Significance
Most vaccines protect from infections by eliciting a long-lived antibody response. B-cell CLL/lymphoma 6 (Bcl6) is a transcriptional repressor that is essential for the generation of long-lived antibody responses by promoting the differentiation of T follicular helper (Tfh) and germinal center B cells. Currently, the mechanisms by which Bcl6 directs Tfh cell differentiation is unclear. We show that mutations in the middle domain of Bcl6 prevent full differentiation of Tfh cells by loss of repression of key target genes. This study reveals a role for the middle domain of Bcl6 in the differentiation and function of Tfh cells.
Keywords: T follicular helper cells, germinal centers, B-cell help
Abstract
T follicular helper (Tfh) cells are essential providers of help to B cells. The transcription factor B-cell CLL/lymphoma 6 (Bcl6) is a lineage-defining regulator of Tfh cells and germinal center B cells. In B cells, Bcl6 has the potential to recruit distinct transcriptional corepressors through its BTB domain or its poorly characterized middle domain (also known as RDII), but in Tfh cells the roles of the Bcl6 middle domain have yet to be clarified. Mimicked acetylation of the Bcl6 middle domain (K379Q) in CD4 T cells results in significant reductions in Tfh differentiation in vivo. Blimp1 (Prdm1) is a potent inhibitor of Tfh cell differentiation. Although Bcl6 K379Q still bound to the Prdm1 cis-regulatory elements in Tfh cells, Prdm1 expression was derepressed. This was a result of the failure of Bcl6 K379Q to recruit metastasis-associated protein 3 (MTA3). The loss of Bcl6 function in Bcl6 K379Q-expressing CD4 T cells could be partially rescued by abrogating Prdm1 expression. In addition to Prdm1, we found that Bcl6 recruits MTA3 to multiple genes involved in Tfh cell biology, including genes important for cell migration, cell survival, and alternative differentiation pathways. Thus, Bcl6 middle domain mediated repression is a major mechanism of action by which Bcl6 controls CD4 T-cell fate and function.
B-cell CLL/lymphoma 6 (Bcl6) is a transcriptional repressor that is required for the differentiation of T follicular helper (Tfh) cells. Tfh cells are a specific subtype of helper CD4 T cells specialized in providing help for B cells (1). Interactions between Tfh and B cells in germinal centers (GCs) are necessary for the production of high-affinity antibodies and long-lived plasma cells. Although Bcl6 is essential for Tfh cell differentiation (2–5), the mechanisms by which Bcl6 controls gene expression in CD4 T cells are not well understood (1).
Bcl6 is thought to be exclusively a transcriptional repressor (6). Furthermore, its repressive capability, at least in B cells, is dependent on association with a variety of corepressors (7). One key function of Bcl6 in T and B cells is the repression of Prdm1, the gene encoding Blimp1. In B cells, Blimp1 induces plasma cell differentiation and blocks the GC B-cell differentiation program by repressing Bcl6 (8). This reciprocal antagonism also exists in T cells, as Blimp1 directly inhibits Bcl6 expression and supports differentiation of non-Tfh effector cells (2, 9–12).
Bcl6 consists of a bric-a-brac, tramtrack, broad-complex (BTB) domain, a middle domain (also known as RDII), and a zinc finger domain consisting of six Kruppel-like zinc fingers (1). Each of these domains can associate with specific corepressor complexes in B cells and macrophages. The Bcl6 DNA binding zinc fingers are required for Bcl6 activity in CD4 T cells in cell culture (4, 12). The Bcl6 BTB domain participates in Tfh cell differentiation (13), most likely by interacting with BCOR (14) and perhaps other corepressors. The middle domain is the largest domain of Bcl6. Nevertheless, the Bcl6 middle domain is not well characterized. This domain contains a lysine KKYK motif that is necessary for binding metastasis-associated protein 3 (MTA3). MTA3 is a cell type-specific component of the NuRD HDAC complex (15, 16). In B-cell lymphoma lines, acetylation of Bcl6 lysine 379 by p300 prevents MTA3 from binding and abrogates the repressive ability of Bcl6. The Bcl6 middle domain also contains PEST motifs that regulate the degradation of Bcl6 when phosphorylated (17).
In this study, we have examined the role of the middle domain of Bcl6 in the context of Tfh cell differentiation. By using a mutation that mimics acetylation of Bcl6 Lys379 (Bcl6 K379Q), we show that modification of this single residue results in significantly impaired Tfh cell differentiation. The Bcl6 K379Q mutant CD4 T cells were severely compromised in B-cell help. Prdm1 expression was increased in Bcl6 K379Q+ Tfh cells, and Tfh cell differentiation was partially restored in the absence of Blimp1. Finally, many Tfh cell-associated gene targets were identified that are specifically repressed by a Bcl6 middle domain-dependent mechanism.
Results
Acetylation of the Bcl6 Middle Domain Inhibits Tfh Differentiation.
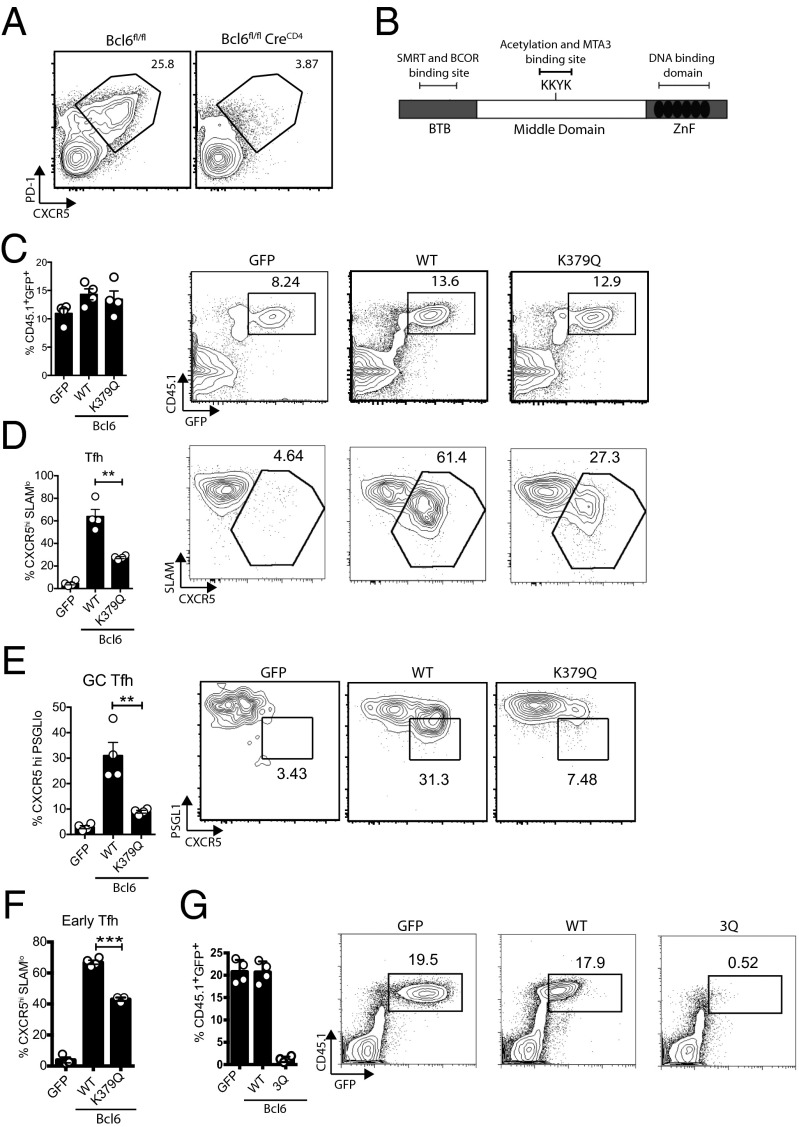
Bcl6fl/fl CreCD4 mice (18) do not generate Tfh cells following acute lymphocytic choriomeningitis virus (LCMV) infection (Fig. 1A). To explore the role of the Bcl6 middle domain in T cells, we determined if expression of Bcl6 mutants in Bcl6-deficient cells could rescue Tfh cell development. We generated a K379Q mutation in the Bcl6 middle domain, which sterically mimics acetylation of this lysine residue (Fig. 1B) (15, 16). LCMV-specific Bcl6fl/fl CreCD4 SMARTA (LCMV GP66-77 I-Ab specific) T-cell receptor (TCR) transgenic CD4 T cells were reconstituted with Bcl6 WT, Bcl6 K379Q, or an empty GFP retroviral vector (RV) and transferred to Bcl6fl/fl CreCD4 hosts. At 7 d following an acute LCMV infection, GFP+, Bcl6+, and Bcl6 K379Q+ SMARTA cells expanded equivalently (Fig. 1C). Control GFP+ Bcl6fl/fl CreCD4 SMARTA cells failed to differentiate into Tfh cells, confirming the absolute requirement of Bcl6 in CD4 T cells for Tfh development (Fig. 1D). When Tfh cells enter the GC, they become biologically distinct GC Tfh cells identifiable as CXCR5hiBcl6hiPD1hiGL7hiMafhiSAPhiPSGL1lo (1, 10, 19–21). Ectopic expression of WT Bcl6 was sufficient to rescue the development of Tfh and GC Tfh cells. However, Bcl6 K379Q was unable to support appropriate Tfh cell (P = 0.0012) and GC Tfh cell (P = 0.0057) differentiation (Fig. 1 D and E).
Fig. 1.
Acetylation of Bcl6 middle domain inhibits Tfh cell development. (A) Bcl6-null CD4 T cells do not develop into Tfh cells. Bcl6fl/fl and Bcl6fl/fl CreCD4 mice were infected with LCMV. Tfh cell development was analyzed 7 d following infection. CD44hi CD4+ T cells are shown. (B) Schematic of Bcl6 domains and acetylation motif KKYK in the middle domain. (C–E and G) Bcl6fl/fl CreCD4 CD45.1+ SMARTA (SM) cells were retrovirally transduced with empty GFP vector, Bcl6 WT, Bcl6 K379Q, or Bcl6 3Q, then transferred to Bcl6fl/fl CreCD4 mice and analyzed 7 d following acute LCMV infection. (C) CD45.1+GFP+ SMARTA cell frequencies. (D) LCMV-specific SMARTA Tfh cells (CXCR5hiSLAMlo). (E) LCMV-specific SMARTA GC Tfh cells (CXCR5hi PSGL1lo). (F) Early Tfh cells (CXCR5hiSLAMlo) among transduced Bcl6fl/fl CreCD4 SMARTA cells at 3 d following LCMV infection. (G) CD45.1+GFP+ SMARTA cell frequencies. Data shown are representative of at least three independent experiments. G is representative of more than six independent experiments (*P< 0.05, **P< 0.01, and ***P< 0.001).
Bcl6-dependent Tfh cell differentiation begins within 48 h in vivo (11). Therefore, to determine whether Bcl6 acetylation may impact early Tfh cell differentiation, we examined Tfh cell development at 3 d following viral infection. We observed a significant reduction in early Tfh cell differentiation among Bcl6 K379Q+ cells compared with WT Bcl6+ cells (P < 0.0001; Fig. 1F). Thus, Bcl6 middle-domain functions are required for optimal early Tfh cell differentiation.
The third lysine in this motif (K379) is the physiological target for acetylation (15). However, some studies have also used triple lysine mutations (16, 22). To this end, we generated a Bcl6-RV expression construct with three lysines mutated to glutamines (3Q). Bcl6 3Q+Bcl6fl/fl CreCD4 SMARTA CD45.1 cells were transferred into Bcl6fl/fl CreCD4 hosts, followed by infection with LCMV. Bcl6 3Q+ CD4 T cells failed to survive (Fig. 1G). Thus, as physiological Bcl6 acetylation is known to occur only at K379, we performed no additional studies with the nonphysiological 3Q mutation. In summary, we conclude that acetylation of Lys379 specifically inhibits Bcl6 activity and impairs the full development of Tfh cells in vivo.
Dysregulated Blimp1 Expression.
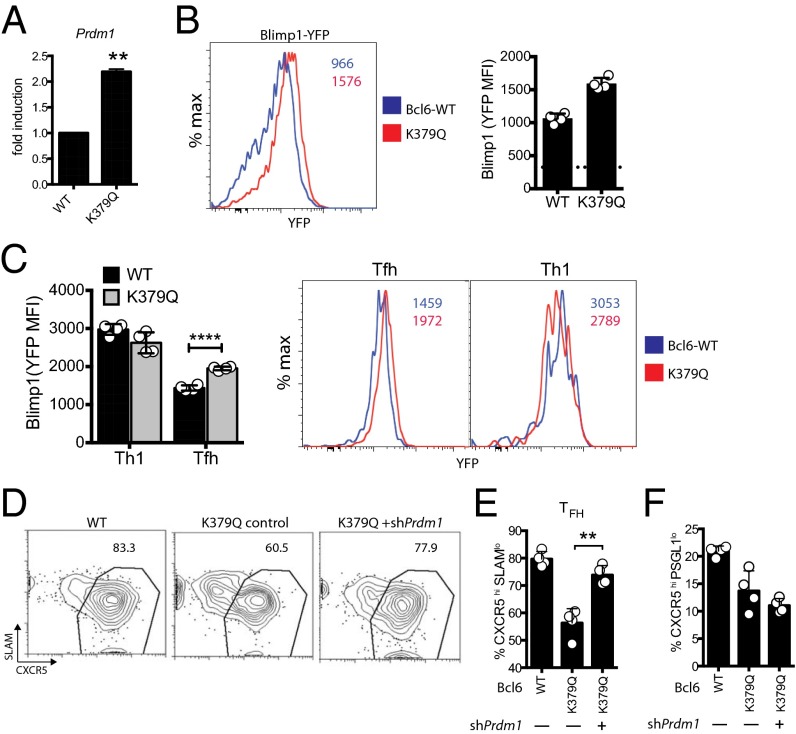
Bcl6 has been shown to be an important inhibitor of the gene Prdm1 during cell fate decisions in T and B lymphocytes. In B cells, acetylation of Lys379 prevents association of Bcl6 with the corepressor MTA3. The MTA3-containing complex mediates repression of key target genes in B cells, including Prdm1 (16). To determine if acetylation of Lys379 regulates Bcl6 repression of Prdm1 in CD4 T cells, Prdm1 gene expression was assessed in GFP+, Bcl6+, or Bcl6 K379Q+Bcl6fl/fl CreCD4 SMARTA CD45.1 cells. RT-PCR analysis revealed derepressed Prdm1 mRNA expression in Bcl6 K379Q+ cells compared with WT Bcl6 (P = 0.0018; Fig. 2A). A Blimp1-YFP reporter also showed Blimp1 derepression in Bcl6 K379Q+Bcl6fl/fl CreCD4 CD4 T cells (Fig. 2B). Blimp1-YFP reporter expression was preferentially increased in Bcl6 K379Q+ Tfh cells (Fig. 2C). ShRNAmir-RV constructs can be used to inhibit gene expression in CD4 T cells in vivo, including Prdm1 (23). To determine if Prdm1 is a major target of the Bcl6 middle domain, we performed a double transduction of Bcl6 K379Q-RV and shCd8-RV (control) or shPrdm1-RV vectors into Bcl6fl/fl CreCD4 SMARTA CD45.1 cells. Double-positive cells were sorted and transferred into Bcl6fl/fl CreCD4 hosts, and Tfh cell populations were analyzed at 6 d following LCMV infection (Fig. 2C). Addition of shPrdm1 rescued Tfh cells (P = 0.0014, CXCR5hiSLAMlo; Fig. 2 D and E). Interestingly, GC Tfh cell differentiation was not rescued (CXCR5hiPSGL1lo; Fig. 2F), indicating that additional gene targets require repression via the Bcl6 middle domain for complete GC Tfh cell differentiation. In summary, inhibition of Prdm1 is one function facilitated by the Bcl6 middle domain.
Fig. 2.
Acetylation of middle domain diminishes the inhibition of Blimp-1 by Bcl6. (A) Transduced Bcl6fl/fl CreCD4 CD45.1+ SMARTA cells were transferred to B6 mice. At 7 d following LCMV infection, RNA was isolated from transduced cells and analyzed for Prdm1 transcript levels. (B and C) Bcl6fl/fl CreCD4 Blimp1-YFP+ SMARTA cells were transduced with GFP, Bcl6, or K379Q RV, and total SMARTA CD4+ T cells (B) or SMARTA Th1 (SLAM+CXCR5−) and Tfh (CXCR5+SLAMlo) cells (C) were analyzed for Blimp1-YFP expression 7 d following acute LCMV infection. Dotted line in the bar graph represents mean fluorescence intensity (MFI) of the YFP-negative control. (D–F) Bcl6fl/fl CreCD4 CD45.1+ SMARTA cells were transduced with GFP, Bcl6, or K379Q RV (GFP) with or without Prdm1shRNA-RV (Ametrine), then transferred to B6 mice and analyzed 7 d following LCMV infection. (E) Tfh cell differentiation (CXCR5hiSLAMlo). (F) GC Tfh cell differentiation (CXCR5hi PSGL1lo). Data shown are representative of at least two independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001).
Acetylation of Middle Domain Inhibits Generation of Tfh Cells Following Protein Immunization.
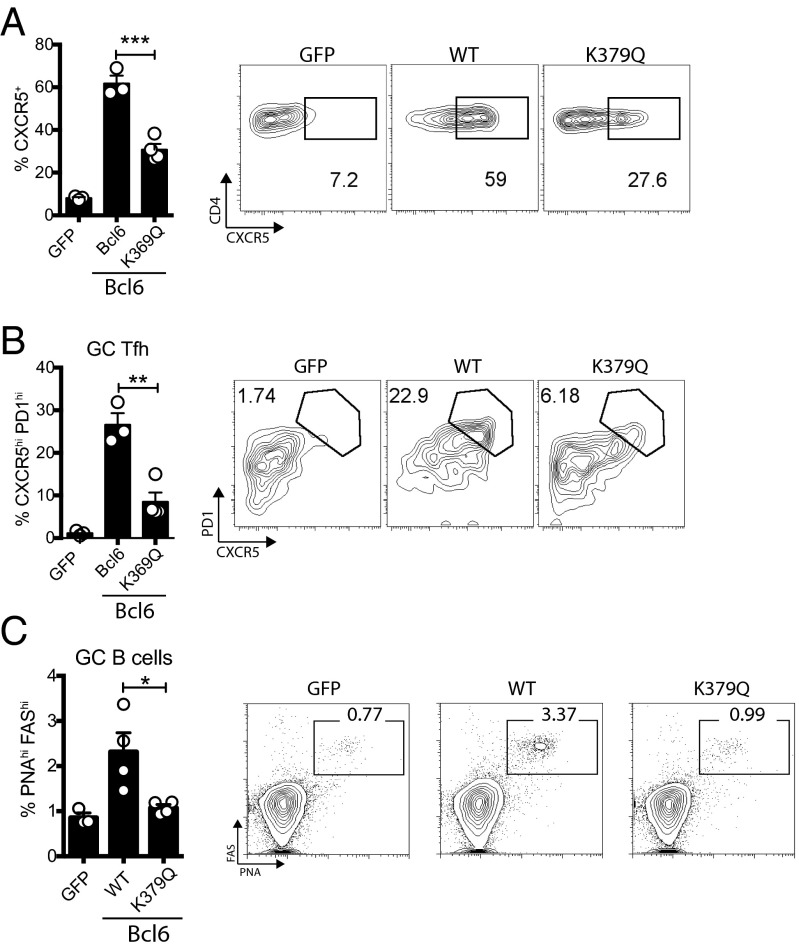
Blimp1 is strongly up-regulated in CD4+ T cells in response to viral infection (2, 11, 24). Following protein immunization, however, Blimp1 is generally minimally induced. Therefore, a protein immunization provides an experimental setting in which Prdm1-independent functions of Bcl6 can be more readily explored. To examine Prdm1-independent Bcl6 function, WT Bcl6-RV+, GFP-RV+, and Bcl6 K379Q-RV+ SMARTA cells were transferred into Bcl6fl/fl CreCD4 hosts immunized with KLH-GP61 in alum. There was a significant decrease in CXCR5+ SMARTA cells (P = 0.0009) as well as GC Tfh cells (P = 0.0032) in the Bcl6 K379Q+ group compared against the WT Bcl6+ group (Fig. 3 A and B). Furthermore, the mice receiving the Bcl6 K379Q+ cells were severely deficient in generating GC B cells (P = 0.0013; Fig. 3C and Fig. S1). Transfer of Bcl6 K379Q+ cells minimally increased the generation of GC B cells compared with mice receiving GFP-RV+ cells, suggesting that the few Tfh cells present are not functional. Together these data indicate that, in addition to repression of Blimp1, acetylation of Bcl6 also likely abrogates the ability of Bcl6 to repress other target genes necessary for Tfh cell differentiation and functions.
Fig. 3.
Acetylation of middle domain inhibits generation of Tfh cells following immunization. Bcl6fl/fl CreCD4 CD45.1+ SMARTA cells were transduced with the indicated RV, transferred into Bcl6fl/fl CreCD4 mice, and analyzed 10 d after immunization with KLH-GP61 in alum. (A) CXCR5+ SMARTA cells. (B) GC Tfh SMARTA cells (CXCR5hiPD-1hi). (C) GC B-cell frequency (FashiPNAhi of CD19+). Data are pooled from four experiments (n = 17–20 per group), normalized to the GFP condition (GFP = 1). Data shown are representative of at least three independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001).
Fig. S1.
Bcl6 middle-domain function in Tfh cells is necessary to support GC formation. Bcl6fl/fl CreCD4 CD45.1+ SMARTA cells were transduced with the indicated RV, transferred into Bcl6fl/fl CreCD4 mice, and analyzed by histology 10 d after immunization with KLH-GP61 in alum. Draining lymph node sections were stained with B220 (blue) and GL-7 (green) antibodies. Representative sections of the lymph nodes are shown. Data are representative of three or four mice per group.
Constitutive Deacetylation of Bcl6 Does Not Augment Tfh Cell Development.
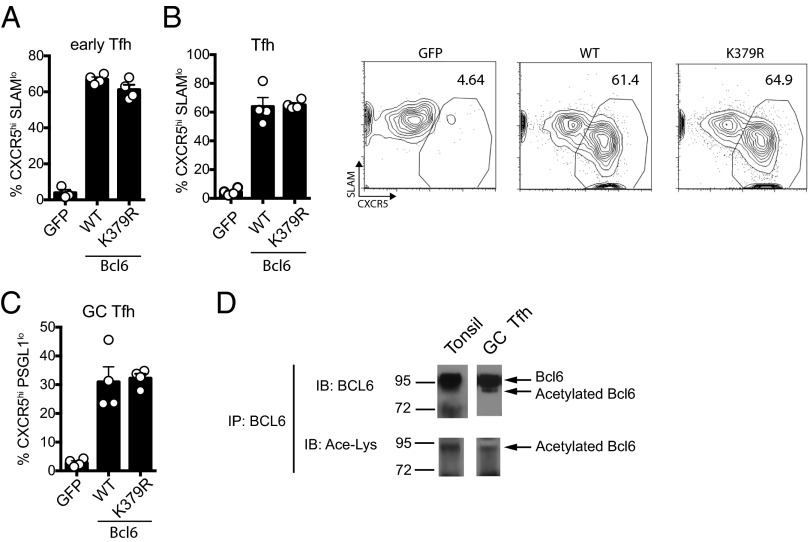
In B-cell lymphoma lines, transcriptional activity of constitutively deacetylated Bcl6 does not differ from endogenous Bcl6, suggesting that the majority of Bcl6 in B cells is deacetylated (15). To determine if this was the case for primary CD4 T cells in vivo, we engineered an acetylation-resistant mutant of Bcl6 in which Lys379 was replaced with arginine (K379R). Bcl6fl/fl CreCD4 SMARTA CD45.1 cells were transduced with WT Bcl6, Bcl6 K379R, or GFP and transferred to Bcl6fl/fl CreCD4 recipients followed by an acute LCMV infection. No differences in day 3 early Tfh cells (Fig. 4A) or day 7 Tfh or GC Tfh cells (Fig. 4 B and C) were observed between WT Bcl6 and acetylation-resistant K379R. These results indicate that the majority of Bcl6 in CD4 T cells is deacetylated and active in vivo. We directly tested whether BCL6 was acetylated in GC Tfh cells in vivo. BCL6 was immunoprecipitated from tonsillar PD-1+ GC Tfh cells followed by probing with anti-BCL6 or anti-acetylated lysine antibodies. A small fraction of BCL6 is acetylated in GC Tfh cells at steady state (Fig. 4D). Thus, at steady state, the majority of Bcl6 remains deacetylated and able to repress gene expression.
Fig. 4.
Constitutive deacetylation does not augment Tfh cell development. (A–C) Bcl6fl/fl CreCD4 CD45.1+ SMARTA cells were retrovirally transduced with empty GFP vector, WT-Bcl6, or K379R-Bcl6 (constitutive deacetylation), then transferred to B6 mice and analyzed 3 d (A) or 7 d (B and C) following acute LCMV infection. (A) Early Tfh cells (CXCR5hiSLAMlo). (B) Tfh cells (CXCR5hiSLAMlo). (C) GC Tfh cells (CXCR5hiPSGL-1lo). (D) BCL6 was immunoprecipitated from total tonsil cells or PD-1+ GC Tfh cells isolated from tonsil followed by immunoblot analysis with anti-BCL6 and anti-acetylated lysine antibodies. Data shown are representative of two (A) or three (B–D) independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001).
Bcl6 K379Q Fails to Recruit the Corepressor MTA3 in Tfh Cells.
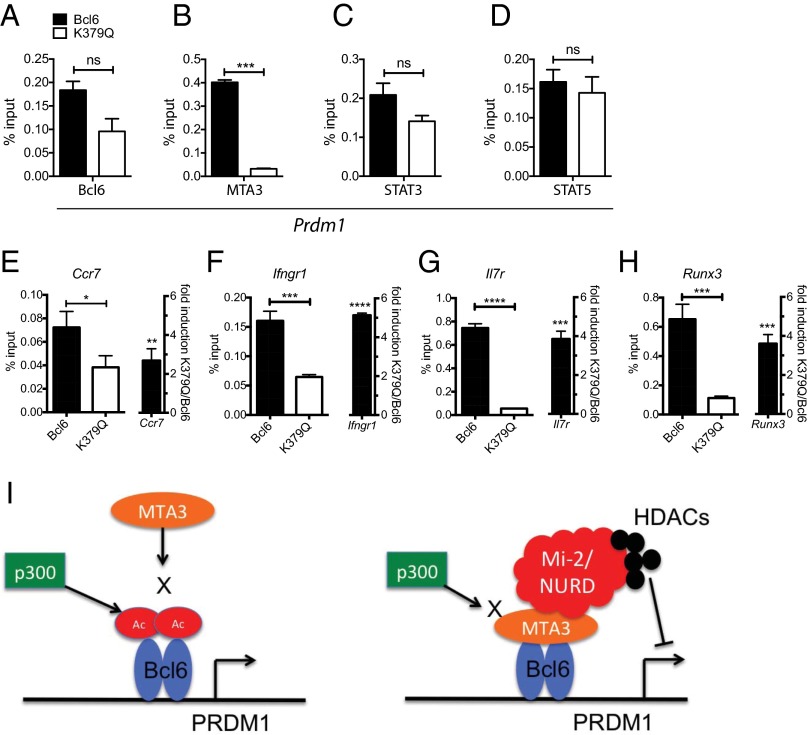
MTA3 is a component of the Mi2/NuRD transcriptional corepressor complex (25, 26). In B cells, this complex can associate with Bcl6 through interaction of MTA3 with the Bcl6 middle domain and represses target genes such as Prdm1 via histone deacetylation activity (16, 27). It was unknown whether Bcl6 acetylation affects the binding of Bcl6 to cis-regulatory elements in target genes, and it was unknown if Bcl6 and MTA3 interact in T cells. To this end, we performed ChIP for Bcl6 and MTA3 occupancy at a conserved Bcl6 response element (BRE) in intron 5 of the Prdm1 gene (28). Modest differences in Bcl6 enrichment between WT Bcl6+ and Bcl6 K379Q+ cells were observed at this locus (P = 0.055; Fig. 5A). However, there was almost a complete loss of MTA3 enrichment in K379Q+ cells (P = 0.0004; Fig. 5B). These results suggest a failure of Bcl6 K379Q to recruit MTA3. Loss of MTA3 recruitment to Prdm1 in K379Q+ cells is most likely not a consequence of differential expression of MTA3 in CD4+ T cells. RNA-Seq of CD4+ T cells 3 d after LCMV infection revealed comparable expression MTA3 in Tfh and Th1 cells (Table S1). Thus, Bcl6 acetylation preferentially abrogates its ability to functionally associate with the corepressor MTA3.
Fig. 5.
Acetylation of Bcl6 prevents association with MTA3. Bcl6 represses Blimp-1 through recruitment of MTA3 and the Mi-2/NURD histone deacetylase complex. (A–F) Bcl6fl/fl CreCD4 SMARTA cells were retrovirally transduced with Bcl6-WT and Bcl6-K379Q, transferred in B6 mice, and isolated by FACS 7 d after LCMV infection. (A–D) Chromatin was prepared and ChIP analyses were performed for Bcl6, MTA3, STAT3, and STAT5 at the Prdm1 promoter. (E–H) (Left) ChIP for MTA3 at Bcl6-binding sites for Ccr7, Ifngr1, Il7r, and Runx3. (Right) qPCR for Ccr7, Ifngr1, Il7r, and Runx3. Data are shown as fold induction of K379Q to WT transduced cells. (I) Model: acetylation of Bcl6 by p300 prevents association with the corepressor MTA3. If Bcl6 is deacetylated, MTA3 is able to bind to the middle domain and recruit the Mi-2/NURD complex. This complex mediates the repression of target genes such as Prdm1 through an HDAC-dependent mechanism. Although MTA3 has been shown to directly bind Bcl6 in B cells, it remains formally possible that the association between MTA3 and Bcl6 in Tfh cells is mediated by an intermediary protein, not represented in the figure. Data shown are representative of at least two independent experiments.
Table S1.
MTA3 expression in CD4 T cells 3 d after LCMV infection
| Gene | Mean Tfh | Mean Th1 | Fold change (Th1/Tfh) | P Value | Adjusted P Value |
| Bcl6 | 1,891.00128 | 306.4076252 | 0.162034594 | 1.81E-18 | 4.57E-16 |
| Mta3 | 1,065.354912 | 1,572.90477 | 1.476413872 | 0.012892308 | 0.126449032 |
| Cxcr5 | 7,203.39112 | 1,278.224348 | 0.177447584 | 2.57E-22 | 1.02E-19 |
| Il2ra | 577.7624698 | 6,922.381051 | 11.98136157 | 3.05E-05 | 0.000911389 |
Gene expression of MTA3 and related genes of interest in Tfh and Th1 SMARTA cells at day 3 after acute LCMV infection. Data from ref. 36.
There has been some evidence of competition between Bcl6 and STATs at cis-regulatory elements, including the Prdm1 gene locus (29, 30). Prdm1 expression is driven by STAT5 in CD4 T cells (9, 31). To determine if Bcl6 acetylation affects STAT competition, STAT3 and STAT5 ChIP assays were performed at Prdm1 BRE. Our results show no significant differences in enrichment of STAT3 or STAT5 between WT Bcl6+ and K379Q+ cells (Fig. 5 C and D), indicating that acetylation of Bcl6 and absence of MTA3 does not affect binding competition between Bcl6 and STATs.
These results suggest that middle-domain acetylation may modulate other important gene targets in Tfh cells in addition to Prdm1. By using ChIP sequencing (6) and gene-expression microarray data (32) from human tonsillar Tfh cells, we identified several Tfh-associated genes that are bound by Bcl6 in Tfh cells: Ccr7, Ifngr1, Il7r, and Runx3. We examined MTA3 occupancy at Bcl6-bound loci for each gene (Fig. 5 E–H). MTA3 occupancy at the BRE of Ccr7, Ifngr1, Il7r, and Runx3 was substantially decreased in Bcl6 K379Q+ antigen-specific CD4 T cells. Furthermore, expression of each of these Bcl6 target genes was increased in Bcl6 K379Q+ CD4 T cells, with Ifngr1, Il7r, and Runx3 exhibiting greater than threefold derepression (Fig. 5 E–H), indicating that these genes are repressed via the activity of the middle domain of Bcl6 (model, Fig. 5I). CCR7 is known to be repressed by Bcl6 (3), and this is required for entry of CD4 T cells into the B-cell follicle (33). IL7R is a critical receptor for survival signals as part of a program to form memory precursors (10), and low IL7R expression may help limit Tfh cell proliferation in GCs. Runx3 and Ifngr1 are both associated with Th1 differentiation. Thus, the Bcl6 middle domain is a regulator of numerous important genes in Tfh cells.
Discussion
Bcl6 is essential for Tfh differentiation (2, 4, 5). A reciprocal antagonistic relationship between Bcl6 and Blimp1 has been revealed in CD4+ T cells, CD8+ T cells, and GC B cells (34). Given the importance of Blimp1 repression in the development of Tfh cells, and that it stands out as a gene highly repressed in Tfh and GC B cells, we asked whether this gene or others are regulated by the Bcl6 middle domain in CD4+ T cells. Bcl6 K379Q+ CD4 T cells lost the majority of their capacity for Tfh cell differentiation and Tfh function in multiple in vivo contexts at all time points examined, clearly demonstrating the importance of the Bcl6 middle domain in the generation of Tfh cells.
Although Blimp1 has been shown to be a repression target of the middle domain in B cells, it is unknown whether the Bcl6 middle domain modulates Blimp1 expression in CD4 T cells. Loss of Bcl6 middle-domain function inhibits the generation of Tfh and GC Tfh cells even in an environment in which Blimp1 is not a major factor. This suggests that, although Blimp1 repression is mediated by the Bcl6 middle domain, there are other gene targets regulated by the Bcl6 middle domain, possibly genes encoding B-cell help molecules in addition to genes involved in Tfh cell differentiation. Indeed, the lack of GC B cells in mice receiving Bcl6 K379Q+ CD4 T cells suggests that even the few Tfh cells that are present are unable to provide help to B cells for GC formation. Thus, Tfh cell function is modulated by the Bcl6 middle domain, as well as Tfh cell differentiation. A recent study showed a defect in GC B-cell formation in Bcl6 middle domain mutant B cells, but no defect in Tfh cell differentiation was observed in Bcl6 middle-domain mutant CD4 T cells (22). However, that study used the triple mutation (i.e., 3Q) in the acetylation motif of the middle domain. In the present study, we demonstrated that Bcl6 3Q+ CD4 T cells failed to survive and proliferate in vivo. This observation, coupled with the original observation that K379 is the primary acetylation target in the middle domain (15), led us to conclude that the 3Q mutation is not a physiological representation of Bcl6 acetylation or function. Recently published work demonstrated that, in the absence of the SWI/SNF complex subunit Srg3, Bcl6 repression of Prdm1 is reduced and Tfh cell differentiation is impaired (35). Importantly, the SWI/SNF complex is known to interact with the members of the NuRD complex. Interestingly, the absence of Srg3 led to a small (twofold) increase in Blimp1 expression, resulting in a larger defect (three- to fourfold reduction) in Tfh cell differentiation. We have observed a similarly moderate dysregulation of Blimp1 expression linked with a substantially larger impairment in Tfh cell differentiation here and in a study of Lef1 and Tcf7 (36). These data suggest that small alterations in Prdm1 expression can substantially affect Tfh cell differentiation. Alternatively, small alterations in Prdm1 expression are indicative of broader changes of Bcl6-regulated genes. We have also shown evidence that Bcl6 middle domain mediates repression of genes other than Prdm1 in Tfh cells. Of the five Bcl6-bound genes that were chosen, all were up-regulated in K379Q+ CD4 T cells, and MTA3 occupancy was greatly reduced. Additionally, these genes represent an array of biological pathways, including migration, cell survival, and Th1 cell differentiation. The derepression of genes associated with the Th1 program (Runx3 and Ifngr1), as well as high expression of SLAM and PSGL-1 in K379Q+ CD4 T cells, indicates that the middle domain of Bcl6 is required to block the Th1 differentiation pathway allowing for CD4 T cells to become Tfh cells.
Taken together, it is concluded that acetylation of the middle domain of Bcl6 prevents Bcl6 association with the corepressor MTA3 and inhibits differentiation and function of Tfh cells by derepression of Prdm1 and other key target genes. When Bcl6 is acetylated, MTA3 is unable to associate with the middle domain (model, Fig. 5I). However, if Bcl6 remains deacetylated, MTA3 is able to associate with Bcl6 and then recruit the Mi-2/NuRD complex, which mediates repression of target genes in an HDAC-dependent manner. Our data indicate that, at steady state, the majority of BCL6 remains deacetylated and able to repress gene expression. Given the importance of Tfh cells in the generation of affinity-matured antibodies and their association with HIV broadly neutralizing antibodies (32), understanding the detailed molecular mechanisms by which Bcl6 governs Tfh cell differentiation and B-cell help may prove valuable for development of new and effective vaccines.
Materials and Methods
Mice.
C57BL/6J (B6) and CreCD4 mice were purchased from the Jackson Laboratory. Bcl6fl/fl (18), CD45.1 congenic, Blimp1-YFP reporter (11), and SMARTA TCR transgenic mice (SM, specific for LCMV gp66-77 on I-Ab) were on a fully B6 background and were bred at the La Jolla Institute (LJI). All animal experiments were conducted in accordance with approved animal protocols by the LJI Institutional Animal Care and Use Committee (IACUC), protocol AP006-SC1-0415.
RVs, Transductions, and Cell Transfers.
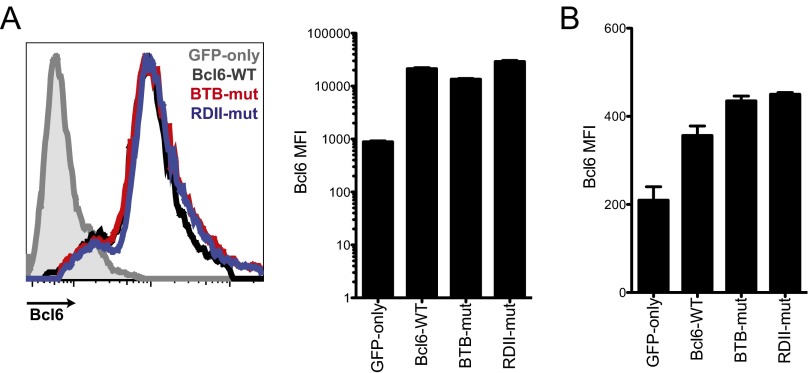
Bcl6 was cloned into the pMIG vector, which contains an IRES-eGFP. Mutant Bcl6 RVs were generated by using site-directed mutagenesis of the WT Bcl6 construct. Middle-domain mutant Bcl6 RV (K379Q) was generated by mutating Lys379 to Gln within the KKYK acetylation motif (amino acids 376–379), resulting in a KKYQ motif that mimics acetylation (15). The acetylation-resistant mutant (K379R) was generated by replacing Lys379 with Arg within the KKYK acetylation motif (amino acids 376–379), resulting in a KKYR motif that cannot be acetylated. Overexpression of Bcl6 was tested in the Plat-E cell line 48 h after transduction and in SMARTA CD4+ T cells 8 d after LCMV infection. Flow-cytometry analysis of Bcl6-overexpressing cells revealed equal overexpression of the WT Bcl6 and of the middle-domain mutant Bcl6 in Plat-E and SMARTA CD4+ T cells (Fig. S2).
Fig. S2.
Mutant Bcl6 RV expression. (A) Plat-E cells were transfected with GFP-only, Bcl6-WT, BTB middle domain mutant (BTB-mut), or RDII middle domain mutant (RDII-mut) plasmid DNA. Bcl6 expression was measured by intracellular staining 48 h after transfection. Histogram overlay shows relative Bcl6 expression. Quantification shows Bcl6 MFI. (B) SMARTA CD4+ T cells were transduced with GFP-only, Bcl6-WT, BTB-mut, or RDII-mut RV. Transduced cells were adoptively transferred into C57BL/6J mice that were subsequently infected with LCMV. Splenocytes were analyzed 8 d after infection. Bcl6 expression in SMARTA TCR transgenic CD4+ T cells was measured by intracellular staining. Quantification shows Bcl6 MFI. Error bars in all graphs depict SEM.
Virions were produced by using the Plat-E cell line as previously described (2). TCR transgenic CD4+ T cells were purified from the splenocytes of naive mice by magnetic bead negative selection (cat. no. 130–090-861; Miltenyi) and resuspended in D-10 [DMEM plus 10% (vol/vol) FCS, supplemented with 2 mM GlutaMAX (Gibco) and 100 U/mL penicillin/streptomycin (Gibco)] with 2 ng/mL recombinant human IL-7 and 50 μM β-mercaptoethanol. A total of 2 × 106 cells per well were stimulated in 24-well plates precoated with 8 μg/mL anti-CD3 (clone 17A2; BioXcell) and anti-CD28 (clone 37.51; BioXcell). After 24 h, cells were transduced then sorted as described previously (2).
Cell transfers into host mice were performed as described previously (2) by i.v. injection via the retro-orbital sinus. Transferred cells were allowed to rest in host mice for 3–5 d before infection or immunization. A total of 5 × 105 transduced SMARTA cell were transferred into each mouse for day 3 analysis, and 25 × 103 transduced SMARTA cells were transferred into each mouse for day 7 analysis. For protein immunization, 5 × 105 cells were transferred into each mouse.
Infections and Protein Immunizations.
Infections were performed by i.p. injection of 0.5–2 × 105 pfu of LCMV Armstrong per mouse. GP61-keyhole limpet hemocyanin (KLH) was prepared in alum and injected as described previously (10). Briefly, LCMV gp61 peptide (GLNGPDIYKGVYQFKSVEFD) was conjugated to maleimide-activated KLH following the manufacturer’s protocol (Pierce). A total of 20 μg gp61-KLH was resuspended in alum for bilateral footpad injections.
Flow Cytometry.
Single-cell suspensions of spleen were prepared by standard gentle mechanical disruption. Surface staining for flow cytometry was done with monoclonal antibodies against SLAM (CD150; BioLegend) and CD4, CD8, CD44, CD62L, CD25, B220, Fas, and GL7 (eBiosciences; Table S2). Stains were done for 30 min at 4 °C in PBS solution supplemented with 0.5% BSA and 0.1% sodium azide unless specified otherwise. CXCR5 staining was done as described previously (2) by using purified anti-CXCR5 (BD Pharmingen or BioXcell) for 1 h, followed by biotinylated anti-rat IgG (Jackson ImmunoResearch), and then by PE-Cy7-labeled streptavidin (eBioscience) at 4 °C in PBS solution supplemented with 0.5% BSA, 2% FCS, and 2% normal mouse serum. Samples were not fixed and were acquired immediately. Intracellular staining for Bcl6 was performed with an Alexa 647-conjugated monoclonal antibody to Bcl6 (clone K112-91; BD Pharmingen) and the FoxP3 ICS kit buffers and protocol (eBioscience).
Table S2.
Flow cytometry antibodies
| Antibody | Company | Clone |
| CD4–BV510 | BioLegend | RM4-5 |
| CD8–PerCP Cy5.5 | eBioscience | 53–6.7 |
| CD44–efluor 450 | eBioscience | IM7 |
| CD62L–PE | eBioscience | MEL-14 |
| SLAM–PerCP Cy5.5 | Biolegend | TC15-12F12.2 |
| PD1–APC | eBioscience | J43 |
| PSGL1–BV421 | BD | 2PH1 |
| CD45.1–efluor 780 | eBioscience | A20 |
| CD19–PerCP Cy5.5 | eBioscience | eBio1D3 |
| B220–efluor 780 | eBioscience | RA3-6B2 |
| Fas–PE | eBioscience | Jo2 |
| PNA–FITC | Vector | N/A |
| GL7–Alexa Fluor 647 | eBioscience | GL-7 |
Histology.
Lymph nodes were embedded in Tissue Tek optimal cutting temperature compound and frozen at −80 C. Lymph node sections 6 μm thick were cut by using a Microm H505E cryostat. For immunofluorescent staining, sections were blocked with 5% skim milk and stained for 45 min in PBS solution containing 0.1% BSA and 1% normal mouse serum. The following Abs were used: GL7 FITC (BD Pharmingen), B220 APC (Ra3-6B2; eBioscience), and DAPI. Sections were imaged with the Axio Scan.Z1 (Carl Zeiss), and ZEN image analysis tools were used.
Quantitative RT-PCR and ChIP-Quantitative PCR.
Primers are listed in Table S3. MTA3 gene expression in Table S1 are from GSE67336 (36). For ChIP, MCC T cells or primary GC Tfh cells were harvested and then cross-linked with 1% formaldehyde. Chromatin was isolated following sonication. Protein G Dynabeads (Life Technologies) were conjugated to antibodies specific to Bcl6 (clones N-3, C-19) and MTA3 (clone 428C2a; Santa Cruz). Rabbit IgG (Santa Cruz) was used as a control. Chromatin was immunoprecipitated by using the conjugated beads, eluted, and reverse cross-linked by using 0.3 M NaCl at 65 °C overnight. Quantitative PCR (qPCR) was performed as described earlier, and sample values were given as a percentage of input.
Table S3.
Primers Used in the Study
| Gene | Forward | Reverse |
| qPCR primers | ||
| Ccr7 | TGT ACG AGT CGG TGT GCT TC | GGT AGG TAT CCG TCA TGG TCT TG |
| Ifngr1 | CTG GCA GGA TGA TTC TGC TGG | GCA TAC GAC AGG GTT CAA GTT AT |
| IL7r | GCG GAC GAT CAC TCC TTC TG | AGC CCC ACA TAT TTG AAA TTC CA |
| Prdm1 | ACA TAG TGA ACG ACC ACC CCT | CTT ACC ACG CCA ATA ACC TCT |
| Runx3 | CAG GTT CAA CGA CCT TCG ATT | GTG GTA GGT AGC CAC TTG GG |
| ChIP primers | ||
| Ccr7 | ACA GCT CAT CTC TCT CAC ACA TT | TTC TCC ACG TCC CTG ACA GT |
| Ifngr1 | AAA AGG GGT GCC AAC GGA TA | TAG GGA CTG GAA GGG CAT GA |
| IL7r | GGG ATT TCC ACT GGT GTG CT | AGG GAA GTT CTT TGC CTG CT |
| Prdm1 | CCA GTA GGC CTT TCA TGG CT | TGC TCA GGT TGA GAA AGC AGT |
| Runx3 | TGA GGT CCA GGT ACA GGG TA | AAG TGT GGC ACC ATG CAC TC |
Statistical Analysis.
Statistical tests were performed by using Prism 5.0 (GraphPad). P values were calculated by two-tailed unpaired Student t tests with a 95% confidence interval. Error bars in figures depict the SEM.
Acknowledgments
We gratefully acknowledge the La Jolla Institute flow cytometry core for cell sorting services. This work was supported by National Institutes of Health National Institute of Allergy and Infectious Diseases Grants R01 AI063107 and U19 AI109976 (to S.C.) and a Fonds de Recherche Santé Québec Postdoctoral Training Award (to S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507312112/-/DCSupplemental.
References
- 1.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroenke MA, et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188(8):3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325(5943):1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31(3):457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Hatzi K, et al. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J Exp Med. 2015;212(4):539–553. doi: 10.1084/jem.20141380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatzi K, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Reports. 2013;4(3):578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 9.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209(2):243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi YS, et al. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol. 2013;190(8):4014–4026. doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi YS, et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34(6):932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13(4):405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nance JP, Bélanger S, Johnston RJ, Takemori T, Crotty S. Cutting edge: T follicular helper cell differentiation is defective in the absence of Bcl6 BTB repressor domain function. J Immunol. 2015;194(12):5599–5603. doi: 10.4049/jimmunol.1500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang JA, Tubo NJ, Gearhart MD, Bardwell VJ, Jenkins MK. Cutting edge: Bcl6-interacting corepressor contributes to germinal center T follicular helper cell formation and B cell helper function. J Immunol. 2015;194(12):5604–5608. doi: 10.4049/jimmunol.1500201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32(4):606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 16.Fujita N, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119(1):75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12(13):1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaji T, et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J Exp Med. 2012;209(11):2079–2097. doi: 10.1084/jem.20120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yusuf I, et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185(1):190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinstein JS, et al. Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood. 2014;124(25):3719–3729. doi: 10.1182/blood-2014-06-582700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poholek AC, et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185(1):313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C, et al. The BCL6 RD2 domain governs commitment of activated B cells to form germinal centers. Cell Reports. 2014;8(5):1497–1508. doi: 10.1016/j.celrep.2014.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R, et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity. 2014;41(2):325–338. doi: 10.1016/j.immuni.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahey LM, et al. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J Exp Med. 2011;208(5):987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: Multiple complexes for many purposes. Biochim Biophys Acta Gene Struct Exp. 2004;1677(1-3):52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Fujita N, et al. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113(2):207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 27.Jaye DL, et al. The BCL6-associated transcriptional co-repressor, MTA3, is selectively expressed by germinal centre B cells and lymphomas of putative germinal centre derivation. J Pathol. 2007;213(1):106–115. doi: 10.1002/path.2199. [DOI] [PubMed] [Google Scholar]
- 28.Tunyaplin C, et al. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173(2):1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 29.Arguni E, et al. JunD/AP-1 and STAT3 are the major enhancer molecules for high Bcl6 expression in germinal center B cells. Int Immunol. 2006;18(7):1079–1089. doi: 10.1093/intimm/dxl041. [DOI] [PubMed] [Google Scholar]
- 30.Diehl SA, et al. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol. 2008;180(7):4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nurieva RI, et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J BiolChem. 2012;287(14):11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locci M, et al. International AIDS Vaccine Initiative Protocol C Principal Investigators Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haynes NM, et al. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179(8):5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 34.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11(2):114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi J, Jeon S, Choi S, Park K, Seong RH. The SWI/SNF chromatin remodeling complex regulates germinal center formation by repressing Blimp-1 expression. Proc Natl Acad Sci USA. 2015;112(7):E718–E727. doi: 10.1073/pnas.1418592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi YS, et al. LEF-1 and TCF-1 orchestrate TFH differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]