required for heart regeneration, NF-κB activity is also induced in response to injury in poorly regenerative tissues such as the adult mammalian heart. Interestingly, the mammalian heart up-regulates a fetal gene expression program with stress, but unlike zebrafish, does not undergo widespread cardiomyocyte proliferation or regeneration (46). One possibility to reconcile this discrepancy is that the accessible targets to NF-κB signaling are different in fish and mammals. Another possibility is that the level and timing of signaling may be important to facilitate efficient regeneration. For example, early inhibition of Stat3 can promote satellite muscle cell expansion, but prolonged inhibition impairs regeneration by limiting progenitor cell differentiation (47). Similarly, prolonged NF-κB activity may be maladaptive for the mammalian heart by promoting cardiomyocyte atrophy and fetal reprogramming of cardiomyocytes (22). Ultimately, the different physiologic states of zebrafish and mammalian hearts may contribute to differential NF-κB signaling with injury. Future investigations into the relative amount of NF-κB activity and sites of NF-κB occupancy in cardiomyocytes in zebrafish and mammals may prove important in reconciling regenerative capacity among species.
Materials and Methods
Zebrafish.
Wild-type or transgenic zebrafish of the EK/AB strain were used for all experiments. βactin2:loxp-mCherry-STOP-loxp-DTA, cmlc2:CreER, NF-κB:eGFP, gata4:eGFP, gata5:eGFP and tcf21:nuceGFP transgenic fish have been described (5, 7, 25, 29, 34, 48). Procedures involving animals were approved by the Institutional Animal Care and Use Committee at Duke University.
Generation of Transgenic Zebrafish.
To generate gata4:DsRed2 zebrafish, DsRed2 cDNA was cloned downstream of the 14.8-kb gata4 promoter (34). The entire construct was flanked with I-SceI sites for transgenesis. To generate βact2:RS-IκBSR, a bicistronic construct encoding mTag-BFP (Evrogen) and IκBSR (Addgene, plasmid 12329) separated by a viral 2A peptide was ligated into the βactin2:loxp-DsRed-STOP-loxp-eGFP plasmid after excision of eGFP with AgeI/NotI. To generate βact2:BS-gata4 zebrafish, gata4 was amplified from 7 dpf zebrafish embryo cDNA and cloned in frame to pFA6a-6xGLY-FLAG (Addgene; plasmid 20751) resulting in a C-terminal FLAG tagged protein. The resulting gata4-FLAG construct was then digested with XmaI/PspOMI and cloned into the βactin2:loxp-mTagBFP-STOP-loxp-eGFP plasmid after excision of eGFP with AgeI/NotI. Each construct was coinjected into one cell-stage wild-type embryos with I-SceI. One founder was isolated and propagated. The full names for these transgenic lines are Tg(gata4:DsRed2)pd28, Tg(βactin2:loxP-mCherry-STOP-loxP-mTagBFP-2A-IκBSR)pd114, and Tg(βactin2:loxP-mTagBFP-STOP-loxP-gata4)pd115.To induce recombination in adult fish, cmlc2:CreER; βact2:RS-IκBSR fish and cmlc2:CreER; βact2:BS-gata4 fish were bathed in 4 μM 4-HT (Sigma) for 24 h.
Histological Analysis and Imaging.
Probes for nfkbiaa, nfkbiab, nfkb1, crel, relb, and p65 were generated from 6 dpf zebrafish cDNA by using the primer sequences in Table S1. In situ hybridization probes were prepared and hybridizations were performed with the aid of an InSituPro robot (Intavis) as described (25). Primary and secondary antibody staining for immunofluorescence was performed as described in ref. 7. Primary antibodies used in this study were anti-PCNA (mouse; Sigma) at 1:200, anti-Mef2 (rabbit; Santa Cruz Biotechnology) at 1:75, anti-troponinT (mouse; Thermo) at 1:150, anti-NF-κB1 (rabbit; Abcam) at 1:50, and anti-FLAG (mouse; Sigma) at 1:100. Confocal imaging was performed using a Zeiss LSM 700 or Leica SP8 upright confocal microscope. AFOG staining was performed as described in ref. 4. Mef2/PCNA imaging to assess cardiomyocyte proliferation was performed and quantified as described in ref. 7. Regenerates were scored on a scale from 1 to 3 after staining for Tnnt as described in ref. 50.
Table S1.
Primers used to generate in situ hybridization probes
| Gene | Forward primer | Reverse primer |
| nfkbiaa | atggatttacacagagccgcaat | ctatctcccacagagccggatg |
| nfkbiab | atggagctttaccgaggcacca | ctactgccccatcactttaatatcatc |
| nfkb1 | aaacctcgcgttgatctcga | gacttgcggttcttctcgct |
| P65 | ctgtcgaggaggagacgaga | agtttctgtcgggtgctctg |
| crel | tgcggttcttcactcagacc | tccgagttgaccaatggagc |
| relb | aggacatcccataccgtcca | caagcactggaggaggaagg |
To quantify sarcomeric integrity, hearts were stained with anti-Tnnt and imaged by using identical imaging parameters. Images were processed by using the EBImage package of R/Bioconductor (51). To quantitatively assess sarcomere integrity, we imaged each heart and ascertained the sum intensity for Tnnt staining and divided by the calculated muscle area. For each heart, three different sections were used. A Student’s t test was used to determine a significant difference between groups.
Epicardial density was performed on 10 μM cryosections by taking confocal images across 2-tiles and z-stacks at a 2 μM step at 20× magnification. For each heart, three maximal intensity projection images corresponding to the largest injury were imaged. Images were then coded by using a random number generator. For each image, a blinded reader traced and measured wound area using ImageJ. The number of tcf21:nuceGFP nuclei within the wound was then counted and recorded. For each heart, the average number of nuclei per square μm was determined and used for calculations. A Student’s t test was used to determine a significant difference between groups.
Quantitative PCR.
RNA was extracted by using whole ventricles from ZCAT fish and quantitative PCR was performed by using Roche Master Mix, Roche upl probes, and Roche LightCycler 480 system as described (49). Primer efficiency was tested, and only primers with an efficiency between 1.95 and 2.05 were used. Primer sequences and their corresponding upl are provided in Table S2.
Table S2.
qPCR primers
| Gene | Forward primer | Reverse primer | upl |
| nfkbiaa | agtcatgccagagagcgaat | cagagccggatgtcatcata | 91 |
| nfkbiab | cagctcggcgcagatataa | tcgatgagaagtttgaccatgt | 68 |
| nfkb2 | catatgtcccacacaatcaagac | agccaccataatgatctggaa | 144 |
| nfkb1 | cgcaagtcctacccacaagt | accagactgtgagcgtgaag | 89 |
| traf2b | tgtggtcatgaaaggcaaata | ccaacagcatcaatgtcacc | 41 |
| ef1α | cctctttctgttacctggcaaa | cttttcctttcccatgattga | 73 |
Chromatin Immunoprecipitation.
Hearts were extracted and whole ventricles were dissected in PBS with heparin (100 U/mL). Hearts were rinsed in PBS containing protease inhibitors (Roche), cross-linked in 1% formaldehyde for 10 min, and rinsed again in PBS. After homogenization in lysis buffer, chromatin was sonicated with a Bioruptor (Diagenode). ChIP was then performed by using an antibody against NF-κB1 (Rabbit; Abcam) or Rabbit-IgG (Invitrogen) at 1 μg of antibody/5 μg of chromatin by using the Magnify ChIP System (Invitrogen). ChIP was performed per manufacturer protocol with the exception that incubation with primary antibody was performed overnight. To detect binding to the gata4 promoter, 5′ CCCCCTGCGTGATATGTTCT 3′ and 5′ TACGTTTTTCGGCGCGTAAC 3′ primers were used for PCR.
Acknowledgments
We thank J. Rawls for sharing transgenic fish; J. Burris, N. Lee, T. Thoren, and S. Davies for zebrafish care; and K.D.P. laboratory members for comments on the manuscript. R.K. is supported by NIH Mentored Clinical Scientist Award K08-HL116485, and this work was supported by NIH Grant R01-HL081674 and the Howard Hughes Medical Institute (K.D.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1511209112/-/DCSupplemental.
References
- 1.Mozaffarian D, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics--2015 update: A report from the American Heart Association. Circulation. 2015;131(4):e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 5.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464(7288):601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta V, et al. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr Biol. 2013;23(13):1221–1227. doi: 10.1016/j.cub.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi K, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20(3):397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127(3):607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Cao J, Dickson AL, Poss KD. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature. 2015;522(7555):226–230. doi: 10.1038/nature14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Y, et al. Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc Natl Acad Sci USA. 2013;110(33):13416–13421. doi: 10.1073/pnas.1309810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han P, et al. Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res. 2014;24(9):1091–1107. doi: 10.1038/cr.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jopling C, Suñé G, Faucherre A, Fabregat C, Izpisua Belmonte JC. Hypoxia induces myocardial regeneration in zebrafish. Circulation. 2012;126(25):3017–3027. doi: 10.1161/CIRCULATIONAHA.112.107888. [DOI] [PubMed] [Google Scholar]
- 14.Kyritsis N, et al. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science. 2012;338(6112):1353–1356. doi: 10.1126/science.1228773. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Yan B, Shi YQ, Zhang WQ, Wen ZL. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J Biol Chem. 2012;287(30):25353–25360. doi: 10.1074/jbc.M112.349126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrie TA, Strand NS, Yang CT, Rabinowitz JS, Moon RT. Macrophages modulate adult zebrafish tail fin regeneration. Development. 2014;141(13):2581–2591. doi: 10.1242/dev.098459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aurora AB, et al. Macrophages are required for neonatal heart regeneration. J Clin Invest. 2014;124(3):1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavine KJ, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci USA. 2014;111(45):16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci USA. 2013;110(23):9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 21.Hayden MS, Ghosh S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012;26(3):203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier HJ, et al. Cardiomyocyte-specific IκB kinase (IKK)/NF-κB activation induces reversible inflammatory cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2012;109(29):11794–11799. doi: 10.1073/pnas.1116584109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell NH, et al. Activation of NF-kappa B is required for hypertrophic growth of primary rat neonatal ventricular cardiomyocytes. Proc Natl Acad Sci USA. 2001;98(12):6668–6673. doi: 10.1073/pnas.111155798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freund C, et al. Requirement of nuclear factor-kappaB in angiotensin II- and isoproterenol-induced cardiac hypertrophy in vivo. Circulation. 2005;111(18):2319–2325. doi: 10.1161/01.CIR.0000164237.58200.5A. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138(16):3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timmers L, et al. Targeted deletion of nuclear factor kappaB p50 enhances cardiac remodeling and dysfunction following myocardial infarction. Circ Res. 2009;104(5):699–706. doi: 10.1161/CIRCRESAHA.108.189746. [DOI] [PubMed] [Google Scholar]
- 27.Santos DG, et al. Nuclear Factor (NF) kappaB polymorphism is associated with heart function in patients with heart failure. BMC Med Genet. 2010;11:89. doi: 10.1186/1471-2350-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou B, et al. Functional polymorphism of the NFKB1 gene promoter is related to the risk of dilated cardiomyopathy. BMC Med Genet. 2009;10:47. doi: 10.1186/1471-2350-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanther M, et al. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. 2011;141(1):197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274(5288):787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 31.Espín-Palazón R, et al. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell. 2014;159(5):1070–1085. doi: 10.1016/j.cell.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan G. 2014. TFBSTools: Software package for transcription factor binding site (TFBS) analysis), R package version 1.4.0.
- 34.Heicklen-Klein A, Evans T. T-box binding sites are required for activity of a cardiac GATA-4 enhancer. Dev Biol. 2004;267(2):490–504. doi: 10.1016/j.ydbio.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 35.Forbes SJ, Rosenthal N. Preparing the ground for tissue regeneration: From mechanism to therapy. Nat Med. 2014;20(8):857–869. doi: 10.1038/nm.3653. [DOI] [PubMed] [Google Scholar]
- 36.Ruckh JM, et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell. 2012;10(1):96–103. doi: 10.1016/j.stem.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204(5):1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Correa RG, et al. Characterization of NF-kappa B/I kappa B proteins in zebra fish and their involvement in notochord development. Mol Cell Biol. 2004;24(12):5257–5268. doi: 10.1128/MCB.24.12.5257-5268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274(5288):782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 40.Franzoso G, et al. Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 1997;11(24):3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iotsova V, et al. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nat Med. 1997;3(11):1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 42.Grossmann M, et al. The combined absence of the transcription factors Rel and RelA leads to multiple hemopoietic cell defects. Proc Natl Acad Sci USA. 1999;96(21):11848–11853. doi: 10.1073/pnas.96.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong Y, et al. Brg1 governs a positive feedback circuit in the hair follicle for tissue regeneration and repair. Dev Cell. 2013;25(2):169–181. doi: 10.1016/j.devcel.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Schwitalla S, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1-2):25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 45.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152(1-2):157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 46.Oka T, Xu J, Molkentin JD. Re-employment of developmental transcription factors in adult heart disease. Semin Cell Dev Biol. 2007;18(1):117–131. doi: 10.1016/j.semcdb.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tierney MT, et al. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat Med. 2014;20(10):1182–1186. doi: 10.1038/nm.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao L, et al. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci USA. 2014;111(4):1403–1408. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. eLife. 2015;4:4. doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahmoud AI, et al. Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev Cell. 2015;34(4):387–399. doi: 10.1016/j.devcel.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage--an R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26(7):979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]