Significance
Colonization of land by plants was a critical event for the emergence of extant ecosystems. The innovations that allowed the algal ancestor of land plants to succeed in such a transition remain unknown. Beneficial interaction with symbiotic fungi has been proposed as one of these innovations. Here we show that the genes required for this interaction appeared in a stepwise manner: Some evolved before the colonization of land by plants and others first appeared in land plants. We thus propose that the algal ancestor of land plants was preadapted for interaction with beneficial fungi and employed these gene networks to colonize land successfully.
Keywords: symbiosis, plant evolution, algae, plant–microbe interactions, phylogeny
Abstract
Colonization of land by plants was a major transition on Earth, but the developmental and genetic innovations required for this transition remain unknown. Physiological studies and the fossil record strongly suggest that the ability of the first land plants to form symbiotic associations with beneficial fungi was one of these critical innovations. In angiosperms, genes required for the perception and transduction of diffusible fungal signals for root colonization and for nutrient exchange have been characterized. However, the origin of these genes and their potential correlation with land colonization remain elusive. A comprehensive phylogenetic analysis of 259 transcriptomes and 10 green algal and basal land plant genomes, coupled with the characterization of the evolutionary path leading to the appearance of a key regulator, a calcium- and calmodulin-dependent protein kinase, showed that the symbiotic signaling pathway predated the first land plants. In contrast, downstream genes required for root colonization and their specific expression pattern probably appeared subsequent to the colonization of land. We conclude that the most recent common ancestor of extant land plants and green algae was preadapted for symbiotic associations. Subsequent improvement of this precursor stage in early land plants through rounds of gene duplication led to the acquisition of additional pathways and the ability to form a fully functional arbuscular mycorrhizal symbiosis.
The colonization of land by plants 450 Mya created a major transition on Earth, causing the burial of large amounts of carbon, with resultant decreases in atmospheric CO2 leading to a dramatically altered climate. This transition provided the foundation for the majority of extant terrestrial ecosystems (1, 2). The terrestrial environment that these early plants colonized must have presented many challenges, primary among them being the acquisition of mineral nutrients. It has been suggested that the appearance of the arbuscular mycorrhizal (AM) symbiosis and other beneficial associations with fungi such as Mucoromycotina facilitated this colonization of land by improving plants’ ability to capture nutrients.
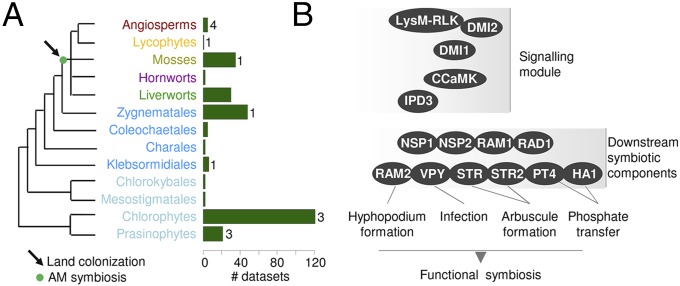
Based on recent phylogenetic analyses, Zygnematales, one of the paraphyletic “advanced charophytes” (i.e., Coleochaetales, Charales, and Zygnematales), has been identified as the closest green algal relative to land plants, whereas the chlorophytes diverged much earlier (Fig. 1A) (3, 4). On the other side of this transition, bryophyte lineages (i.e., liverworts, mosses, and hornworts) are considered to be the earliest diverging land plants, although their branching order remains debated (5–7). Key innovations present in bryophytes but not in advanced charophytes thus are good candidates for understanding the basis of land colonization by plants. In support of previous suppositions, one innovation that discriminates bryophytes from charophytes is the ability to develop beneficial associations with AM fungi (8–11). The discovery of 450-My-old fossilized spores similar to the spores of extant AM fungi supports their presence in the environment encountered by the first land plants (9). Remnants of intracellular fungal structures, likely belonging to ancestral AM fungi and to fungi belonging to the closely related Mucoromycotina, were found in the oldest plant macrofossils and suggest that one or both of these associations were instrumental for the colonization of lands (8, 12). Today, these two types of fungi also are found in, and provide a growth benefit to, liverworts and hornworts (11, 13, 14). However, although Mucoromycotina fungi are restricted to a few plant species and likely evolved independently in various lineages, AM fungi colonize more than 80% of extant land plants, and their absence in a few taxa results from independent losses (15). Taken together, these lines of evidence strongly support the notion that AM fungi played an important role in the persistent colonization of land by the first Embryophyta.
Fig. 1.
The green lineage and the AM symbiosis. (A) Schematic representation of the green lineage. Light blue: chlorophytes and basal charophytes; dark blue: advanced charophytes. Green bars on the left indicate the actual number of transcriptomes and genomes included in the extended dataset used in this study. (B) Schematic representation of the symbiotic genes.
Genetic and physiological studies in model legumes such as Medicago truncatula allowed the characterization of the steps leading to functional symbiosis in angiosperms (Fig. 1B). At the onset of the AM association, both partners initiate a chemical communication, with strigolactone signals from the plant inducing the production of a mix of diffusible signals called “Myc factors” from the fungus (16–18). The host plants detect these Myc factors that activate part of the symbiotic responses (17). The plant then allows penetration and subsequent colonization by the fungus. When the fungus reaches cortical cells in vascular plants or the inner parenchyma in liverworts (10), highly branched fungal structures surrounded by the plant plasma membrane, known as “arbuscules,” are formed (19). This specialized interface improves the transfer of nutrients and water from the AM fungi to the plant in exchange for carbohydrates (10, 19). Genetic and transcriptomic analyses in angiosperms led to the characterization of genes controlling these steps (19, 20). At the plasma membrane, a receptor complex composed of lysin motif receptor-like kinase (LysM-RLK) proteins and a malectin-like domain (MLD)-RLK (DMI2) perceives the Myc factor signals (17, 21, 22) and activates a nuclear envelope-localized potassium channel (DMI1) that initiates and maintains rapid oscillations of calcium within and around the nucleus (23). These calcium “spikes” are decoded by a calcium- and calmodulin-dependent protein kinase (CCaMK), a specific calcium-dependent protein kinase (CDPK) also regulated by calmodulin (24). Downstream transcription factors (TFs), including interacting protein of DMI3 (IPD3), which is a direct target of CCaMK, and a network of four GRAS TFs [reduced for arbuscular mycorrhization 1 (RAM1), required for arbuscule development 1 (RAD1), nodulation signaling pathway (NSP) 1, and NSP2] then activate the plant symbiotic program (25–28). Components of this program play roles at various steps: RAM2, a glycerol-3-phosphate acyl transferase (GPAT), is required for the induction of the fungal penetration structure (29). VAPYRIN controls penetration of the epidermis and arbuscule formation (30). The half–ATP-binding cassette (ABC) transporters stunted arbuscule (STR) and STR2 are critical for arbuscule development, and finally, the H+-ATPase HA1 and the phosphate transporter PT4 allow the uptake of phosphate in arbuscule cells (31–33). In angiosperms, loss of AM symbiosis results in the consistent loss of these genes, whose function seems to be linked specifically to AM symbiosis (15). Thus, we hypothesized that tracing the origin of these symbiotic genes would allow us to investigate the link between the appearance of AM symbiosis and land colonization.
Results
Extensive Phylogeny Indicates the Presence of Symbiotic Signaling Genes in Algae.
A current limitation in the study of the transition from algae to embryophytes is the limited number of datasets available. To decipher the evolution of the symbiotic genes at this transition, we first combined an extended dataset spanning the entire green lineage (Fig. 1A and Dataset S1). In addition to publicly available genomes and transcriptomes, this dataset includes the newly generated deep transcriptomes of a liverwort (Lunularia cruciata) and an advanced charophyte complex (Closterium peracerosum–strigosum–littorale, hereafter Closterium), the draft genome of another advanced charophyte (Spirogyra sp.), and 249 transcriptomes of green algae, liverworts, mosses, and hornworts generated through the 1,000 Plants (1KP) project (https://sites.google.com/a/ualberta.ca/onekp/). This extended dataset was mined for homologs of the symbiotic genes, starting with genes belonging to the “signaling module”: LysM-RLK, DMI2, DMI1, CCaMK, and IPD3. In line with previous studies using PCR-based approaches, we found LysM-RLK, DMI1, DMI2, CCaMK, and IPD3 orthologs in the bryophyte assemblies (Fig. 2 and SI Appendix, Figs. S1–S6) (10, 34). Much more surprising is the presence of homologs of most of these genes in several species across the charophyte portion of the tree (Fig. 2 and SI Appendix, Figs. S1–S6). LysM-RLKs are present in advanced charophytes together with IPD3 (SI Appendix, Figs. S1 and S6A). These orthologs displayed the specific features of these proteins, and phylogenetic analyses recovered them as direct orthologs of the angiosperm proteins (SI Appendix, Figs. S6B and S7). Homologs of DMI2 also were present in a single charophyte clade, with the actual DMI2 clade and its paralogs originating from various rounds of gene duplication (SI Appendix, Figs. S2 and S3). Finally, potential homologs of DMI1 and CCaMK were found in charophytes and even chlorophytes (Fig. 2 and SI Appendix, Figs. S4 and S5). Taken together these results indicate that, with the exception of clear DMI2 orthologs, the symbiotic signaling genes are present in extant charophytes and predate the colonization of land by plants. The presence of chlorophyte sequences with high similarity to DMI1 and CCaMK suggests that these two genes may have originated even before the divergence of the chlorophytes.
Fig. 2.
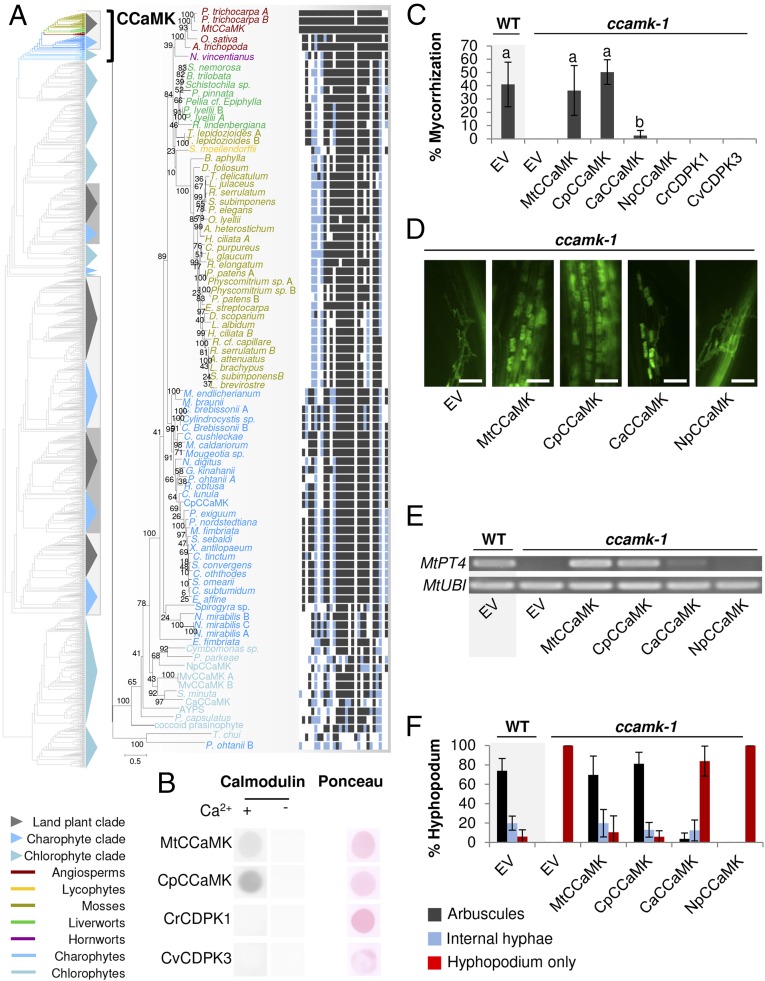
Stepwise evolution of CCaMK in the green lineage. (A) Maximum-likelihood phylogenetic tree of CCaMK extracted from the CDPK tree on the left. A high-resolution image of the CDPK tree is available in SI Appendix, Fig. S5. Colored boxes on the right indicate the conservation of the CBD within the CCaMK clade compared with MtCCaMK. Black, blue, and white squares indicate identical, homologous, and different amino acids, respectively. (B) Calmodulin-binding activity of MtCCaMK, CpCCaMK, and two chlorophyte CDPKs (CrCDPK1 and CvCDPK3) in the presence or absence of calcium. This assay is positive for MtCCaMK and CpCCaMK (dark spot) and negative for CrCDPK1 and CvCDPK3. The original plots are available in SI Appendix, Fig. S20. (C) Transcomplementation assays of the Mtccamk-1 TRV25 mutant with various CCaMKs and CDPKs including NpCCaMK and CaCCaMK. Letters indicate statistically supported groups according to ANOVA and Tukey’s honestly significant difference tests (P < 0.01). EV, empty vector. (D) Representative pictures of wheat germ agglutinin-FITC–stained AM fungi colonizing plant roots in the transcomplementation assay. (Scale bars, 100 μm.) (E) Amplification by RT-PCR of the symbiotic phosphate transporter MtPT4 and ubiquitin (MtUBI). (F) Quantification of the hyphopodia leading to internal hyphae and arbuscules.
The Key Components DMI1 and CCaMK Coevolved in Charophytes.
To understand better the evolutionary steps that led to the appearance of the signaling module, we looked at DMI1 and CCaMK homologs in charophytes and chlorophytes in more detail. DMI1 displays a very specific filter domain (ADAGNHA) compared with less specialized and broadly distributed potassium channels (GYGD) (23). In land plants, this sequence is highly conserved, with mutations seen only when a duplicated version compensates for the decreased activity (SI Appendix, Fig. S4) (35). Similarly, the advanced charophyte sequences also are extremely conserved with a filter domain identical to the one from M. truncatula DMI1 (MtDMI1) (SI Appendix, Fig. S4). In contrast, the few chlorophyte homologs displayed a different amino acid composition (SI Appendix, Fig. S4). This result suggests that DMI1 evolved in charophytes from a precursor stage present in chlorophytes.
Among the chlorophyte and charophyte proteins showing high homology with CCaMK, we found previously identified CDPKs (SI Appendix, Fig. S5) (36). One specific feature of CCaMKs compared with CDPKs is the presence of a calmodulin-binding domain (CBD) overlapping with the autoinhibitory domain (37). We compared the CBD of angiosperm CCaMKs with the corresponding region of the homologs from chlorophytes, charophytes, and, bryophytes. This domain was highly conserved in most land plants, advanced charophytes, and a few chlorophytes (Prasinophytes), all belonging to the CCaMK clade (Fig. 2A and SI Appendix, Fig. S5). However, several chlorophyte CDPKs belonging to other clades displayed a high level of conservation of the amino acids of the CBD known to be critical for calmodulin binding (38, 39) (SI Appendix, Fig. S5). To test whether CBD evolved only once in the CDPK family (i.e., in the CCaMK clade) or is a shared feature of algal CDPKs, we purified Escherichia coli-expressed M. truncatula CCaMK (MtCCaMK), chlorophyte CDPKs [Chlamydomonas reinhardtii CDPK1 (CrCDPK1)/C. reinhardtii Cre33.g782750 and Chlorella variabilis CDPK3 (CvCDPK3)/C. variabilis Cva20239], and a charophyte protein from the CCaMK clade [C. peracerosum CCaMK (CpCCaMK)/Closterium c24502] and then tested their ability to bind calmodulin. As expected, MtCCaMK was able to bind calmodulin in the presence of calcium. Interestingly, the protein from Closterium also displayed a strong calmodulin-binding activity in the presence of calcium (Fig. 2B). In contrast, CDPKs from the two chlorophytes were not able to bind calmodulin (Fig. 2B).
In angiosperms, the ability of CCaMK to bind both calcium and calmodulin is a key feature allowing this protein to decode calcium spikes (38). At basal calcium concentrations, CCaMK autophosphorylates its Thr-271, leading to the inhibition of its activity by the formation of a predicted hydrogen-bound network (38). Upon perception of repetitive calcium increases, calmodulin is activated, binds CCaMK CBD, and overrides autophosphorylation to activate CCaMK (38). Because of its ability to bind calmodulin, Closterium c24502 might behave like an actual CCaMK. To test this hypothesis, we first looked at the position corresponding to Thr-271. Closterium c24502 harbored a serine at that specific location (Ser-242) and at the following residue (Ser-243), both of which could be phosphorylated (SI Appendix, Fig. S8B). Additionally, we investigated whether Closterium c24502 may exhibit hydrogen-bonding behavior similar to that predicted for MtCCaMK by building a structural model following the computational protocol outlined in ref. 34 (SI Appendix, Fig. S8C). Only Ser-243 forms a hydrogen bond to the CBD as was observed in MtCCaMK Thr-271 (SI Appendix, Fig. S8C). However, given the structural uncertainty of the homology model, we studied the effects of phosphorylation at both serines. Phosphorylation of Ser-242 formed only one hydrogen bond with Arg-295, a residue equivalent to Medicago Arg-323 (SI Appendix, Fig. S8C). Ser-243, however, formed three hydrogen bonds to Arg-295 and Cys-215, thus mimicking the behavior observed in Medicago and suggesting that a similar mechanism is involved (SI Appendix, Fig. S8C). To test this hypothesis further, we assayed the ability of Closterium c24502 to autophosphorylate in the presence or absence of calcium. The addition of calcium led to strong autophosphorylation, but very low autophosphorylation activity was detected in the absence of calcium (SI Appendix, Fig. S8D). The addition of calmodulin was found to inhibit this activity, suggesting that Closterium c24502 may be a bona fide CCaMK (CpCCaMK) (SI Appendix, Fig. S8E).
Together with their individual biochemical properties, evolving the ability to participate and fulfill their function in specific signaling pathways is a critical innovation of signaling components. Symbiotic signaling events are abolished in a Medicago ccamk-1 mutant (24). If CpCCaMK is a bona fide CCaMK, expressing it in the ccamk-1 mutant should rescue its symbiotic defects. To test this hypothesis, CpCCaMK, the endogenous MtCCaMK gene, and two CDPKs from Chlorophytes (CvCDPK3 and CrCDPK1) were expressed in the ccamk-1 mutant using Agrobacterium-mediated root transformation (40). Wild-type and ccamk-1 seedlings transformed with empty expression vectors were used as controls. After 6 wk of incubation with the AM fungus Rhizophagus irregularis, none of the ccamk-1 roots transformed with the empty vector or the chlorophyte CDPKs were colonized (Fig. 2C). In contrast, ccamk-1 roots transformed with CpCCaMK or MtCCaMK were colonized at a level similar to that seen in to wild-type plants transformed with the empty vector (Fig. 2C). Internal hyphae, vesicles, and arbuscules were present in these roots, and expression of the symbiotic phosphate transporter MtPT4 was detected, suggesting a full complementation of the mutant phenotype (Fig. 2 D–F). Taken together these results confirm that an advanced charophyte protein, CpCCaMK, is fully functional in a symbiotic context.
Interestingly, a few chlorophytes (prasinophytes) and basal charophyte sequences also were recovered in the CCaMK clade (Fig. 2A). To determine if a functional CCaMK evolved even before the advanced charophytes, we expressed two of them in the ccamk-1 mutant and conducted mycorrhization assays. Roots transformed with CCaMK, from the prasinophyte Nephroselmis pyriformis (NpCCaMK), did not display any colonization event, and the expression of MtPT4 was not induced (Fig. 2 C–E). In contrast, roots expressing CaCCaMK from the basal charophyte Chlorokybus atmophyticus showed events of internal colonization and even arbuscules (Fig. 2 C and D). However, the colonization rate was extremely low, and the expression of MtPT4 was induced only slightly (Fig. 2 C and E). Closer inspection confirmed that most hyphopodia that formed on the ccamk-1 roots expressing CaCCaMK aborted, whereas they led to internal hyphae and arbuscules in roots transformed with MtCCaMK or CpCCaMK (Fig. 2F). The partial complementation of the ccamk-1 mutant achieved by CaCCaMK suggests that an additional layer of functionalization occurred after the divergence of the basal charophytes. Even if the CBD of CaCCaMK is predicted to be functional, and the key amino acids are present, we cannot exclude the possibility that the ability to bind calmodulin is affected. The CBD of CpCCaMK was swapped with the CBD from CaCCaMK, and the ability of the resulting chimeric protein (Cp-CaCBD) to bind calmodulin was assayed as previously. Calmodulin binding of Cp-CaCBD was reduced compared with the native CpCCaMK but was not abolished (SI Appendix, Fig. S9 A and B). Additionally, when expressed in a ccamk-1 mutant, this chimeric gene was able to complement mycorrhizal defects to a level close to that achieved with CpCCaMK (SI Appendix, Fig. S9 C–F). Thus, even though the difference in calmodulin-binding activity may explain part of the differences observed between CaCCaMK and CpCCaMK, other innovations must be present in CCaMK from advanced charophytes. In addition to the conservation of the CBD, the formation of the hydrogen-bond network might be affected in basal charophyte and chlorophyte CCaMKs. To test this hypothesis, we first determined the conservation of the autophosphorylation site. Although land plant CCaMKs and most advanced charophyte CCaMKs display an amino acid that can be phosphorylated at the position orthologous to Thr-271 (either threonine or serine), the basal charophyte and chlorophyte sequences from the CCaMK clade displayed other amino acids (SI Appendix, Fig. S10A). Because nearby residues could play a similar role, we built structural models for the CCaMK from two basal charophyte species, CaCCaMK and Mesostigma viride CCaMK (MvCCaMK). Neither the predicted CaCCaMK nor the MvCCaMK structure has serine or threonine in the region of interest, and thus neither can be phosphorylated (SI Appendix, Fig. S10 C and D).
Taken together, these results strongly suggest that a fully functional CCaMK evolved in the common ancestor of charophytes and land plants in a three-step process: (i) duplication of an ancestral CDPK before the divergence of the chlorophyte–streptophyte lineages; (ii) evolution of a CBD in one of the paralogs; and (iii) emergence of additional layers of regulation, including the autophosphorylation site, that allow the formation of an inhibitory hydrogen-bond network. Further experiments with additional CCaMK from Prasinophytes and basal charophytes would help elucidate the remaining uncertainty in this proposed model. Interestingly, the evolution of CCaMK mirrored the evolution of DMI1, which acquired its elaborate filter domain in advanced charophytes. Together with the presence of LysM-RLK and IPD3 in advanced charophytes, these data support the evolution of the signaling module before colonization of land by embryophytes.
Downstream Symbiotic Genes Evolved in Land Plants.
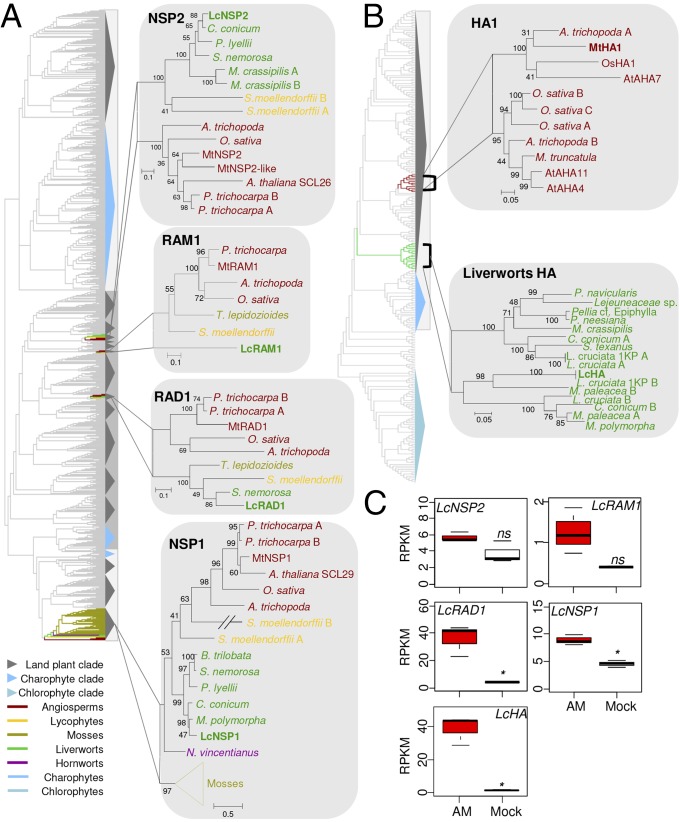
In addition to the signaling module, establishing AM symbiosis in angiosperms requires a network of downstream GRAS transcription factors (NSP1, NSP2, RAM1, and RAD1) and six other components involved at various stages during the mycorrhization process (RAM2, VAPYRIN, STR and STR2, HA1, and PT4). Given the presence of the signaling module in charophytes, we hypothesized that downstream genes also might predate land colonization. We searched for potential orthologs in the extended datasets. GRAS proteins were found only in advanced charophytes and liverworts (Fig. 3A and SI Appendix, Fig. S11). A maximum-likelihood tree was built with the recovered sequences to distinguish between homologs and orthologs of the four symbiotic GRAS proteins (SI Appendix, Fig. S11). Based on this reconstruction, three clades, supported by low bootstrap values, encompassing charophyte and land-plant sequences can be recovered (SI Appendix, Fig. S11). However, the number of GRAS proteins per species is much higher (Dataset S2), suggesting the occurrence of multiple rounds of duplication at various levels. The presence of bryophyte sequences at the base of the NSP1, NSP2, RAM1, and RAD1 clades suggests that they originate from duplication events that occurred in the last common ancestor of land plants (Fig. 3A). Half-ABC transporters of the G subfamily were found in chlorophytes and charophytes, but direct orthologs of STR and STR2, which function in the cells of angiosperms containing arbuscules, evolved in land plants only via duplication (SI Appendix, Figs. S12 and S13A). No VAPYRIN or GPAT homologs were found in either chlorophytes or charophytes, but clear orthologs were found in liverworts (SI Appendix, Figs. S14–S16). Finally, orthologs of PT4 and HA1 were found only in vascular plants (Fig. 3B and SI Appendix, Figs. S17–S19). In addition to their symbiotic function identified by genetics, NSP1, RAM1, RAD1, RAM2, STR, and STR2 are up-regulated during symbiosis in angiosperms, whereas NSP2 is down-regulated (25, 26, 29, 32, 41). To test if their orthologs identified in liverworts displayed the same expression pattern, which would suggest a conserved function in symbiosis, we sequenced the transcriptomes of Lunularia in association with the AM fungus R. irregularis and compared them with noncolonized Lunularia thalli. A total of 509 contigs were found to be significantly up-regulated (fold change >2, false-discovery rate <0.05) (Dataset S3), including the orthologs of NSP1, RAD1, RAM2, STR, and STR2 (Fig. 3C and SI Appendix, Fig. S12B). The ortholog of RAM1 was slightly, but not significantly, up-regulated, and the expression of LcNSP2 was not affected (Fig. 3C).
Fig. 3.
Multiple paths for the evolution of symbiotic genes in basal land plants. (A) Maximum-likelihood phylogenetic tree of NSP2, RAM1, RAD1, and NSP1 extracted from the GRAS tree on the left. These four clades originate from land plant-specific duplications. (B) Maximum-likelihood phylogenetic tree of HA1 and liverworts HA extracted from the H+-ATPase tree (available in SI Appendix, Fig. S17). (C) Differential expression of LcNSP2, LcRAM1, LcRAD1, LcNSP1, and LcHA in mock-treated (Mock) or colonized (AM) Lunularia cruciata. *P < 0.05. AM-inducible H+-ATPase evolved independently in angiosperms and liverworts.
Taken together, these results indicate that the downstream symbiotic components very likely appeared in first land plants via two processes, one being the duplication of existing gene families, with one of the paralogs being recruited for symbiotic function—as suggested by the expression pattern—and the other being the appearance of new proteins such as VAPYRIN and RAM2. Interestingly, these two proteins display domains found in many other protein families (29, 30, 42, 43) and thus likely originate from domain shuffling. However, several components and regulations are missing in basal land plants, such as the orthologs of PT4 and HA1 and the down-regulation of NSP2 during mycorrhizae. In legumes, down-regulation of NSP2 is mediated by microRNA 171h (mir171h) and allows plants to control the level of root infection (41). This microRNA and the corresponding target sequence in NSP2 are not present in basal land plants, providing an explanation for the absence of down-regulation (41). The absence of mir171h suggests that clade-specific innovations evolved after the appearance of AM symbiosis to fine-tune the association in a new developmental context (gametophyte vs. root sporophyte). The absence of PT4 and HA1 is more surprising, given that the transfer of phosphate to the host plant is an ancestral feature of the association (11). To determine if a nonorthologous pair of symbiotic PT and H+-ATPase could be present in liverworts, we mined the Lunularia transcriptome and found one PT (LcPT) and one HA (LcHA) strongly up-regulated during mycorrhizae (Fig. 3C and SI Appendix, Fig. S19B).
Discussion
Identifying the innovations that led to the colonization of land by plants has been a long-lasting effort in plant and evolutionary biology. It requires genomic and transcriptomic data from a large and phylogenetically diverse panel of species across charophyte green algae and land plants. The charophyte and bryophyte transcriptomes generated by the 1KP project together with the deep transcriptomes generated for Closterium and Lunularia and the draft genome of Spirogyra sp. allowed us to address such questions. It must be noted that, until multiple additional genomes from charophyte and chlorophyte species have been sequenced, the presence of genes not detected in transcriptomes cannot be ruled out. However, when a similar dataset for liverworts and other bryophytes with an even lower coverage of the species space was used, all the symbiotic genes were recovered, in line with results obtained in the study of other processes (44–46) and lending support to our conclusions.
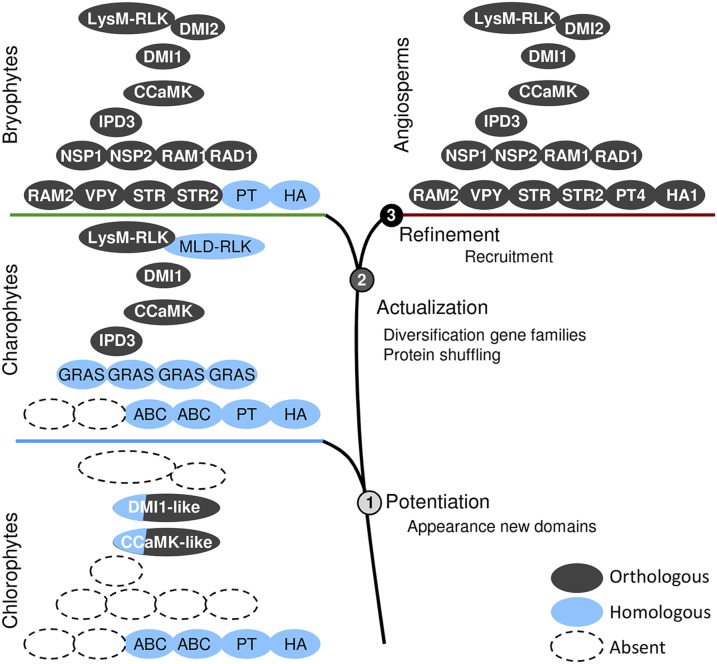
Experimental evolution and phylogenetic studies have identified three steps toward the stable acquisition of a trait: potentiation, actualization, and refinement (47, 48). Here, we identified the genetic innovations linked to these steps that lead to functional AM symbiosis in land plants. We propose that potentiation occurred in a common ancestor of charophytes and land plants via mutations in CDPK and DMI1 precursors present before the divergence of chlorophytes (Fig. 4). MLD-RLKs, LysM-RLKs, IPD3, and nonsymbiotic GRAS TFs also evolved in the streptophyte lineage from as yet unknown ancestors. So far, there is no evidence of symbiotic associations between AM fungi charophytes. These data argue for a clear absence of correlation between the appearance of the signaling module in an ancestor of charophytes and land plants and the appearance of AM symbiosis in the first land plants. Strigolactones produced by plants to activate AM fungal metabolism also are present in advanced charophytes (49). In addition to their symbiotic function, these compounds are plant hormones controlling numerous developmental traits, and a hormonal role seems conserved in charophytes (49, 50). Similarly, a nonsymbiotic function of the signaling module in charophytes cannot be ruled out. Given that green algae can form specific associations with bacteria and fungi (51, 52), another exciting hypothesis would be that this module might be required for symbiotic associations in charophytes and would have been recruited for AM symbiosis by the first land plants. In early land plants, actualization took place with the diversification of GRAS transcription factors and half-ABC transporters via gene duplication together with the acquisition of the other downstream symbiotic genes. Whether these steps occurred together or as successive events remains an open question. Finally, refinement is an often ongoing process, with multiple paths for optimization of a trait (47, 48) linked to specificity of every organism. Independent recruitments of symbiotic nutrient transport systems in liverworts and angiosperms reflect this process (Fig. 4).
Fig. 4.
Stepwise acquisition of symbiotic genes in the green lineage. The appearance of new domains in advanced charophytes led to the emergence of new protein families and to the evolution of the symbiotic signaling module via the neofunctionalization of existing proteins (DMI1 and CCaMK). Most of the downstream symbiotic genes evolved in basal land plants via gene duplication in these newly evolved gene families (i.e., STR) or by combining existing domains in new proteins (i.e., VAPYRIN). Finally, genes originating from lineage-specific duplications were recruited independently to fulfill their symbiotic functions (PT and H+-ATPase).
Other gene regulatory networks playing major roles in vascular plants are thought to have originated in early land plants (53). Mining advanced charophyte genomes and transcriptomes is a powerful approach to identify precursor stages of these networks and to confirm the role played by charophyte ancestors in the colonization of land. As we showed for symbiotic genes, actualization of these networks in early land plants, potentially via massive gene duplications, could have been a major enhancer of the successful colonization of land by plants.
Materials and Methods
Details of materials and methods are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank Neil D. Clarke for providing sequences; Joanna Maki, Ajay Shah, and Wesley Kohls for technical assistance; and Matthew Hartley, Paul Fretter, and Chris Bridson for assistance. This research was supported by National Science Foundation Grant IOS-0701846 (to J.-M.A. and M.R.S.); by bridge funds from the University of Wisconsin-Madison (to J.-M.A. and P.-M.D.); by the OpenPlant initiative; by the Bill and Melinda Gates Foundation (G.E.D.O.); and by the High Performance Computing Resources of the Norwich BioScience Institute Partnership Computing infrastructure for Science group. The 1KP initiative, led by G.K.-S.W., is funded by the Alberta Ministry of Innovation and Advanced Education, Alberta Innovates-Technology Futures Innovates Centre of Research Excellence, Musea Ventures, and Beijing Genomics Institute-Shenzhen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI database (accession nos. SRR1027885, PRJNA296352, and PRJNA290461).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515426112/-/DCSupplemental.
References
- 1.Beerling D. The Emerald Planet. How Plants Changed Earth’s History. Oxford Univ Press; Oxford: 2007. [Google Scholar]
- 2.Lal R. Soil carbon sequestration impacts on global climate change and food security. Science. 2004;304(5677):1623–1627. doi: 10.1126/science.1097396. [DOI] [PubMed] [Google Scholar]
- 3.Timme RE, Bachvaroff TR, Delwiche CF. Broad phylogenomic sampling and the sister lineage of land plants. PLoS One. 2012;7(1):e29696. doi: 10.1371/journal.pone.0029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wodniok S, et al. Origin of land plants: Do conjugating green algae hold the key? BMC Evol Biol. 2011;11:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu YL, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103(42):15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickett NJ, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA. 2014;111(45):E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox CJ, Li B, Foster PG, Embley TM, Civán P. Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Syst Biol. 2014;63(2):272–279. doi: 10.1093/sysbio/syt109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remy W, Taylor TN, Hass H, Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91(25):11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redecker D, Kodner R, Graham LE. Glomalean fungi from the Ordovician. Science. 2000;289(5486):1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 10.Humphreys CP, et al. Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat Commun. 2010;1:103. doi: 10.1038/ncomms1105. [DOI] [PubMed] [Google Scholar]
- 11.Field KJ, et al. Contrasting arbuscular mycorrhizal responses of vascular and non-vascular plants to a simulated Palaeozoic CO2 decline. Nat Commun. 2012;3:835. doi: 10.1038/ncomms1831. [DOI] [PubMed] [Google Scholar]
- 12.Strullu-Derrien C, et al. Fungal associations in Horneophyton ligneri from the Rhynie Chert (c. 407 million year old) closely resemble those in extant lower land plants: Novel insights into ancestral plant-fungus symbioses. New Phytol. 2014;203(3):964–979. doi: 10.1111/nph.12805. [DOI] [PubMed] [Google Scholar]
- 13.Field KJ, et al. First evidence of mutualism between ancient plant lineages (Haplomitriopsida liverworts) and Mucoromycotina fungi and its response to simulated Palaeozoic changes in atmospheric CO2. New Phytol. 2015;205(2):743–756. doi: 10.1111/nph.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. Symbiotic options for the conquest of land. Trends Ecol Evol. 2015;30(8):477–486. doi: 10.1016/j.tree.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Delaux PM, et al. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 2014;10(7):e1004487. doi: 10.1371/journal.pgen.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besserer A, et al. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006;4(7):e226. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maillet F, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469(7328):58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 18.Genre A, et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198(1):190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 19.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6(10):763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 20.Delaux PM, Séjalon-Delmas N, Bécard G, Ané JM. Evolution of the plant-microbe symbiotic ‘toolkit’. Trends Plant Sci. 2013;18(6):298–304. doi: 10.1016/j.tplants.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Op den Camp R, et al. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science. 2011;331(6019):909–912. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- 22.Miyata K, et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014;55(11):1864–1872. doi: 10.1093/pcp/pcu129. [DOI] [PubMed] [Google Scholar]
- 23.Ané JM, et al. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science. 2004;303(5662):1364–1367. doi: 10.1126/science.1092986. [DOI] [PubMed] [Google Scholar]
- 24.Lévy J, et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science. 2004;303(5662):1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- 25.Delaux PM, Bécard G, Combier JP. NSP1 is a component of the Myc signaling pathway. New Phytol. 2013;199(1):59–65. doi: 10.1111/nph.12340. [DOI] [PubMed] [Google Scholar]
- 26.Xue L, et al. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 2015;167(3):854–871. doi: 10.1104/pp.114.255430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gobbato E, et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol. 2012;22(23):2236–2241. doi: 10.1016/j.cub.2012.09.044. [DOI] [PubMed] [Google Scholar]
- 28.Singh S, Katzer K, Lambert J, Cerri M, Parniske M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 2014;15(2):139–152. doi: 10.1016/j.chom.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Wang E, et al. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol. 2012;22(23):2242–2246. doi: 10.1016/j.cub.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 30.Pumplin N, et al. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010;61(3):482–494. doi: 10.1111/j.1365-313X.2009.04072.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang E, et al. A H+-ATPase That Energizes Nutrient Uptake during Mycorrhizal Symbioses in Rice and Medicago truncatula. Plant Cell. 2014;26(4):1818–1830. doi: 10.1105/tpc.113.120527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Blaylock LA, Harrison MJ. Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell. 2010;22(5):1483–1497. doi: 10.1105/tpc.110.074955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 2007;104(5):1720–1725. doi: 10.1073/pnas.0608136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B, et al. Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 2010;186(2):514–525. doi: 10.1111/j.1469-8137.2009.03137.x. [DOI] [PubMed] [Google Scholar]
- 35.Venkateshwaran M, et al. The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell. 2012;24(6):2528–2545. doi: 10.1105/tpc.112.098475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valmonte GR, Arthur K, Higgins CM, MacDiarmid RM. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014;55(3):551–569. doi: 10.1093/pcp/pct200. [DOI] [PubMed] [Google Scholar]
- 37.Singh S, Parniske M. Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol. 2012;15(4):444–453. doi: 10.1016/j.pbi.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Miller JB, et al. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell. 2013;25(12):5053–5066. doi: 10.1105/tpc.113.116921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimoda Y, et al. Rhizobial and fungal symbioses show different requirements for calmodulin binding to calcium calmodulin-dependent protein kinase in Lotus japonicus. Plant Cell. 2012;24(1):304–321. doi: 10.1105/tpc.111.092197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boisson-Dernier A, et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact. 2001;14(6):695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 41.Lauressergues D, et al. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012;72(3):512–522. doi: 10.1111/j.1365-313X.2012.05099.x. [DOI] [PubMed] [Google Scholar]
- 42.Murray JD, et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 2011;65(2):244–252. doi: 10.1111/j.1365-313X.2010.04415.x. [DOI] [PubMed] [Google Scholar]
- 43.Feddermann N, et al. The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J. 2010;64(3):470–481. doi: 10.1111/j.1365-313X.2010.04341.x. [DOI] [PubMed] [Google Scholar]
- 44.Bennett T, et al. Paralogous radiations of PIN proteins with multiple origins of noncanonical PIN structure. Mol Biol Evol. 2014;31(8):2042–2060. doi: 10.1093/molbev/msu147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sayou C, et al. A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science. 2014;343(6171):645–648. doi: 10.1126/science.1248229. [DOI] [PubMed] [Google Scholar]
- 46.Li FW, et al. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc Natl Acad Sci USA. 2014;111(18):6672–6677. doi: 10.1073/pnas.1319929111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489(7417):513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quandt EM, Deatherage DE, Ellington AD, Georgiou G, Barrick JE. Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc Natl Acad Sci USA. 2014;111(6):2217–2222. doi: 10.1073/pnas.1314561111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delaux PM, et al. Origin of strigolactones in the green lineage. New Phytol. 2012;195(4):857–871. doi: 10.1111/j.1469-8137.2012.04209.x. [DOI] [PubMed] [Google Scholar]
- 50.Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H. The biology of strigolactones. Trends Plant Sci. 2013;18(2):72–83. doi: 10.1016/j.tplants.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438(7064):90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 52.Hom EF, Murray AW. Plant-fungal ecology. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science. 2014;345(6192):94–98. doi: 10.1126/science.1253320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langdale JA. Evolution of developmental mechanisms in plants. Curr Opin Genet Dev. 2008;18(4):368–373. doi: 10.1016/j.gde.2008.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.