Significance
In pollen grains, the known function of the vegetative cell is to extend a pollen tube to transport the two sperm cells to the embryo sac for fertilization. Because the vegetative cell is much larger and more metabolically active than sperm, it has been assumed that the vegetative cell also might supply various components to sperm, but there was no direct evidence to support this hypothesis. We used deletion and promoter exchange constructs to provide direct evidence that the vegetative cell provides protein-encoding transcripts to sperm cells, highlighting a previously unidentified role of the vegetative cell.
Keywords: pollen, sperm, mRNA transport, protein phosphatase 2C, vegetative nucleus
Abstract
An Arabidopsis pollen grain (male gametophyte) consists of three cells: the vegetative cell, which forms the pollen tube, and two sperm cells enclosed within the vegetative cell. It is still unclear if there is intercellular communication between the vegetative cell and the sperm cells. Here we show that ABA-hypersensitive germination3 (AHG3), encoding a protein phosphatase, is specifically transcribed in the vegetative cell but predominantly translated in sperm cells. We used a series of deletion constructs and promoter exchanges to document transport of AHG3 transcripts from the vegetative cell to sperm and showed that their transport requires sequences in both the 5′ UTR and the coding region. Thus, in addition its known role in transporting sperm during pollen tube growth, the vegetative cell also contributes transcripts to the sperm cells.
Pollen grains are derived by stereotypical cell divisions (1, 2). Each male meiotic product (microspore) undergoes an asymmetric mitotic division, which generates a bicellular pollen grain composed of a vegetative cell and a generative cell in which the generative cell is engulfed inside the cytoplasm of the vegetative cell. The generative cell undergoes a second mitosis to generate two sperm cells. The vegetative cell forms the pollen tube that delivers the sperm to the embryo sac. One sperm cell fertilizes the egg to produce the zygote, and the second sperm cell fuses with the central cell to produce the endosperm (3).
Intercellular communication plays an important role in the regulation of plant development (4). Plasmodesmata, microscopic channels that traverse the cell walls of most plant cells, are usually the conduit for intercellular transport in plants (5). Plant sperm are surrounded by their own plasma membrane and by an endomembrane of vegetative cell origin; there is a thin polysaccharide extracellular matrix between these two membranes, but there is no true cell wall comprised of cellulose and callose (6). Although pollen grains lack bona fide plasmodesmata, plasmodesmata-like connections between the sperm and vegetative cell cytoplasm were reported in Nicotiana alata pollen grains (6). In addition, there is a cytoplasmic projection that connects one sperm cell with the vegetative cell nucleus, first observed in cotton (7) and then described in other species (reviewed in ref. 2). Moreover, the two sperm cell membranes are connected to each other through a tetraspanin-enriched microdomain (8). Although all these physical connections presumably ensure that the vegetative nucleus and the sperm cells move in the pollen tube as a unit (known as the “male germ unit”), they also may provide a route for intercellular communication. It has been proposed that small RNAs move from the vegetative cell to sperm cells (9); however, this notion has been challenged (10). Moreover, the reported mechanism of mRNA movement and small RNA movement in sporophytic tissues is different (11, 12). Thus, to date there is no unequivocal evidence of intercellular mRNA communication between the vegetative cell and the sperm cells during pollen development.
In this study we investigated if there is transport between the vegetative cell and sperm cells. While studying ABA-hypersensitive germination3 (AHG3), which encodes a protein phosphatase 2C, we unexpectedly found that the promoter of AHG3 was transcriptionally active in the vegetative cell, whereas a translational fusion protein, AHG3-GFP, driven by the same AHG3 promoter, was localized in sperm. These different localizations suggested that AHG3 transcripts or the AHG3 protein could move from the vegetative cell to sperm cells. Here we provide evidence that AHG3 transcripts move from the vegetative cell to sperm cells and that the transport of AHG3 transcripts requires sequences in both the 5′ UTR and coding region. Our results thus document an additional role for the vegetative cell in providing transcripts to the sperm cells.
Results
The Pollen Transcription Pattern of AHG3 Is Different from Its Protein Pattern.
Protein phosphorylation and dephosphorylation are important mechanisms for modulating protein activity. In the course of experiments to study protein phosphorylation during pollen development, we became interested in a PP2C type of protein phosphatase, AHG3, whose transcripts accumulated in sperm cells (13). AHG3 is a negative regulator of the abscisic acid (ABA) pathway in sporophytic parts of the plant (14, 15). AHG3 expression was ABA-inducible in roots, leaves, inflorescences, and siliques, as judged by ProAHG3:GUS lines (14). According to microarray analysis (13), the expression value for AHG3 was about 10 times higher in sperm cells than in mature pollen. Because other sperm-specific genes (15, 16) exhibited similar expression ratios in microarray experiments, we predicted that in pollen AHG3 might be restricted to sperm cells (i.e., not expressed in the vegetative cell).
AHG3 expression was assayed by quantitative RT-PCR (qRT-PCR) in unicellular microspores, mature pollen, and sperm cells. No AHG3 transcripts were detected by qRT-PCR in unicellular microspores, and their levels in mature pollen were substantially lower than in sperm cells (Fig. 1A). Therefore, in mature pollen, AHG3 transcripts accumulate mainly in sperm cells. To confirm the expression pattern of AHG3 in pollen development, we generated transgenic plants with two constructs, a transcriptional fusion construct, ProAHG3:NLS-3xGFP (native promoter driving three copies of a GFP reporter gene with a nuclear localization sequence), and a translational GFP fusion construct, ProAHG3:geAHG3-GFP (the native promoter sequence driving the AHG3 genomic coding sequence fused to a C-terminal GFP). We obtained 24 T1 lines with the promoter fusion; each mimicked the expression pattern previously reported (14, 15) in sporophytic tissues (Figs. S1 A–F and S2 A and B), indicating that the promoter in this construct was functional. During pollen development, NLS-3xGFP was not detected at unicellular or bicellular stages (Fig. 1 B–E) but was detected in mature pollen and pollen tubes in all 24 T1 lines only in the vegetative cell nucleus and not in sperm cell nuclei (Fig. 1 F–I and Fig. S3A). We obtained 40 T1 lines with the geAHG3-GFP protein fusion construct. To test if the promoter and AHG3-GFP in this construct were functional, ahg3; ProAHG3:geAHG3-GFP and ahg3 control seeds were germinated for 5 d on Murashige and Skoog (MS) medium containing 0.3 uM ABA. The ProAHG3:geAHG3-GFP construct suppressed the ABA hypersensitive phenotype of ahg3 seeds (Fig. S4), showing that the promoter fragment did confer correct expression of AHG3 in sporophytic tissues. No GFP signal was detected in roots, leaves, or inflorescences under normal conditions (Fig. S1 G–R), but geAGH3-GFP expression was observed in nuclei in roots, leaves, and in some flower tissues upon ABA treatment, so AHG3 is an ABA-inducible protein localized in the nucleus (Fig. S2 C–H). In mature pollen and pollen tubes, geAHG3-GFP expression was detected in sperm cell nuclei (Fig. 1 J–Q and Fig. S3B). In addition to the strong signal in sperm cell nuclei, a weak geAHG3-GFP signal was detected in the vegetative cell nucleus of some pollen grains upon prolonged exposure (Fig. S3 C and D), but no signal was detected in sperm cell nuclei for the NLS-3xGFP fusion upon similar prolonged exposure (Fig. S3 E and F). To conclude, in pollen the site of AHG3 transcription is different from AHG3 protein localization.
Fig. 1.
AHG3 transcriptional activity is different from AHG3 protein localization. (A) qRT-PCR analysis of AHG3 transcript levels in unicellular microspores (UNM), mature pollen, and sperm cells (SC). (B–Q) Representative images showing the ProAHG3:NLS-3xGFP (B–I) or the ProAHG3:geAHG3-GFP expression patterns (J–Q) during pollen development. (B, C, J, and K) Unicellular microspores. (D, E, L, and M) Bicellular pollen. (F, G, N, and O) Mature pollen. (H, I, P, and Q) Pollen tubes. B, D, F, J, L, and N are DAPI images. H and P are brightfield images. C, E, G, I, K, M, O, and Q are GFP images. (Scale bars, 10 μm.)
Fig. S1.
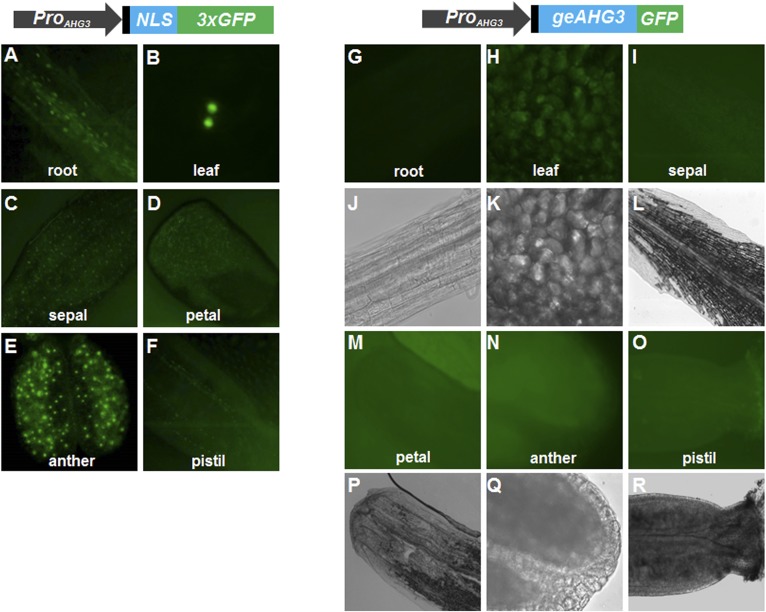
Expression pattern of ProAHG3:NLS-3xGFP and ProAHG3:geAHG3-GFP in different tissues. The 5′ UTR is shown in black. (A–F) Representative images of ProAHG3:NLS-3xGFP expression in root, leaf, and various flower parts. (G–R) Representative images of ProAHG3:geAHG3-GFP expression in root, leaf, and various flower parts. A–F, G–I, and M–O are GFP images. J–L and P–R are brightfield images.
Fig. S2.
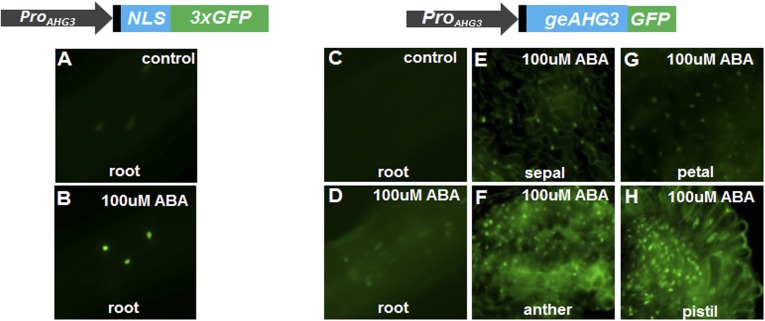
AHG3 expression is ABA-inducible. Seedlings with ProAHG3:NLS-3xGFP or ProAHG3:geAHG3-GFP constructs were cultivated on MS medium containing 100 mM ABA. The 5′ UTR is shown in black. (A and B) ProAHG3:NLS-3XGFP expression in root. (C–H) ProAHG3:geAHG3-GFP expression in root (C and D), sepal (E), anther (F), petal (G), and pistil (H).
Fig. S3.
Localization of geAHG3-GFP and NLS-3xGFP in mature pollen. (A and B) Representative images of pollen expressing ProAHG3:NLS-3xGFP (A) and ProAHG3:geAHG3-GFP (B). (C and E) Representative long-exposure (6-s) image of pollen expressing ProAHG3:NLS-3xGFP (C) or ProAHG3:geAHG3-GFP (E). (D and F) DAPI images corresponding to C and E, respectively.
Fig. S4.
ProAHG3:geAHG3-GFP can suppress the ahg3 ABA hypersensitive seed germination phenotype. (A and B) ahg3 seeds germinate but have no cotyledons (A) or have tiny white cotyledons (B). (C and D) ahg3; ProAHG3:geAHG3-GFP seedlings with green cotyledons. (E and F) Col seedling with green cotyledons. (Scale bars, 1 mm.)
A Combination of Sequences in the 5′ UTR and Coding Region Are Necessary for Transport to Sperm Cells.
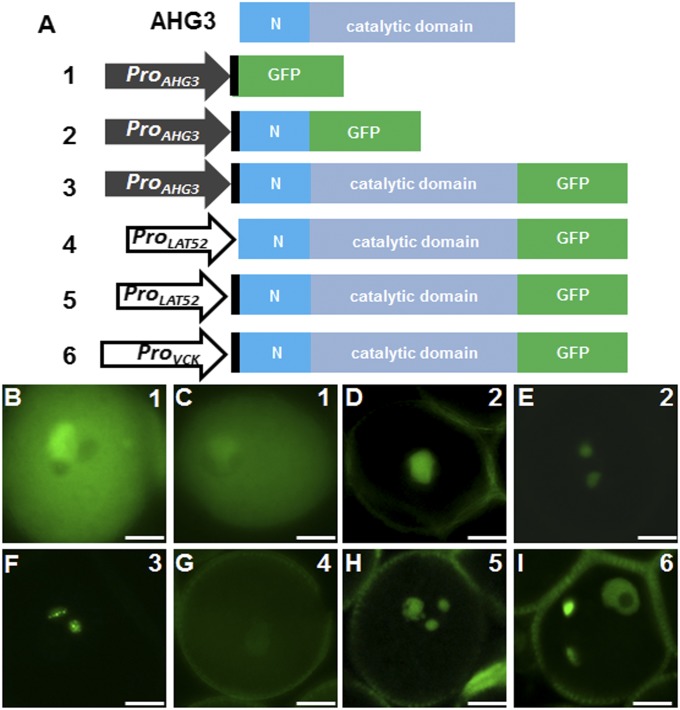
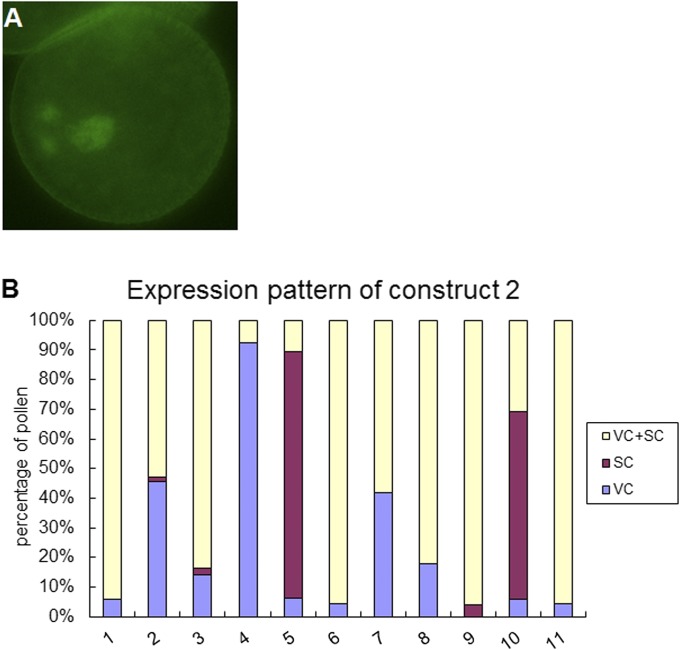
The different expression patterns of ProAHG3:NLS-3xGFP and ProAHG3:geAHG3-GFP in mature pollen suggested transport of transcripts or protein from the vegetative cell to sperm cells. To delimit the regulatory regions required for the transport, we generated two constructs corresponding to different regions of AHG3 (Fig. 2A). In all 12 T1 lines with construct 1, which includes only the promoter and 5′ UTR sequence, the GFP signal was detected in the vegetative cell cytoplasm but not in the sperm cell cytoplasm (Fig. 2 B and C and Table 1). This localization was the same as that seen with ProAHG3:NLS-3xGFP (Fig. 1G), because the two constructs represent equivalent transcriptional fusions. In the 11 T1 lines with construct 2, which includes the sequence of construct 1 and the sequence encoding the N-terminal region of AHG3, the GFP signal was detected in the vegetative cell nucleus (Fig. 2D), in both the vegetative cell and sperm cell nuclei (Fig. S5A), or only in the two sperm cell nuclei (Fig. 2E). In more than half the lines all three patterns could be observed (Fig. S5B). We therefore concluded that transport of AHG3 transcripts required the coding region, that the sequence encoding the N-terminal region in construct 2 was sufficient to assure partial transport, and that the sequence encoding the catalytic domain in construct 3 enhanced transport from the vegetative cell to the sperm cells.
Fig. 2.
A combination of 5′ UTR and coding sequences is necessary for the transport of AHG3 transcripts to sperm cells. (A) AHG3 protein domains and diagram of constructs. The 5′ UTR is shown in black, the specific N terminal sequence is shown in blue, and the catalytic C-terminal domain is shown in gray. (B–I) Representative pollen grains expressing construct 1 (B and C), construct 2 (D and E), construct 3 (F), construct 4 (G), construct 5 (H), and construct 6 (I). (Scale bars, 5 μm.)
Table 1.
Pollen expression pattern of GFP lines
| Name | No. of plants | VC | SC |
| Construct 1 | 12 | +++ | − |
| Construct 2 | 8 | + | ++ |
| 3 | +++ | + | |
| Construct 3 | 40 | − | +++ |
| Construct 4 | 20 | +++ | − |
| Construct 5 | 10 | +++ | ++ |
| Construct 6 | 10 | +++ | ++ |
GFP signals in vegetative cells (VC) and sperm cells (SC) are noted as +, ++, and +++, representing weak, medium, and strong signals, respectively. For construct 2, two different expression patterns observed in transgenic lines are noted on separate lines.
Fig. S5.
Localization of AHG3N-GFP in mature pollen. (A) Representative pollen grains expressing construct 2 in which signal could be detected in both the vegetative cell and the sperm cells. (B) Percentage of localization patterns in pollen of 11 lines expressing construct 2. SC, sperm cells; VC, vegetative cells.
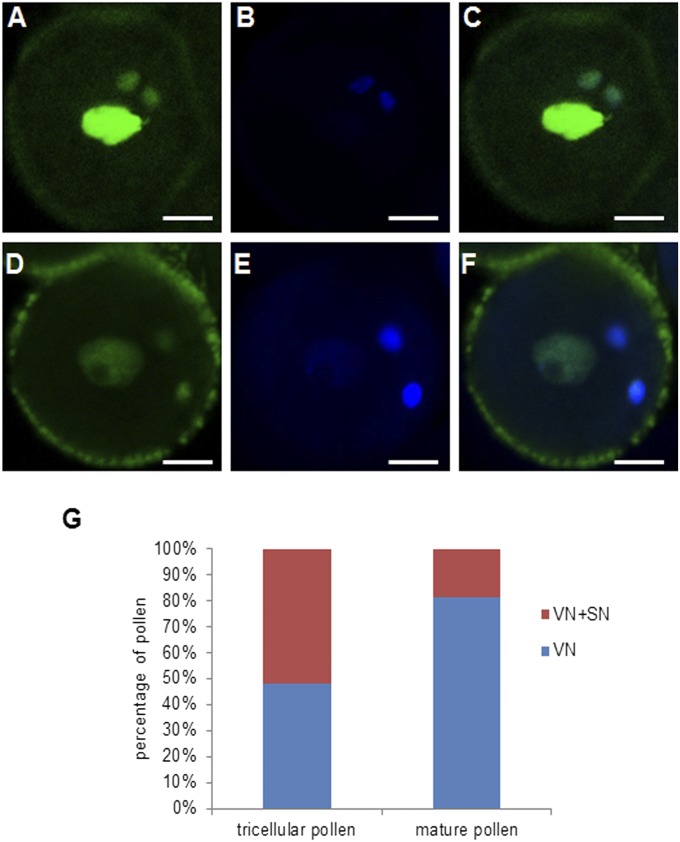
To determine if the coding sequence of AHG3 was sufficient for transport, we first used the LAT52 promoter, which is expressed predominantly in the vegetative cell (10, 17), to drive expression of the AHG3 genomic coding sequence (ProLAT52:geAHG3-GFP; construct 4 in Fig. 2A). The GFP signal in these lines was observed only in the vegetative cell nucleus (Fig. 2G and Table 1) but not in sperm cell nuclei. Thus, the coding DNA sequence (CDS) or genomic sequence alone was not sufficient to drive AHG3 accumulation in sperm cells. Because the AHG3 5′ UTR was not included in construct 4 (Fig. 2A), we tested the requirement of the 5′ UTR for transport. We generated a LAT52 promoter-driven construct that included both the AHG3 5′ UTR and the genomic coding sequence (ProLAT52:5′UTR-geAHG3-GFP, construct 5 in Fig. 2A). Unlike the constructs lacking the 5′ UTR, AHG3-GFP in this line was localized in both the vegetative cell nucleus and the sperm cell nuclei (Fig. 2H). However, because the LAT52p promoter was reported to drive weak expression at the unicellular microspore stage (10), the signal detected potentially could result from carry over from early stages of pollen development to sperm cells. To examine this possibility, we used the VCK promoter, a vegetative cell-specific promoter expressed first in late bicellular pollen (10), to drive expression of AHG3 5′ UTR plus the genomic coding sequence (ProVCK:5UTR-geAHG3-GFP, construct 6 in Fig. 2A). The pattern of GFP in this line was similar to that of lines with construct 5, i.e., AHG3-GFP was localized in both the vegetative cell nucleus and in the sperm cell nuclei. In mature pollen (from open flowers), about 20% of the pollen showed AHG3-GFP in both the vegetative cell nucleus and the sperm cell nuclei (Fig. S6 A–G), whereas at an earlier developmental stage (tricellular pollen from closed flowers) about half of the pollen had the AHG3-GFP signal in both the vegetative cell nucleus and the sperm cell nuclei (Fig. 2M and Fig. S6 D–G). The high proportion of pollen that had sperm cell localization in construct 9 lines strongly supports the hypothesis that sequences in the 5′ UTR are important for AHG3 transcript transport. Together, these data suggest that AHG3 transcripts and not AHG3 proteins are transported from the vegetative cell to sperm cells and that this transport requires sequences in both the 5′ UTR and the coding region of AHG3.
Fig. S6.
Localization patterns of ProVCK:5UTR-geAHG3-GFP. (A–C) Representative images of mature pollen from open flowers. (D–F) Representative images of tricellular pollen from closed flowers. A and D are GFP images. B and E are DAPI images. C and F are merged images. (Scale bars, 5 μm.) (G) Percentage of expression patterns in tricellular and mature pollen of ProVCK:5UTR-geAHG3-GFP–expressing lines. VN, vegetative cell nucleus; SN, sperm cell nuclei.
AHG3 Transcripts Move from the Vegetative Cell to Sperm Cells.
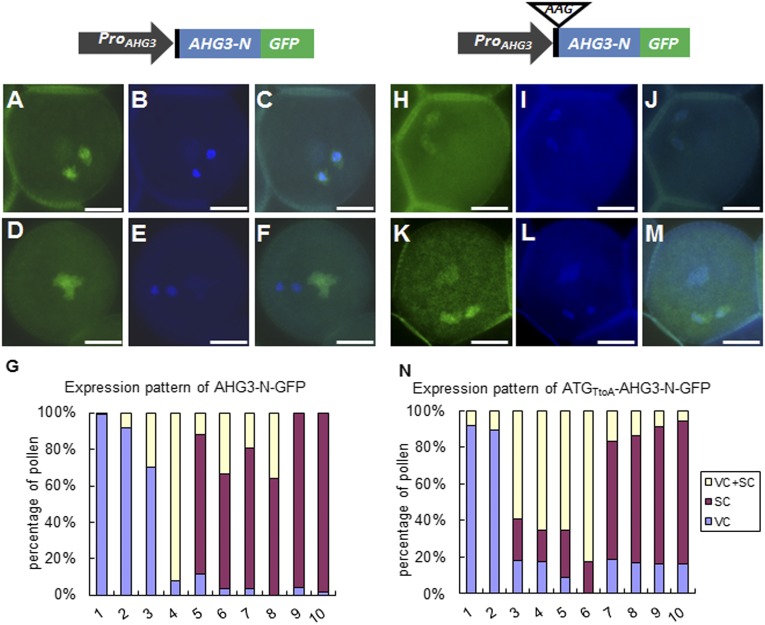
To test the hypothesis that AHG3 transcripts could move from the vegetative cell to sperm cells, we generated two additional constructs in which the AHG3 promoter sequence drove expression of AHG3 (referred to as “AHG3-N”) comparable to construct 2 in Fig. 2A or a version with a mutated AHG3 start codon (referred to as “ATGTtoA-AHG3-N”) (Fig. 3). As shown in Fig. 2A, 225 bp of the coding region of AHG3-N conferred partial ability for transcript movement from the vegetative cell to sperm cells. Because there is no other ATG codon in frame with the GFP in the N-terminal AHG3 region, mutating the ATG codon of AHG3-N will result in the translation of only the GFP, even though both region N and GFP would be transcribed by the AHG3 promoter. The expression pattern was observed in 10 T1 lines (each of the ProAHG3:AHG3-N-GFP and ProAHG3:ATGTtoA-AHG3-N-GFP constructs). In 6 of 10 ProAHG3:AHG3-N-GFP lines, more than 50% of the pollen had only a sperm cell signal, whereas the remaining pollen had a signal either in the vegetative cell or in both the vegetative cell and the sperm cells (Fig. 3G). In three other lines more than 70% of the pollen had a signal only in the vegetative cell, whereas the remaining pollen had a signal in the vegetative cell and in the sperm cells. In the remaining line more than 90% of the pollen had a signal in both the vegetative cell and the sperm cells. In pollen with the ProAHG3:ATGTtoA-AHG3-N-GFP construct, four lines had a cytoplasmic GFP signal in both the vegetative cell and the sperm cells, and four lines had a cytoplasmic GFP signal predominantly in sperm cells (Fig. 3 H–N and Fig. S7 A and B); only two lines showed exclusive vegetative cell expression (Fig. S7C). Although the percentage of pollen with complete sperm cell localization was less than that in pollen expressing ProAHG3:AHG3-N-GFP, the ProAHG3:ATGTtoA-AHG3-N-GFP construct had the capacity to confer sperm cell localization of GFP (Fig. 3N). Together, these data strongly support our hypothesis that AHG3 transcripts are transported from the vegetative cell to sperm cells, where they are locally translated.
Fig. 3.
GFP localization patterns of AHG3-N-GFP and ATGTtoA-AHG3-N-GFP in mature pollen. The 5′ UTR is shown in black, GFP is shown in green, and AHG3 is shown in blue. The construct on the left contains the AHG3 native promoter driving an AHG3 genomic sequence with a C-terminally fused GFP (ProAHG3:AGH3-N-GFP); the construct on the right contains the same sequence except that the start codon in AHG3 was mutated to AAG (ProAHG3: ATGTtoA-AGH3-N-GFP). (A–F) Representative pollen grains expressing ProAHG3:AHG3-N-GFP. (A and D) GFP images. (B and E) DAPI images. (C and F) Merged GFP and DAPI images. (G) Percentage of localization patterns in pollen of 10 independent ProAHG3:AGH3-N-GFP transgenic lines. Each bar shows the distribution of localization patterns in one transgenic line. (H–M) Representative pollen grains expressing ProAHG3: ATGTtoA-AHG3-N-GFP. (H and K) GFP images. (I and L) DAPI images. (J and M) Merged GFP and DAPI images. (N) Percentage of localization patterns in pollen of 10 ProAHG3: ATGTtoA-AHG3-N-GFP transgenic lines. (Scale bars, 5 μm.) SC, sperm cells; VC, vegetative cells.
Fig. S7.
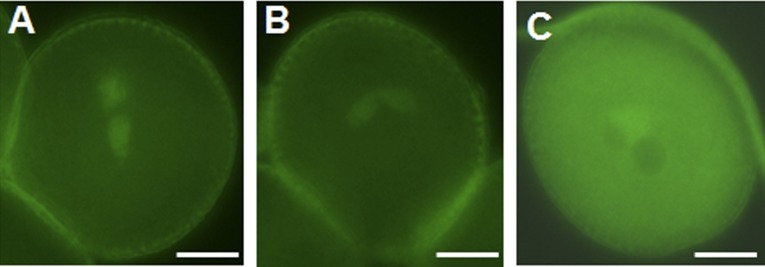
Localization pattern of ATGTtoA-AHG3-A-GFP. (A and B) Representative pollen grains with obvious signal in sperm but no signal in the vegetative cell. (C) Representative pollen grain with a strong signal in the vegetative cell but no signal in the sperm cells. (Scale bars, 5 μm.)
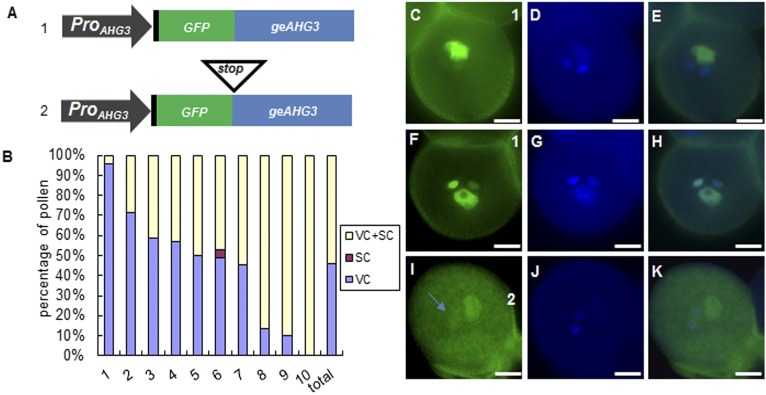
To test this idea further, we generated two constructs in which the AHG3 promoter sequence, including the 5′ UTR, drove expression of GFP in front of the AHG3 genomic sequence (Fig. 4A). The first construct should produce GFP-AHG3, but the second construct should produce only GFP because of a stop codon introduced between the GFP and AHG3 genomic sequences (GFP-stop-AHG3). In the majority of the ProAHG3:GFP-AHG3 T1 lines (9 of 10), we observed a GFP signal in the vegetative cell nucleus or in both the vegetative cell nucleus and sperm cell nuclei (Fig. 4 B and C–H). Interestingly, in pollen that had GFP signals in vegetative cell and sperm cell nuclei, the intensity of the GFP signal in sperm cells was comparable to the sperm cell signal seen in pollen expressing ProAHG3:geAHG3-GFP (Fig. 2F), suggesting that transport of GFP-AHG3 to sperm cell nuclei was not substantially impaired. However, the strong GFP signal in the vegetative cell nucleus implies that the GFP-AHG3 mRNA is transcribed in the vegetative cell nucleus and translated in the cytoplasm; then the protein moves back to the vegetative cell nucleus. This signal clearly differs from the sperm-specific GFP signal in the lines expressing ProAHG3:geAHG3-GFP (Fig. 2F), suggesting that the N-terminal GFP fusion to AHG3 disrupts a signal responsible for translational repression of AHG3 in the vegetative cell. In 10 T1 lines with the GFP-stop-AHG3 construct, the GFP signal was weak, even after prolonged exposure, suggesting that the stop codon triggered nonsense-mediated decay of the transcript (18). Nonetheless, there was some pollen with a weak GFP signal in both the vegetative cell cytoplasm and in sperm cells (Fig. 4 I–K), consistent with the idea that AHG3 transcripts move from the vegetative cell to sperm cells.
Fig. 4.
Localization patterns of GFP-AHG3 and GFP-Stop-AHG3. (A) Diagrams of constructs. The 5′ UTR is shown in black, GFP is shown in green, and AHG3 is shown in blue. (B) Percentage of localization patterns in pollen expressing ProAHG3:GFP-geAHG3. The data represent pollen from 10 independent T1 lines, and the total bar shows the average from 10 T1 plants. SC, sperm cells; VC, vegetative cells. (C–K) Representative images of pollen expressing ProAHG3:GFP-geAHG3 (C–H) or ProAHG3:GFP-Stop-geAHG3 (I–K). C, F, and I are GFP images. D, G, and J are DAPI images. E, H, and K are merged GFP/DAPI images. The arrow in I marks GFP in sperm cell nuclei. (Scale bars, 5 μm.)
Discussion
Here we show that AHG3 transcripts move from the vegetative cell to sperm cells. Although more than 20 mobile endogenous mRNAs have been reported in other tissues in plants (reviewed in ref. 11), intercellular mRNA communication in pollen was not previously documented. We provide multiple lines of evidence to support this claim. (i) The AHG3 protein localized specifically in sperm cells, whereas transcription of AHG3 occurred in the vegetative cell. (ii) Pollen of ProAHG3: ATGTtoA-AHG3N-GFP lines (a nontranslatable fusion of a 225-bp portion of AHG3 to GFP under the control of ProAHG3) also showed predominant sperm cell localization. (iii) Pollen of ProLAT52:geAHG3-GFP lines (only the AHG3 coding sequence under the control of ProLAT52) did not confer sperm cell localization, but pollen of the ProLAT52:5′UTR-geAHG3-GFP and ProVCK:5′UTR-geAHG3-GFP lines (in which the 5′ UTR and AHG3 coding sequence are under the control of ProLAT52 or ProVCK, respectively) had obvious sperm cell localization, showing that the AHG3 coding region alone is not sufficient for mRNA movement but also requires the 5′ UTR sequence. (iv) Both the microarray dataset from sperm cells (13) and our qRT-PCR results (Fig. 1A) indicated substantially higher levels of AHG3 mRNA in sperm cells than in pollen. Together, our data strongly support the idea that the presence of the AHG3 protein in sperm cells is dependent mainly on RNA movement from the vegetative cell to sperm cells and that this mRNA transport depends on both the AHG3 5′ UTR and coding sequence. There is precedent for the requirement of UTRs and coding sequences for RNA movement and subcellular localization. For example, in Saccharomyces cerevisiae, a motif in the 3′ UTR together with three motifs in the coding region are important for movement of ASH1 mRNA (19). Similarly, in Arabidopsis, motifs in both the 3′ UTR and coding region are important for GAI mRNA movement (20).
RNA localization is a mechanism for regulating gene expression posttranscriptionally. In animals, localized RNAs are important determinants of cell fate in eggs and embryos, and such localization allows spatially restricted synthesis of specific proteins in distinct regions of the cytoplasm (21). During animal spermiogenesis, mRNA localization and translation are regulated in a stage-specific manner (22–24). During spermiogenesis, DNA-binding histones are replaced by protamines, resulting in chromatin condensation and cessation of transcription in elongating spermatids (25, 26). Hence, in haploid spermatids, transcription and translation are temporally uncoupled to ensure protein synthesis in transcriptionally silent germ cells. The storage of translationally repressed transcripts is a common phenomenon in haploid spermatids, resulting in temporal differences in the occurrence of mRNAs and their corresponding proteins (27, 28).
In plants, as in animals, the chromatin of sperm cells is highly condensed and contains a male gamete-specific histone H3 that might serve a function similar to that of protamines in animals (29). However, transcriptome studies of isolated sperm cells showed that sperm cells contain thousands of transcripts, consistent with transcriptional activity (13, 30). This idea is supported by the identification of several sperm cell-specific genes, such as GEX1 and DUO1 (31, 32). Nevertheless, the extent of active transcription in plant sperm cells remains to be investigated, because some transcripts identified in the sperm cell transcriptome might have been transported from the vegetative cell during pollen development. By analyzing the incorporation of 5-BrUra into RNA in Hyacinthus pollen, sperm cell nuclei were shown to be transcriptionally active, although at a markedly lower level than in the vegetative cell nucleus (33). AHG3 might represent a class of genes that are transcribed but translationally repressed in the vegetative cell in tricellular pollen and whose mRNAs are transported to sperm cells where they are locally translated. This view is strongly supported by our finding that an N-terminal fusion of GFP to AHG3 caused the accumulation of GFP-AHG3 in the vegetative cell nucleus, possibly by disrupting elements between the 5′ UTR and translational start codon required for the translational repression of AHG3 in the vegetative cell. The mechanisms responsible for translational repression are currently unknown, as are the intercellular routes promoting transport within the male germ unit in pollen. However, AHG3 is most likely not an exceptional case, because it has been shown that ribonucleoprotein particles in pollen contain translationally silent mRNAs (34). These ribonucleoprotein complexes have been interpreted to serve as long-term storage of mRNAs that are transported to the pollen tube tip, along with the translational machinery, during pollen tube growth. Analogously, a subset of these mRNAs might move to and be translated in sperm cells. The mechanisms for mRNA and small RNA movement in sporophytic tissues reported so far differ (11, 12); therefore, we cannot make any assumptions about mechanisms for the transport of small RNAs or mRNAs between the vegetative cell and sperm cells; these mechanisms must be addressed using different experimental strategies. In conclusion, our findings document that the vegetative cell provides protein-encoding transcripts to sperm cells.
Materials and Methods
Seed and Plant Growth Conditions.
Seeds of the ahg3 mutant (CS851888) were obtained from the Arabidopsis Biological Resource Center. Plants were grown in a greenhouse in a 4:1:1 mix of Fafard 4P:perlite:vermiculite under an 18-h light/6-h dark cycle at 21 °C. For ABA treatments of the ProAHG3:NLS-3xGFP and ProAHG3:geAHG3-GFP lines, seeds were germinated on MS medium for 7 d and then were transferred to medium containing 100 μM ABA for 2 d. For complementation tests of the ahg3 mutant, ahg3;ProAHG3:geAHG3-GFP and ahg3 control seeds were germinated on MS medium containing 0.3 μM ABA for 5 d (35). For ABA treatment of inflorescences, inflorescences were cultured as described in ref. 36. Briefly, inflorescences were cut from the plants, and all open flowers were removed. The inflorescences were inserted immediately through a hole in the lid of a microtiter plate into MS medium containing 100 μM ABA. The plates were incubated in a 22 °C growth chamber for 4 d.
Plasmid Constructions.
AHG3 was amplified from Col-0 genomic DNA and cloned into pENTR-D/TOPO, then transferred into a plant expression destination vector modified from pB7FWBG2 (37) (35S promoter removed). The AHG3 CDS was amplified from pollen cDNA with the primers specified in Table S1 and cloned into pENTR-D/TOPO (Invitrogen), then transferred into a plant expression destination vector modified from pZY06 (38). The resulting plasmid was introduced into qrt1 plants (39) by Agro-infiltration (40). For the AHG3 promoter, a 1,472-bp fragment upstream of the ATG codon was subcloned into pENTR-D/TOPO and then transferred into the plant expression vector pGII-NLS3XGFP (15). To generate ProLAT52:5UTR-geAHG3-GFP and ProVCK:5UTR-geAHG3-GFP, we replaced the 35S promoter in pB7FWG2.0 with the VCK promoter (identical to that used in reference 10) and then introduced the 5′ UTR and AHG3 genomic sequence into this vector and the pZY06 vector. To generate ProAHG3:GFP-AHG3 and ProAHG3:GFP-Stop-AHG3, we replaced the 35S promoter in pB7FWG2.0 with the AHG3 promoter and then introduced a wild-type AHG3 genomic sequence or a mutated AHG3 genomic sequence without ATG into this vector. To generate ProAHG3:AGH3-N-GFP and ProAHG3:-AHG3-N-GFP, we introduced a wild-type region N or a mutated (without ATG) region N into the vector.
Table S1.
Primers for cloning
| ProAHG3:NLS-3xGFP and construct 1 | CACCGAACTTAACCCAAATGCCCAC |
| TTGATCTCTAACAAAACTTCTCCAA | |
| ProAHG3:geAHG3-GFP | CACCGAACTTAACCCAAATGCCCA |
| AGACGACGCTTGATTATTCCTC | |
| Construct 1 | CACCGAACTTAACCCAAATGCCCA |
| TTGATCTCTAACAAAACTTCTCCAA | |
| Construct 2 | CACCGAACTTAACCCAAATGCCCA |
| CACATTCGAATCCAGATCTAGA | |
| construct 4 | CACCATGGCTGGGATTTGTTGC |
| AGACGACGCTTGATTATTCCTC | |
| ProAHG3:GFP-GW | CACCGAGCTCGAACTTAACCCAAATGCCCA |
| ACTAGTTTGATCTCTAACAAAACTTCTCCAA | |
| ProAHG3:GFP-AHG3 | CACCATGTGATAAGCTGGGATTTGTTGC |
| AGACGACGCTTGATTATTCCTC | |
| ProAHG3:GFP-Stop-AHG3 | CACCTAGTGATAAGCTGGGATTTGTTGC |
| AGACGACGCTTGATTATTCCTC | |
| ProAHG3:GW-GFP | CACCGAGCTCGAACTTAACCCAAATGCCCA |
| ACTAGTTTGATCTCTAACAAAACTTCTCCAA | |
| ProAHG3:AGH3-N-GFP | CACCATGGCTGGGATTTGTTGC |
| CACATTCGAATCCAGATCTAGA | |
| ProAHG3: ATGTtoA -AGH3-N-GFP | CACCAAGGCTGGGATTTGTTGC |
| CACATTCGAATCCAGATCTAGA | |
| VCK promoter | CACCGAGCTCCCAATCCTTAAGGCATGAGTA |
| CCATACTAGTTTCGTCTCTCTATCGCTTTCTC | |
| 5′ UTR and geAHG3 | CACCTAACCCTTCCATGCGAAAAT |
| AGACGACGCTTGATTATTCCTC | |
| AHG3 Q-PCR | GCTTCTCTTAGACGGAGGTTAG |
| CACTACTGTCTCACGCTTCT | |
| HTA10 Q-PCR | CGAAGAAGGCAACAACAAGAAG |
| GAGAACGGCGGCTAAGTAAA | |
| ahg3 genotyping | GTCCGCAATGTGTTATTAAGTTG (LB) |
| TTCCCCAGCCTGAATTAAGAG (LP) | |
| TTTGGTTGATTTTAGGTTGCG (RP) |
LB, left T-DNA border primer; LP, left genomic primer; RP, right genomic primer.
Microscopic Analysis of Transgenic Lines.
Pollen from closed buds and open flowers of transgenic T1 plants were collected as described in ref. 41. Images were acquired with an Axiovert microscope (Zeiss) using the epifluorescence channel, an AxioCamRM camera, and AxioVision 4.3 software, and a Zeiss confocal microscope LSM 780. Images were processed using Adobe Photoshop 7.
Real-Time PCR.
Purified FACS samples from unicellular microspores, pollen, and sperm cells were isolated as described in ref. 42. Total RNA was extracted using the Qiagen RNeasy Micro Kit according to the manufacturer’s instructions. Expression levels were analyzed using qRT-PCR, using 10 ng of amplified cDNA resulting from aliquots of cRNA synthesized following the Ambion WT Expression kit protocol for hybridization with Affymetrix GeneChip Whole Transcript Expression Arrays. All real-time PCR reactions were performed using the ABI7900 Sequence Detection System (Applied Biosystems), and the amplifications were done using the SYBR Green FastMix ROX (Quanta BioSciences). The relative quantification in gene expression was determined using the 2−ΔΔCt method (43); fold changes in gene expression were normalized to the internal control (HTA10) and relative to the calibrator sample (pollen). There were three biological replicates in three independent experiments.
Acknowledgments
We thank Sarah Chan and Wonsang Tahk, participants in the University of California, Berkeley undergraduate research apprentice program, for technical assistance and Binglian Zheng, Guang Wu, and Peng Qin for useful suggestions and comments on the manuscript. This work was supported by US Department of Agriculture-Agricultural Research Service Current Research Information System Grant 5335–21000–030–00D (to S.M.), by a European Research Council Starting Independent Researcher grant (to C.K.), and by Fundação Ciência e Tecnologia Grant EXPL/BIA-PLA/0244/2013 (to L.C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510854112/-/DCSupplemental.
References
- 1.McCormick S. Control of male gametophyte development. Plant Cell. 2004;16(Suppl):S142–S153. doi: 10.1105/tpc.016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCue AD, Cresti M, Feijó JA, Slotkin RK. Cytoplasmic connection of sperm cells to the pollen vegetative cell nucleus: Potential roles of the male germ unit revisited. J Exp Bot. 2011;62(5):1621–1631. doi: 10.1093/jxb/err032. [DOI] [PubMed] [Google Scholar]
- 3.Twell D. Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod. 2011;24(2):149–160. doi: 10.1007/s00497-010-0157-5. [DOI] [PubMed] [Google Scholar]
- 4.Van Norman JM, Breakfield NW, Benfey PN. Intercellular communication during plant development. Plant Cell. 2011;23(3):855–864. doi: 10.1105/tpc.111.082982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zambryski P, Crawford K. Plasmodesmata: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu Rev Cell Dev Biol. 2000;16:393–421. doi: 10.1146/annurev.cellbio.16.1.393. [DOI] [PubMed] [Google Scholar]
- 6.Cresti M, Lancelle SA, Hepler PK. Structure of the generative cell wall complex after freeze substitution in pollen tubes of Nicotiana and Impatiens. J Cell Sci. 1987;88:373–378. [Google Scholar]
- 7.Jensen WA, Fisher DB. Cotton embryogenesis: The pollen tube in the stigma and style. Protoplasma. 1970;69:215–235. [Google Scholar]
- 8.Boavida LC, Qin P, Broz M, Becker JD, McCormick S. Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo- and heterodimers when expressed in yeast. Plant Physiol. 2013;163(2):696–712. doi: 10.1104/pp.113.216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136(3):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant-Downton R, et al. Artificial microRNAs reveal cell-specific differences in small RNA activity in pollen. Curr Biol. 2013;23(14):R599–R601. doi: 10.1016/j.cub.2013.05.055. [DOI] [PubMed] [Google Scholar]
- 11.Nazim Uddin M, Kim JY. Intercellular and systemic spread of RNA and RNAi in plants. Wiley Interdiscip Rev RNA. 2013;4(3):279–293. doi: 10.1002/wrna.1160. [DOI] [PubMed] [Google Scholar]
- 12.Brosnan CA, Voinnet O. Cell-to-cell and long-distance siRNA movement in plants: Mechanisms and biological implications. Curr Opin Plant Biol. 2011;14(5):580–587. doi: 10.1016/j.pbi.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Borges F, et al. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 2008;148(2):1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chérel I, et al. Physical and functional interaction of the Arabidopsis K(+) channel AKT2 and phosphatase AtPP2CA. Plant Cell. 2002;14(5):1133–1146. doi: 10.1105/tpc.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura N, et al. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007;50(6):935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- 16.Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2010;24(10):1010–1021. doi: 10.1101/gad.1882810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Twell D, Yamaguchi J, McCormick S. Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development. 1990;109(3):705–713. doi: 10.1242/dev.109.3.705. [DOI] [PubMed] [Google Scholar]
- 18.Reddy AS, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25(10):3657–3683. doi: 10.1105/tpc.113.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9(6):333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 20.Huang NC, Yu TS. The sequences of Arabidopsis GA-INSENSITIVE RNA constitute the motifs that are necessary and sufficient for RNA long-distance trafficking. Plant J. 2009;59(6):921–929. doi: 10.1111/j.1365-313X.2009.03918.x. [DOI] [PubMed] [Google Scholar]
- 21.Gonsalvez GB, Long RM. Spatial regulation of translation through RNA localization. F1000 Biol Rep. 2012;4:16. doi: 10.3410/B4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besse F, Ephrussi A. Translational control of localized mRNAs: Restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9(12):971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 23.Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell. 2009;136(4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percipalle P, Raju CS, Fukuda N. Actin-associated hnRNP proteins as transacting factors in the control of mRNA transport and localization. RNA Biol. 2009;6(2):171–174. doi: 10.4161/rna.6.2.8195. [DOI] [PubMed] [Google Scholar]
- 25.Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: What is the link? Hum Reprod Update. 2007;13(3):313–327. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- 26.Gill-Sharma MK, Choudhuri J, D’Souza S. Sperm chromatin protamination: An endocrine perspective. Protein Pept Lett. 2011;18(8):786–801. doi: 10.2174/092986611795714005. [DOI] [PubMed] [Google Scholar]
- 27.Steger K. Haploid spermatids exhibit translationally repressed mRNAs. Anat Embryol (Berl) 2001;203(5):323–334. doi: 10.1007/s004290100176. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda N, et al. The transacting factor CBF-A/Hnrnpab binds to the A2RE/RTS element of protamine 2 mRNA and contributes to its translational regulation during mouse spermatogenesis. PLoS Genet. 2013;9(10):e1003858. doi: 10.1371/journal.pgen.1003858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada T, Endo M, Singh MB, Bhalla PL. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 2005;44(4):557–568. doi: 10.1111/j.1365-313X.2005.02554.x. [DOI] [PubMed] [Google Scholar]
- 30.Anderson SN, et al. Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: Evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 2013;76(5):729–741. doi: 10.1111/tpj.12336. [DOI] [PubMed] [Google Scholar]
- 31.Engel ML, Holmes-Davis R, McCormick S. Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 2005;138(4):2124–2133. doi: 10.1104/pp.104.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rotman N, et al. A novel class of MYB factors controls sperm-cell formation in plants. Curr Biol. 2005;15(3):244–248. doi: 10.1016/j.cub.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Zienkiewicz K, Suwinska A, Niedojadło K, Zienkiewicz A, Bednarska E. Nuclear activity of sperm cells during Hyacinthus orientalis L. in vitro pollen tube growth. J Exp Bot. 2011;62(3):1255–1269. doi: 10.1093/jxb/erq354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honys D, et al. Cytoskeleton-associated large RNP complexes in tobacco male gametophyte (EPPs) are associated with ribosomes and are involved in protein synthesis, processing, and localization. J Proteome Res. 2009;8(4):2015–2031. doi: 10.1021/pr8009897. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida T, et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140(1):115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnard JL, Yang M, Chen YCS, Leary M, McCormick S. The Arabidopsis gene tardy asynchronous meiosis is required for the normal pace and synchrony of cell division during male meiosis. Plant Physiol. 2001;127(3):1157–1166. [PMC free article] [PubMed] [Google Scholar]
- 37.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, McCormick S. A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104(47):18830–18835. doi: 10.1073/pnas.0705874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preuss D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science. 1994;264(5164):1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- 40.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Johnson-Brousseau SA, McCormick S. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 2004;39(5):761–775. doi: 10.1111/j.1365-313X.2004.02147.x. [DOI] [PubMed] [Google Scholar]
- 42.Borges F, et al. FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods. 2012;8(1):44. doi: 10.1186/1746-4811-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]