Significance
Over 3,500 patients receive life-saving lung transplants every year. Nonetheless, complications due to infection and rejection occur frequently and undermine the long-term benefits of lung transplantation. Although clinicians strive to carefully monitor patients, diagnostic options are often limited. Rejection monitoring currently relies on invasive tissue biopsies, and tests of infection are predominately limited to testing one pathogen at a time. This manuscript describes a noninvasive assay based on sequencing of circulating cell-free DNA that simultaneously enables diagnosis of rejection and broad screening of infections.
Keywords: organ transplantation, cell-free DNA, infection, rejection, diagnosis
Abstract
The survival rate following lung transplantation is among the lowest of all solid-organ transplants, and current diagnostic tests often fail to distinguish between infection and rejection, the two primary posttransplant clinical complications. We describe a diagnostic assay that simultaneously monitors for rejection and infection in lung transplant recipients by sequencing of cell-free DNA (cfDNA) in plasma. We determined that the levels of donor-derived cfDNA directly correlate with the results of invasive tests of rejection (area under the curve 0.9). We also analyzed the nonhuman cfDNA as a hypothesis-free approach to test for infections. Cytomegalovirus is most frequently assayed clinically, and the levels of CMV-derived sequences in cfDNA are consistent with clinical results. We furthermore show that hypothesis-free monitoring for pathogens using cfDNA reveals undiagnosed cases of infection, and that certain infectious pathogens such as human herpesvirus (HHV) 6, HHV-7, and adenovirus, which are not often tested clinically, occur with high frequency in this cohort.
Despite limited success with the procedure until the 1990s, lung transplantation is now routinely offered to patients with a variety of end-stage pulmonary diseases (1). Over 3,500 lung transplants are performed annually worldwide (2). However, clinical outcomes remain poor, with median survival (5.3 y) lagging that of other solid-organ transplants (11 y for heart and 8.5 y for liver) (3–5). Complications frequently occur after lung transplantation and can be broadly divided into four classes (6): (i) complications related to injuries sustained during the transplant procedure, defined as primary graft dysfunction (PGD); (ii) acute cellular rejection, which occurs in ∼35% of recipients within the first year posttransplant (2); (iii) chronic rejection, the leading cause of mortality, clinically defined as chronic lung allograft dysfunction (CLAD); and (iv) infectious complications due to both immunosuppression as well as the direct interface between the transplanted lungs and the external environment (7).
Although clinicians strive to carefully monitor for these complications, diagnostic options are limited. The current gold standard for diagnosis of acute rejection is the transbronchial biopsy, an invasive procedure with limited predictive value (8). Diagnosis of infection requires the treating physician to correctly identify the source and to order a battery of pathogen-specific tests. Finally, posttransplantation infection and rejection often present with similar symptoms, such as shortness of breath, and standard clinical tests often fail to distinguish between these two common complications. With these challenges in mind, we developed and evaluated a diagnostic assay that simultaneously tests for rejection and infection by sequencing of circulating cell-free DNA (cfDNA). Rejection is monitored by quantifying cell-free donor-specific DNA (9) (cfdDNA) in the transplant recipient’s plasma via shotgun sequencing (genome transplant dynamics; GTD). Infectious pathogens are identified by simultaneously identifying nonhuman cfDNA sequences and comparing them with known genomic databases of bacterial, viral, and fungal pathogens.
By using single-nucleotide polymorphisms (SNPs) to discriminate between donor and recipient DNA molecules, GTD provides a noninvasive yet direct measure of graft damage (10). One would expect such an approach to be universally applicable, but few organs have been studied in depth. A previous prospective study by our group determined that GTD accurately detects acute rejection after heart transplantation [area under the curve (AUC) 0.83] (11). In the current work, we extend the application of the GTD assay to lung transplantation, making it the first such test, to our knowledge, for transplant rejection that is applicable to multiple organs. Through comparison with evidence from transbronchial biopsy, we find that cfdDNA is informative of acute and chronic rejection after lung transplantation (AUC 0.9, sensitivity 1.0, and specificity 0.73 at a cfdDNA threshold level of 1%). We moreover demonstrate that cfdDNA levels correlate well with clinical indicators of graft dysfunction, including pulmonary function test results.
Through comparison of donor DNA levels with clinical tests for cytomegalovirus (CMV), we found evidence of significant graft injury during CMV infection, a finding that supports the bidirectional relationship between CMV infection and allograft injury reported in the transplant literature (12, 13): CMV reactivation can be triggered during acute rejection, and CMV infection in turn can trigger allograft rejection. This observation underscores the importance of simultaneous rejection and infection monitoring to guide clinical treatment. We therefore evaluated whether cell-free DNA sequencing data, obtained to measure graft injury through donor-derived DNA, is informative of infection as well as rejection, that is, whether pathogen-derived sequences of cfDNA can be identified. We found that clinical tests for CMV infection correlated well with the level of CMV-derived cell-free DNA (AUC 0.91). We then used hypothesis-free screening of pathogen-derived cell-free DNA to identify undiagnosed cases of infection within our study cohort.
Results
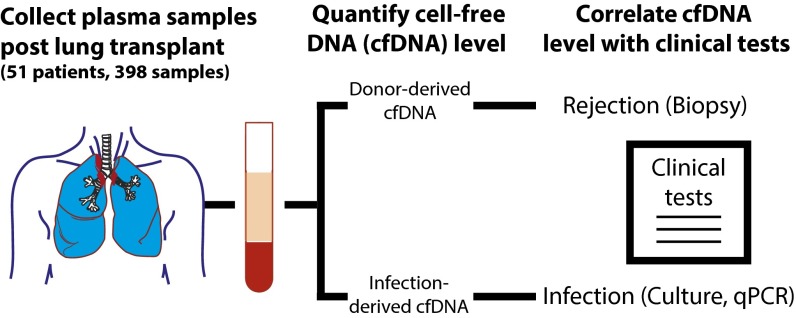
We performed a prospective cohort study to evaluate the performance of sequencing-based analysis of cell-free DNA for the simultaneous monitoring of rejection and infection after lung transplantation. Fifty-one patients were enrolled while awaiting transplantation (Fig. 1 and Table S1), and 398 serial plasma samples were collected at specified time points after the transplant procedure. Following purification and sequencing of cell-free DNA (mean sequencing depth 24 million fragments, 1 × 50 bp or 2 × 100 bp), human- and nonhuman-derived sequences were isolated and analyzed with respect to clinical metrics of graft injury and infection, respectively.
Fig. 1.
Patient recruitment, sample collection, and comparison with clinical indicators of rejection and infection. We performed a prospective cohort study to evaluate the performance of the GTD cell-free DNA assay after lung transplantation with simultaneous infection monitoring. Fifty-one patients were recruited (44 bilateral and 7 single-lung) while awaiting transplantation, and 398 plasma samples were collected longitudinally at prespecified intervals posttransplant. For each sample, human- and nonhuman-derived cell-free DNA sequences were identified and analyzed with respect to clinical metrics of graft rejection (108 transbronchial biopsies, tests of pulmonary function, and diagnosis of CLAD and AMR) and infection, respectively.
Table S1.
Lung transplant recipient demographic characteristics
| Characteristic | |
| Adult recipients, n | 51 |
| Age at time of transplant, y | |
| Mean ± SD | 53 ± 17 |
| Range | 11–77 |
| Male sex, no. (%) | 32 (63) |
| Race or ethnic groups, no. (%) | |
| White | 41 (80) |
| Black | 4 (8) |
| Asian or Pacific Islander | 4 (8) |
| Other | 2 (4) |
| Ethnicity, no. (%) | |
| Hispanic/Latino | 4 (8) |
| Non-Hispanic/non-Latino | 47 (92) |
| Indication for lung transplantation,* no. (%) | |
| COPD | 14 (27) |
| IPF | 13 (25) |
| Cystic fibrosis | 12 (24) |
| UIP | 8 (16) |
| ILD | 8 (16) |
| Bronchiectasis | 5 (10) |
| S/P prior lung transplant | 3 (6) |
| Alpha-1 AD | 1 (2) |
| OB | 1 (2) |
| Sarcoidosis | 1 (2) |
| NSIP | 1 (2) |
| Tobacco use | 1 (2) |
| Other | 3 (6) |
| Type of lung transplanted | |
| Both | 44 (86) |
| Left | 6 (12) |
| Right | 1 (2) |
| Cytomegalovirus status, no. (%) | |
| Donor and recipient positive | 23 (46) |
| Donor and recipient negative | 6 (12) |
| Donor positive and recipient negative | 11 (21) |
| Donor negative and recipient positive | 11 (21) |
| Hospitalization status, no. (%) | |
| Inpatient, intensive care | 6 (12) |
| Inpatient, not intensive care | 6 (12) |
| Outpatient | 39 (76) |
| Hemodialysis, no. (%) | |
| Panel reactive antibodies > 20%, no. (%) | 29 (57) |
| Diabetes at time of transplant, no. (%) | 7 (14) |
| History of hypertension, no. (%) | 7 (14) |
| History of hyperlipidemia, no. (%) | 18 (35) |
| Lung allocation score, no. (%) | |
| 0–20 | 0 |
| 21–40 | 22 (43) |
| 41–60 | 20 (39) |
| 61–80 | 3 (6) |
| 81–100 | 6 (12) |
| Maintenance immunosuppression† | |
| Cyclosporine, no. (%) | 2 (4) |
| Tacrolimus, no. (%) | 51 (100) |
Alpha-1 AD, Alpha 1-antitrypsin deficiency; COPD, chronic obstructive pulmonary disease; ILD, interstitial lung disease; IPF, idiopathic pulmonary fibrosis; NSIP, nonspecific interstitial pneumonia; OB, obliterative bronchiolitis; S/P prior lung transplant, status/post prior lung transplant; UIP, usual interstitial pneumonia.
Some patients were diagnosed with concurrent indications.
Includes all agents used during the study period.
Quantifying Donor-Derived DNA.
The human fraction of cell-free DNA was a mixture of donor- and recipient-derived sequences. These sequences were distinguished based on SNP genotype information (10, 11), which was measured via SNP arrays performed on donor and recipient whole-blood samples collected before transplantation. After counting the number of recipient and donor molecules, the fraction of donor-derived DNA was computed. The rate of erroneous donor or recipient assignments was estimated for each sample independently, and donor levels were corrected based on the measured assignment error rates as described previously (SI Materials and Methods) (11). Samples with high measured technical error rate, indicating technical problems or issues with sample cross-contamination, were excluded from analysis (error rate > 0.5%, n = 5).
Organ-Specific Features in Early and Late Dynamics.
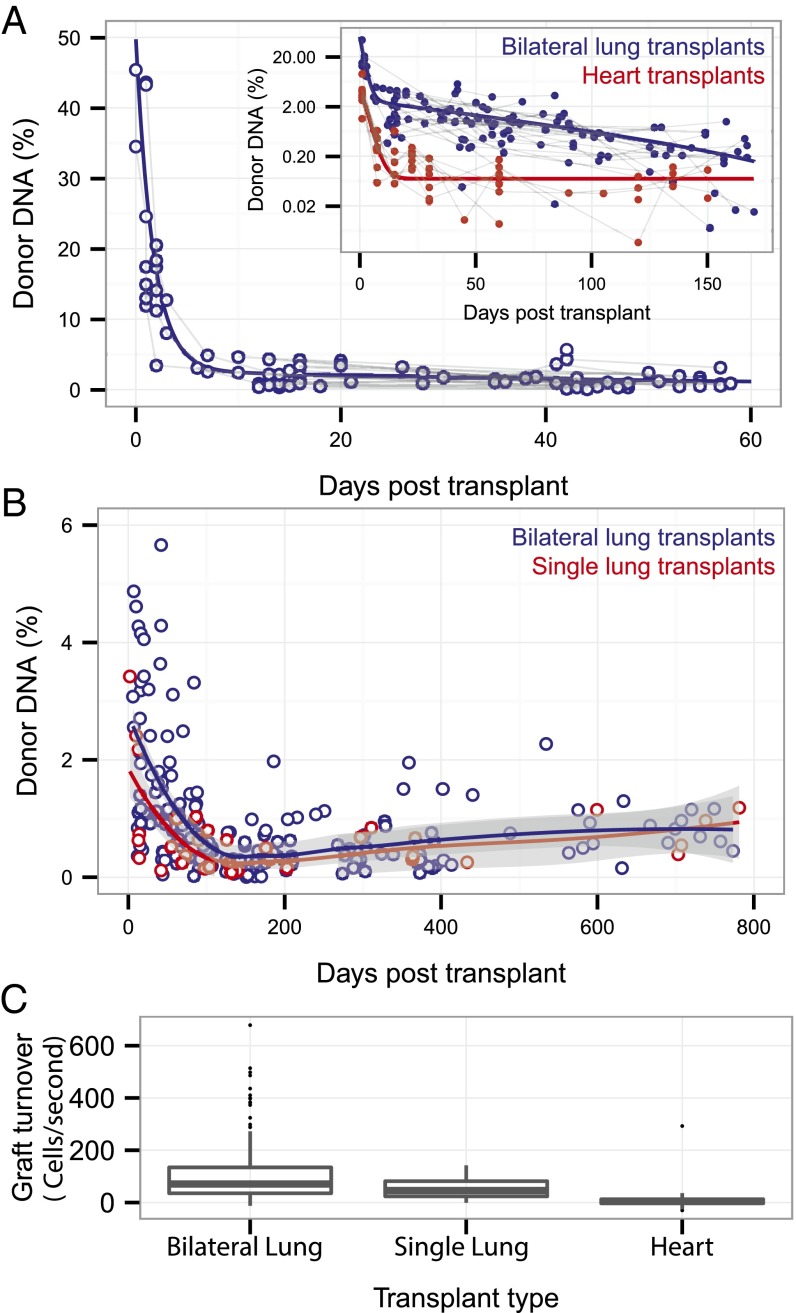
We first examined cfdDNA levels in lung transplant recipients without evidence of allograft rejection or infection (n = 240; Fig. 2). We observed changes in cfdDNA levels throughout the posttransplant time course that differ from the cfdDNA levels seen following heart transplantation (Fig. 2A, Inset; heart transplant data are from ref. 10). After either lung or heart transplantation, highly elevated cfdDNA levels are observed during the first few days posttransplant (26 ± 14% and 3.8 ± 2.3% on the first postoperative day for bilateral lung and heart transplants, respectively), and the levels subsequently decrease over time. Whereas the data for heart transplants were well-described using a model that assumes single-decay kinetics (single exponential), the data for lung transplants were best described by a model that assumes two-step decay kinetics (bilateral transplants, double exponential, R-squared measure of the goodness of fit 0.89 and 0.47 for double- and single-exponential fits, respectively; Fig. S1). cfdDNA levels measured after lung transplantation remained elevated compared with heart transplants throughout the posttransplant course. Moreover, a small but significant increase in cfdDNA levels in the absence of rejection or infection was observed starting at 3 mo posttransplant (Fig. 2B; single and bilateral transplants, ρ = 0.36, P = 3.10−5, Spearman rank correlation). This indicator of ongoing tissue damage is consistent with a gradual loss of lung function, a clinical hallmark of postlung transplant morbidity.
Fig. 2.
Early and late posttransplant cell-free DNA dynamics and organ-specific features. (A) Fraction of donor-derived cell-free DNA during the first 2 mo posttransplant for rejection-free lung transplant recipients. Donor cfDNA levels were higher for lung than for heart transplant recipients. (Inset) Data for heart transplants from ref. 11. (B) Donor cfDNA levels measured for single- and double-lung transplants throughout the posttransplant course in rejection-free patients. Solid lines are fits (local polynomial regression, smoothing parameter alpha 0.75; gray shading indicates 90% confidence interval). (C) Estimate of graft cell-decay rate from measurements of the fraction of donor cfDNA. Data are shown as boxplots. The bottom and top of the box indicate the upper and lower quartile, the band inside the box indicates the median.
Fig. S1.
Model fits of cfdDNA early dynamics. (A) Comparison of double- (blue line) and single-exponential (black line) fit of cfdDNA levels recorded for lung transplant recipients as a function of time posttransplant (blue dots). The double-exponential fit is better at capturing the time dynamics at later time points. (B) Weighted residuals (weighted by the square of the cfdDNA levels) for the single- (black line) and double-exponential model (blue line) as function of time posttransplant. The R-squared measure of the goodness of the fit with weighted residuals is 0.89 for the double-exponential model and 0.47 for the single-exponential model.
Tissue Mass Dependence and Estimates of Tissue Turnover Rate.
We quantified cfdDNA after single and bilateral lung transplantation and observed a correlation between cfdDNA levels and transplant tissue mass. In conjunction with a previously measured median clearance rate [median half-life 22 min (14)] and load (mean 30 ng/µL) of cell-free DNA in plasma, we used the cfDNA measurement to estimate the allograft cellular turnover rate (Fig. 2C). The measured cell turnover rate was higher in bilateral than in single-lung transplants (mean turnover 107 and 58 cells per s for bilateral and single-lung transplants, respectively) and greater for lung than heart transplants (mean turnover rate 8 cells per s for heart transplants, samples collected >14 d after transplant in the absence of rejection), consistent with the larger mass of transplanted lungs relative to hearts and the greater turnover rate of lung versus heart cells (15, 16). In the calculations that follow, we account for differences in bilateral versus single-lung transplant tissue mass by multiplying the cfdDNA level for single-lung transplants by a factor of 2.
cfdDNA Levels During Acute Rejection.
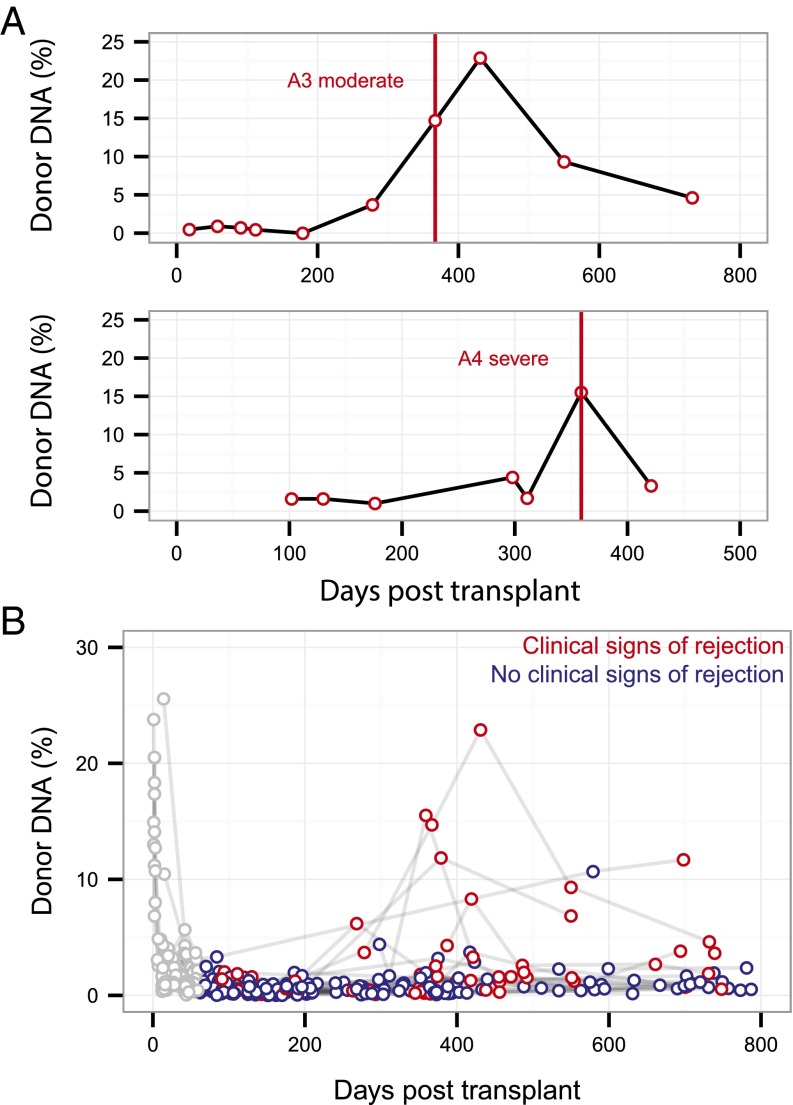
After correcting for allograft tissue mass, we examined cfdDNA levels in the plasma of lung transplant recipients diagnosed with acute or chronic rejection. Representative time courses for individual patients diagnosed with moderate or severe acute cellular rejection by biopsy (Fig. 3A) show cfdDNA levels that are markedly elevated from baseline (cfdDNA 14.7% and 15.2%, respectively) and, in one case, the cfdDNA level continued to increase following diagnosis and rejection treatment (cfdDNA 22.9%, 64 d postdiagnosis). An aggregate of the data collected on 51 lung transplant recipients further demonstrates elevated cfdDNA levels detected during acute rejection events (red points, Fig. 3B) relative to samples collected in the absence of rejection (blue points, Fig. 3B).
Fig. 3.
Cell-free donor DNA at rejection. (A) Donor cfDNA level measured for patients diagnosed with a moderate or severe acute rejection event, with vertical lines indicating rejection diagnosis via transbronchial biopsy. (B) Overview of all data collected for 51 lung transplant recipients (n = 398 samples) highlighting points measured in the absence (blue) and presence (red) of clinical signs of rejection. Data collected during the first 2 mo posttransplant are shown in gray, with early time dynamics excluded from signal analysis.
Analysis of Test Performance.
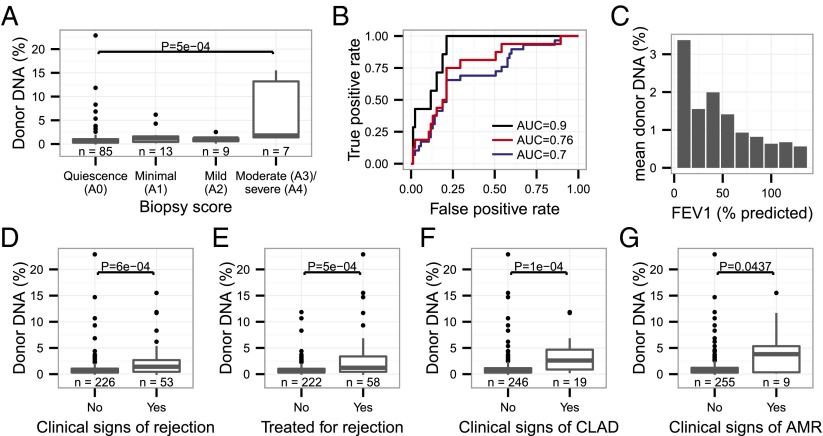
We next evaluated whether cfdDNA levels can predict acute and chronic rejection. Given the uniformly elevated levels of cfdDNA observed during the initial 60 d after lung transplantation for all patients, these early time points were excluded from analysis (gray points, Fig. 3B). We examined the concordance of cfdDNA with transbronchial biopsy grades where available (n = 113). The level of cfdDNA was significantly higher for samples collected at moderate or severe rejection (≥A3) than for samples collected at quiescence (A0) (P = 5.10−4, Mann–Whitney U test; Fig. 4A). A receiver-operating characteristic (ROC) analysis of the performance of cfdDNA in classifying moderate-to-severe acute rejection (≥A3) versus the absence of rejection (grade A0) yielded an area under the curve of 0.9 (sensitivity 1 and specificity 0.73 at a cfdDNA threshold level of 1%; Fig. 4B). An ROC analysis of the performance of cfdDNA in distinguishing rejection-free patients and patients with mild-to-severe rejection yielded an AUC of 0.76. Last, an ROC analysis of the performance of cfdDNA in distinguishing rejection-free and patients with minimal-to-severe rejection yielded an AUC of 0.70.
Fig. 4.
Analysis of the performance of cfdDNA as a marker for lung transplant rejection. (A) cfdDNA levels by transbronchial biopsy score. (B) ROC analysis of the performance of cfdDNA in classifying rejection-free versus rejecting patients (black line, A0 vs. A3–A4, moderate-to-severe rejection, AUC 0.9). Also shown are ROC analyses of the test performance in distinguishing rejection-free patients and patients with mild-to-severe rejection (red line, A0 vs. A2–A4, AUC 0.76) and the performance in distinguishing rejection-free and patients with minimal-to-severe rejection (blue line, A0 vs. A1–A4, AUC 0.7). (C) cfdDNA levels versus FEV1 level (% predicted given age, sex, and body composition). (D–G) cfdDNA levels by (D) the presence or absence of clinical signs of acute rejection, (E) decision to treat for acute rejection, (F) CLAD, and (G) clinical signs of AMR. Data are shown as boxplots in A, D–G. The bottom and top of the box indicate the upper and lower quartile, the band inside the box indicates the median.
We further compared levels of cfdDNA against an array of clinical metrics that are used to guide treatment decisions, including pulmonary function tests (spirometry), clinical signs of rejection, rejection treatment decisions, diagnosis of antibody-mediated rejection, and clinical diagnosis of chronic allograft dysfunction. We found an inverse relationship between the cfdDNA signal and the measurement of the volume of air exhaled by the patient within 1 s during forced breath (FEV1, percentage of predicted for age, height, and body composition; Fig. 4C). cfdDNA was elevated, coinciding with clinical diagnoses of CLAD (P = 1.10−4, Mann–Whitney U test; Fig. 4F) and clinical diagnoses of antibody-mediated rejection (AMR) (P = 0.04, Mann–Whitney U test; Fig. 4G). cfdDNA furthermore correlated with rejection treatment decisions (P = 5.10−4, Mann–Whitney U test; Fig. 4E) and clinical diagnosis of rejection (a diagnosis of exclusion: In the absence of a biopsy and evidence of another cause of a decline in lung function, e.g., infection or heart failure, a presumptive diagnosis of acute rejection is made; P = 6.10−4, Mann–Whitney U test; Fig. 4D).
Cytomegalovirus-Induced Graft Injury.
We next compared cfdDNA levels and clinical indicators of infection. Together with allograft rejection, infectious complications remain among the most important causes of morbidity and mortality after lung transplantation (17), with cytomegalovirus infections posing the most significant known threat (13).
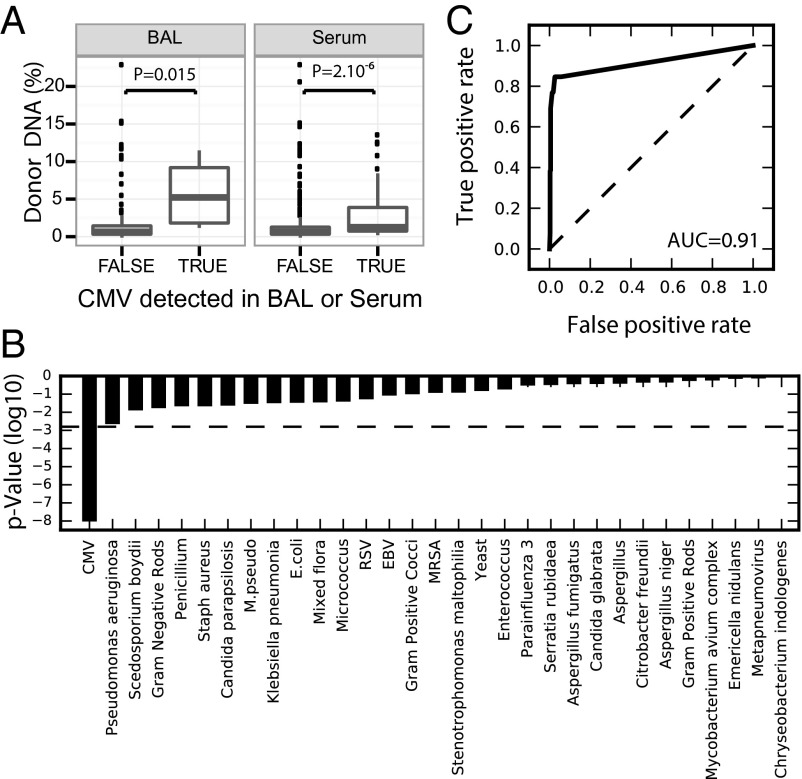
To study the relationship between infection and graft damage, we collected clinical measurements of specific infections performed on 14 specimen types (Fig. S2). Infections with 56 different agents were reported at time points for which plasma samples were available for sequencing, including a large number of rare infections (25 infections were reported only once). We tested whether donor-derived cfDNA levels correlated with the presence or absence of these infections, and observed a significant elevation of cfdDNA for patients who tested positive for CMV in bronchial lavage samples or serum [P << 1e-6 (Fig. 5A); samples collected in the first 2 mo were excluded]. This observation was unique to CMV, as no significant correlation was found for other infectious agents after accounting for multiple comparisons (31 infections for which more than one positive test was reported coinciding with a sample point; Bonferroni-corrected significance threshold, P = 0.05/31), that is, other infections that do not cause direct graft injury did not result in elevations of lung-derived cell-free DNA (Fig. 5B). The observed correlation between CMV diagnosis and cfdDNA is consistent with reports of direct and indirect pathways through which CMV causes allograft injury (12). Our data also indicate that CMV infection does not confound the use of donor DNA as a marker of acute rejection; the AUC for donor DNA versus biopsy is minimally affected (±1%) by the removal/inclusion of clinical CMV positives (CMV tests performed within 10 d of plasma sampling).
Fig. S2.
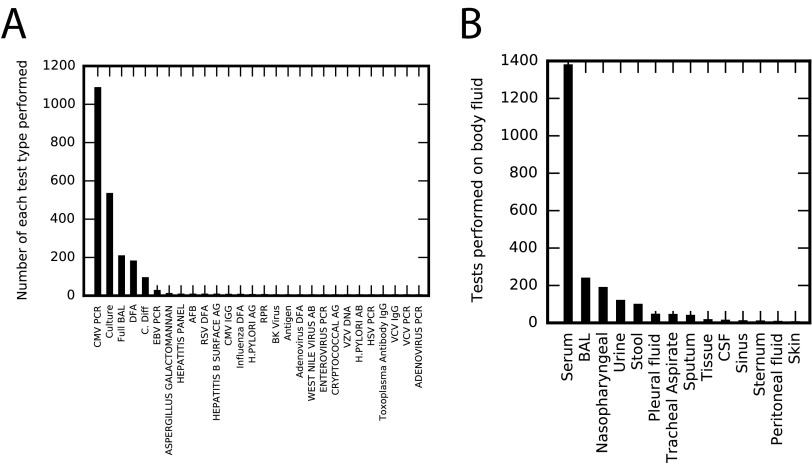
Statistics for collected clinical data on infections. (A) The number of each clinical test type performed. There are 28 different tests performed on the cohort, although 13 are performed only once and CMV PCR represents approximately half the total number of tests performed. (B) Clinical tests were performed on 14 different body fluids, with the majority of tests (∼64%) performed on serum. For each test, we determined the number of unique infectious agents that were measured. In some cases, the measured infection is obvious based upon the test type (e.g., for quantitative PCR tests for a given pathogen). For culture assays, we determined the total number of infectious agents cultured in our cohort (46 unique organisms cultured). Assuming these 46 infections to be detectable for each culture performed, a total of ∼36,000 measurements for specific infections was performed on our cohort. The majority (∼98.5%) of these measurements are negative. Of the 540 positive test results, CMV is the most common.
Fig. 5.
CMV infection-induced allograft injury. (A) Correlation between clinical report of CMV (HHV-5, BAL, and serum) and donor cfdDNA signal matched to clinical test date (P values, Mann–Whitney U test). Data are shown as boxplots. The bottom and top of the box indicate the upper and lower quartile, the band inside the box indicates the median. (B) P values for the correlation between clinical diagnosis of infection and cell-free DNA level (dashed line indicates the Bonferroni-corrected significance threshold, P = 0.05/31) for infections with more than one clinical positive test result (31 infections). (C) ROC curve that tests the performance of CMV-derived cell-free DNA level in CMV-positive and CMV-negative patients (AUC 0.91).
Clinical Monitoring of the Virome.
The high incidence of infections in lung transplantation and the potential benefit of coupling cfdDNA and infection measurements prompted us to examine whether pathogen-derived sequences can be detected and used to monitor infections (18–21). We first quantified reads that map to the CMV genome for each sample (SI Materials and Methods) (10) and observed increased CMV abundance in samples that were clinically positive for infection (P = 7.10−9, Mann–Whitney U test; Fig. 5C); the level of CMV-derived DNA in our samples matched clinical reports of CMV with an AUC of 0.91 (Fig. 5C). These data indicate that CMV surveillance can be performed in parallel with rejection monitoring using the same sequence data, and led us to examine whether other viral infections could be similarly monitored.
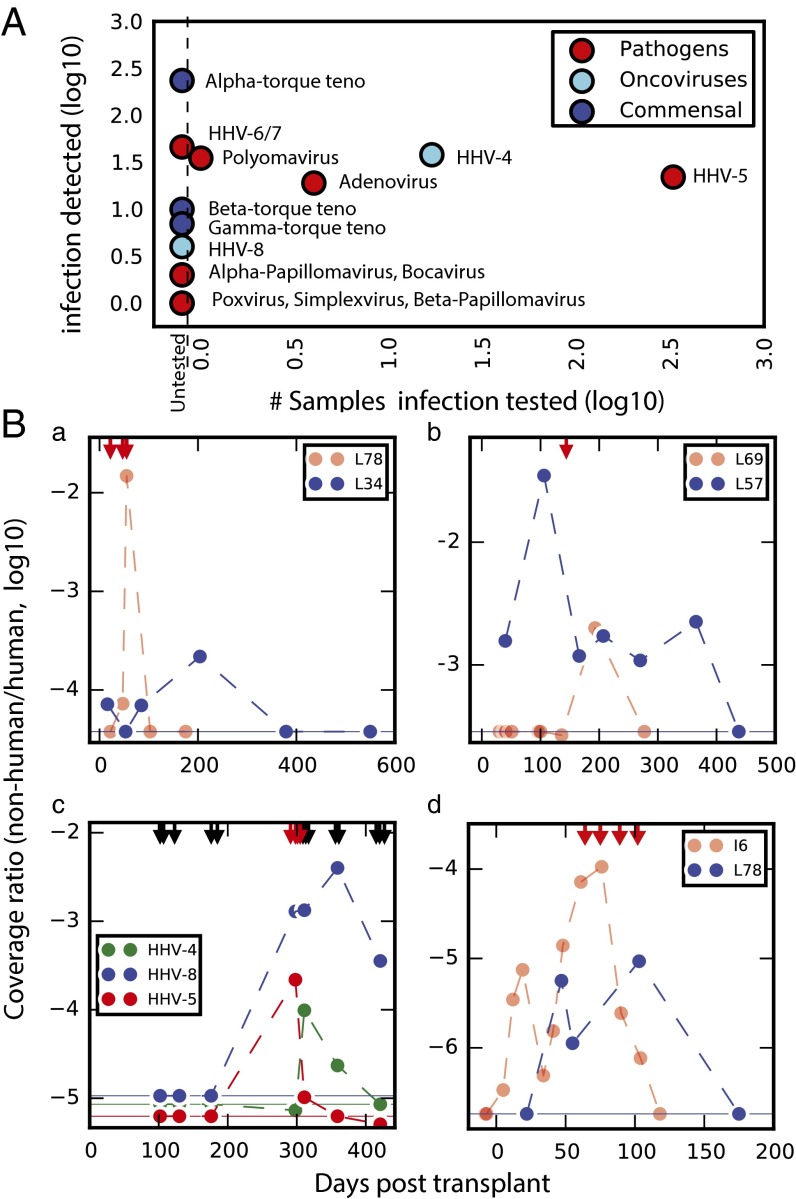
We identified well-characterized pathogens and oncoviruses (Fig. 6A) as well as commensal torque teno viruses (TTVs; Alphatorquevirus genus), which is consistent with previous observations of a link between immunosuppression and TTV abundance (21–24). The frequency of clinical testing for these viruses varied considerably, with frequent surveillance of CMV [human herpesvirus 5 (HHV-5); n = 1,082 tests in our cohort] relative to other pathogens (Fig. 6A). We evaluated the incidence of infection (number of samples in which a given virus is detected via sequencing) relative to clinical screening frequency. Although CMV was screened for most frequently (335 samples), its incidence as determined by sequencing (detected in 22 samples) was similar to that of other pathogens that were not routinely screened, including adenovirus and polyomavirus (clinically tested on four occasions and one occasion, respectively; Fig. 6A).
Fig. 6.
Monitoring the infectome. (A) Clinical testing frequency compared with the incidence of viral infections detected in sequencing. (B) Time-series data for patients who tested positive (red arrows) for specific infections relative to those who were not tested. (a) Coverage ratio for adenovirus in L78 with clinical positives highlighted relative to an untested patient (L34). (b) Coverage ratio for polyomavirus in L69 with one positive test relative to sustained signal in an untested patient (L57). (c) Three herpesvirus infections (HHV-4, -5, and -8) in L58 with both positive (red) and negative (black) tests for CMV (HHV-5) highlighted. (d) Coverage ratio for microsporidia in I6, with four positive tests shown, relative to the signal observed in patient L78, who had symptoms of microsporidiosis but was not tested. In all cases, the logged coverage ratio (organism coverage relative to human coverage for the sample) is reported. Because the data are logged, zero values were replaced with the detection limit of the assay (horizontal lines indicate the coverage ratio obtained for a single read assigned to the organism in the given sample).
Adenovirus is a community-acquired respiratory infection that can cause graft loss in lung transplant recipients and poses a particularly high risk for pediatric patients (25, 26). Samples were collected from one pediatric patient (L78, Fig. 6B, a) who tested positive for adenovirus. This patient had the highest adenovirus-derived DNA load in our cohort. A sustained adenovirus load was furthermore observed in several adult transplant recipients who were not screened clinically (e.g., L34, Fig. 6B, a), as testing is typically restricted to pediatric lung transplant cases.
Polyomavirus is the leading cause of allograft rejection after renal transplantation (26) but is not routinely included in postlung transplant surveillance. We detected polyomavirus in two patients who were not tested for this virus (L57 and L15). In both cases, the clinical records indicated persistent renal insufficiency, which could have resulted from polyomavirus infection. A time series of the polyomavirus load measured for one of these untested patients (L57) is shown in comparison with a time series for a patient who was clinically diagnosed with polyomavirus infection (Fig. 6B, b). In a last example of the potential benefit of broad and hypothesis-free infection screening, we examined a patient who exhibited a high load of HHV-8 (Fig. 6B, c), an oncovirus that can cause complications following solid-organ transplantation (27). This patient (L58) tested positive for two other herpesviruses (HHV-4a and HHV-5) that have the potential to stimulate HHV-8 reactivation (28). Although posttransplant monitoring for HHV-8 is only recommended in particular clinical circumstances (27), use of sequencing enables the identification of the virus in nonsuspect cases that would otherwise go undetected.
Clinical Monitoring of the Microbiome.
In addition to viruses measured in serum, we also observed an agreement between cell-free DNA measurements and fungi and bacteria detected in other body fluids, including Klebsiella pneumoniae infections detected via urine culture (ROC 0.98; Fig. S3A) and fungal infection detected in bronchoalveolar lavage (BAL) (Fig. S3B). We found that the strength of these correlations depended both on the infection type and body site queried. We observed better performance for body sites that have tighter coupling to blood (Fig. S3C). In addition, we found that the assay performance depends on the level of the normal background signal. For example, test performance for the most commonly cultured bacteria (Pseudomonas), detected in over 80% of patient samples, was poor (AUC 0.62), in contrast to test performance for the most commonly detected viral pathogen (CMV), detected in only 6% of our patient samples (AUC 0.91). This highlights an important caveat in the ability of the plasma DNA sequencing to detect increased abundance of commensal organisms (including Pseudomonas), which are part of the normal flora (29, 30), compared with organisms that only have a pathogenic role, which have a low or zero background. In the case of commensal bacteria, the clinical question is not presence or absence but rather presence or absence in particular body sites; cell-free DNA may be of limited use in such cases because it lacks such specificity.
Fig. S3.
Correlations with infections detected at various body sites. (A) ROC curve and time-series data for Klebsiella pneumoniae (687 negative results and 13 positive clinical tests). All positive test results are for patient L24, and (except for one for BAL) are detected in urine. (B) Matched sample–test data for Aspergillus niger (481 clinical negatives, 2 clinical positives). Both positives are in L17 (one BAL and one sputum). The single test for BAL is shown. The percentile plot (Left) indicates that measurements in L24 are high relative to the cohort. (C) Correlations with clinical tests based upon both infection type and body fluid queried by the clinical assay. For each category, the measured cell-free DNA signal is aggregated across all infections and partitioned based upon the result of the matched clinical test, and P values are computed using Mann–Whitney U test.
We conclude with an example of the detection of clinically relevant fungal infection. We detected cell-free DNA derived from microsporidia, a fungus that can cause intestinal infections in immunosuppressed patients (31). We measured a sustained microsporidia load in one patient who exhibited classic symptoms of microsporidiosis (L78, Fig. 6B, d). An adenoviral infection (Fig. 6B, a) was the suspected cause, although endoscopy and sigmoidoscopy results were inconclusive and stool samples tested negative for adenovirus as well as Clostridium difficile, a common cause of diarrhea in immunocompromised hosts. Based on our sequencing data, microsporidiosis was a possible cause of the patient’s symptoms, because the microsporidia signal measured in this patient is similar to that of I6, a patient from an unrelated cohort who tested positive for microsporidia by a clinical assay (Fig. 6B, d).
SI Materials and Methods
Plasma Processing and DNA Extraction.
Plasma was extracted from whole-blood samples as described previously (34) and stored at −80 °C. When required for analysis, plasma samples were thawed and circulating DNA was extracted from 0.5–1 mL plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen).
Sequencing Library Preparation and Sequencing.
Sequencing libraries were prepared from purified plasma DNA using the NEBNext DNA Library Prep Master Mix Set for Illumina with standard Illumina indexed adapters (IDT) or using a microfluidics-based automated library preparation platform (Mondrian ST; Ovation SP Ultralow Library Systems). Libraries were characterized using the Agilent 2100 Bioanalyzer (High Sensitivity DNA Kit) or the AATI fragment analyzer, quantified by quantitative PCR, and sequenced (Illumina HiSeq 2000, HiSeq 2500, or NextSeq 500; 1 × 50 bp or 2 × 100 bp).
Sample Preparation Genotyping.
Whole-blood samples were collected from the donor and recipient. DNA was purified from whole blood (DNeasy Blood and Tissue Kit; Qiagen) and amplified (REPLI-g Mini Kit; Qiagen). Genotyping was performed on Illumina Whole-Genome Arrays (HumanOmni2.5–8 or HumanOmni1).
Quantification of Nonhuman-Derived Cell-Free DNA.
Preprocessing and human subtraction.
Low-quality bases and Illumina-specific sequences were trimmed (Trimmomatic 0.32) (37). For paired-end datasets, reads from short fragments were merged and a consensus sequence of the overlapping bases was determined using FLASH 1.2.7 (38). Reads were aligned (Bowtie 2.1.0; very sensitive mode) (39) against the human reference (UCSC hg19; https://genome.ucsc.edu/). Unaligned reads were extracted and aligned to the bacteriophage phi X174, which is used as a control sequence in the Illumina platform. Unaligned reads were extracted again, and human coverage was calculated using the nonredundant reads (SAMtools 0.1.19 rmdup) (40) for later normalization purposes.
Alignment to infection database.
National Center for Biotechnology Information (NCBI) BLAST (2.2.28+) was used to align cleaned nonhost reads against a custom database of potential pathogens, encompassing viruses, bacteria, and fungi in the NCBI database, with the addition of pathogens: those related to microsporidia (Enterocytozoon bienusi, Nosema bombycis, Nosema ceranae, Hamoltosporidium tvaerminnensis, Nematocida parisii, Vavraia culicis, Vittaforma corneae, Edhazardia aedis, Trachipleistophora hominis, Nosema apis, Anncaliia algeae, Spraguea lophii, Rozella allomycis, Encephalitozoon hellem, Encephalitozoon romaleae, Encephalitozoon hellem, Encephalitozoon intestinalis, Encephalitozoon cuniculi), human herpesvirus 6, and SEN virus. These were added based on their clinical relevance or prior visibility in a more expanded database (RefSeq). Alignments were required to have an identity of at least 90% across 90% of the bases of the query.
Determination of nonhuman load.
Each high-confidence read may align to multiple infections in a given pathogen database. To correctly assign organism abundance based upon this possible mapping redundancy, an algorithm (GRAMMy) (41) was used to compute the most likely organisms that a given read originated from. In turn, the number of reads assigned to each organism per sample was computed as well as the associated organism coverage, which is the read count normalized by the genome size. A ratio of organism coverage relative to human genome coverage is computed for each sample, which corrects for differences in sequencing depth between samples and enables cross-sample comparisons. The relative coverage is used as our reported metric of infectious load.
Discussion
With 10–100 billion fragments per milliliter of plasma, circulating cell-free DNA is an information-rich window into human physiology, with rapidly expanding applications in cancer diagnosis (32), cancer treatment monitoring (33), genetic prenatal diagnosis (34), and monitoring of heart transplant rejection via genome transplant dynamics (11). In this work, we applied the principle of GTD to lung transplantation, a particularly challenging type of solid-organ transplant that is limited by poor survival rates as well as by an inaccurate and invasive test for allograft rejection. We have shown a strong concordance between cfdDNA levels and clinical indicators of rejection, demonstrating that cfdDNA is a broadly applicable marker of graft injury that can potentially improve upon the performance of transbronchial biopsy, the current gold standard.
Using donor DNA as a metric, we found evidence of graft injury during CMV infection. This is in line with previous reports of the direct and indirect immunomodulatory pathways through which CMV causes graft injury (12). Given the high incidence of infectious complications in lung transplant recipients and the bidirectional relationship that frequently exists between rejection and infection, clinical decision making relies on accurate diagnosis of both infection and rejection. We therefore extended the scope of GTD to include infectious disease monitoring. We first demonstrated a good agreement between clinical test results and cfDNA derived from CMV, a leading cause of posttransplant graft injury. We furthermore showed that hypothesis-free infection monitoring revealed numerous untested pathogens, including undiagnosed cases of adenovirus, polyomavirus, HHV-8, and microsporidia infection in patients who had similar microbial cfDNA levels compared with patients with positive clinical test results and associated symptoms.
The assay presented herein has the potential to become an important tool for lung transplant surveillance within the next few years, considering (i) the high incidence of acute rejection and difficult-to-diagnose infectious complications; (ii) the numerous limitations of transbronchial biopsy in rejection surveillance; (iii) the lack of alternative, noninvasive tests of graft injury after lung transplantation; and (iv) the relatively low cost associated with the assay. This study furthermore illustrates the potential benefit of broad, sequencing-based monitoring of infection as opposed to pathogen-specific testing, an approach that will have clinical applications beyond transplantation (35). A number of issues need to be resolved before sequencing-based infection testing can be widely considered as a clinical tool (36), including the establishment of pathogen-specific thresholds to discriminate between colonization, infection, and disease. This approach, however, may be of immediate use in formulating hypotheses for the occurrence and source of an infection. This is of particular relevance in transplantation, where the incidence of infections is high, rejection and infection can occur concomitantly, and symptoms of infection and rejection are often difficult to discriminate.
In summary, we describe a noninvasive diagnostic assay that can be used to simultaneously monitor for acute rejection as well as a broad spectrum of infections in a contemporary lung transplant cohort.
Materials and Methods
The goal of this study was to test the utility of genomic analyses of cell-free DNA for the diagnosis of rejection and infection after lung transplantation. Patients listed for lung transplantation at Stanford University Hospital were enrolled in the study. Multiorgan transplant recipients were excluded. The study was approved by the Stanford University Institutional Review Board (Protocol 17666). All patients provided written informed consent.
Pretransplant blood samples were collected from the donor and recipient for SNP genotyping. Plasma samples were collected longitudinally at the following time points posttransplant: weeks 1, 2, 4, and 6, and months 2, 2.5, 3, 4, 5, 6, 8, 10, 12, 16, 20, and 24. A subset of samples (36/398) was collected at time of clinical change. A subset of lung transplant recipients also had blood samples collected on posttransplant day 1 (up to three samples), day 2 (up to two samples), and day 3. In each case, blood samples were collected before biopsy or bronchoalveolar lavage procedures were performed.
Sequencing and genotyping sample preparation protocols are described in detail in SI Materials and Methods and in ref. 11. The analysis workflow used to quantify the fraction of donor-derived DNA is described in detail in ref. 11. The analysis workflow for the quantification of nonhuman-derived DNA is described in detail in SI Materials and Methods.
Analyses were performed in R 2.15.1 (www.r-project.org/) and Python 2.7 (www.python.org/download/releases/2.7/). Groups were compared by the nonparametric Mann–Whitney U test. A value of P < 0.05 was considered statistically significant.
The sequence data generated in this study have been deposited in the Sequence Read Archive (accession no. PRJNA263522). Other patient data, including patient genotyping data, not included in this manuscript can be shared upon request.
Acknowledgments
We thank G. Mantalas for assistance with the sequencing assays and B. Passarelli for setting up and maintaining the computing infrastructure. This work was supported by NIH Grant RC4AI092673. The S.R.Q. laboratory is supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence data reported in this paper have been deposited in the Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. PRJNA263522).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517494112/-/DCSupplemental.
References
- 1.Arcasoy SM, Kotloff RM. Lung transplantation. N Engl J Med. 1999;340(14):1081–1091. doi: 10.1056/NEJM199904083401406. [DOI] [PubMed] [Google Scholar]
- 2.Yusen RD, et al. International Society for Heart and Lung Transplantation The Registry of the International Society for Heart and Lung Transplantation: Thirtieth adult lung and heart-lung transplant report—2013; focus theme: Age. J Heart Lung Transplant. 2013;32(10):965–978. doi: 10.1016/j.healun.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Stehlik J, et al. The Registry of the International Society for Heart and Lung Transplantation: Twenty-eighth adult heart transplant report—2011. J Heart Lung Transplant. 2011;30(10):1078–1094. doi: 10.1016/j.healun.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Bloom RD, Goldberg LR, Wang AY, Faust TW, Kotloff RM. An overview of solid organ transplantation. Clin Chest Med. 2005;26(4):529–543, v. doi: 10.1016/j.ccm.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Lodhi SA, Lamb KE, Meier-Kriesche HU. Solid organ allograft survival improvement in the United States: The long-term does not mirror the dramatic short-term success. Am J Transplant. 2011;11(6):1226–1235. doi: 10.1111/j.1600-6143.2011.03539.x. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad S, Shlobin OA, Nathan SD. Pulmonary complications of lung transplantation. Chest. 2011;139(2):402–411. doi: 10.1378/chest.10-1048. [DOI] [PubMed] [Google Scholar]
- 7.Charlson ES, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186(6):536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arcasoy SM, et al. Pathologic interpretation of transbronchial biopsy for acute rejection of lung allograft is highly variable. Am J Transplant. 2011;11(2):320–328. doi: 10.1111/j.1600-6143.2010.03382.x. [DOI] [PubMed] [Google Scholar]
- 9.Lo YM, et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998;351(9112):1329–1330. doi: 10.1016/s0140-6736(05)79055-3. [DOI] [PubMed] [Google Scholar]
- 10.Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci USA. 2011;108(15):6229–6234. doi: 10.1073/pnas.1013924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vlaminck I, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6(241):241ra77. doi: 10.1126/scitranslmed.3007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolkoff-Rubin NE, Fishman JA, Rubin RH. The bidirectional relationship between cytomegalovirus and allograft injury. Transplant Proc. 2001;33(1-2):1773–1775. doi: 10.1016/s0041-1345(00)02674-9. [DOI] [PubMed] [Google Scholar]
- 13.Zamora MR. Cytomegalovirus and lung transplantation. Am J Transplant. 2004;4(8):1219–1226. doi: 10.1111/j.1600-6143.2004.00505.x. [DOI] [PubMed] [Google Scholar]
- 14.Lo YM, et al. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64(1):218–224. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kajstura J, et al. Cardiomyogenesis in the adult human heart. Circ Res. 2010;107(2):305–315. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remund KF, Best M, Egan JJ. Infections relevant to lung transplantation. Proc Am Thorac Soc. 2009;6(1):94–100. doi: 10.1513/pats.200809-113GO. [DOI] [PubMed] [Google Scholar]
- 18.Quiñones-Mateu ME, Avila S, Reyes-Teran G, Martinez MA. Deep sequencing: Becoming a critical tool in clinical virology. J Clin Virol. 2014;61(1):9–19. doi: 10.1016/j.jcv.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wylie KM, Weinstock GM, Storch GA. Emerging view of the human virome. Transl Res. 2012;160(4):283–290. doi: 10.1016/j.trsl.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138(1):30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 21.De Vlaminck I, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155(5):1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Focosi D, Macera L, Pistello M, Maggi F. Torque teno virus viremia correlates with intensity of maintenance immunosuppression in adult orthotopic liver transplant. J Infect Dis. 2014;210(4):667–668. doi: 10.1093/infdis/jiu209. [DOI] [PubMed] [Google Scholar]
- 23.Touinssi M, et al. TT virus infection: Prevalence of elevated viraemia and arguments for the immune control of viral load. J Clin Virol. 2001;21(2):135–141. doi: 10.1016/s1386-6532(01)00157-3. [DOI] [PubMed] [Google Scholar]
- 24.Li L, et al. AIDS alters the commensal plasma virome. J Virol. 2013;87(19):10912–10915. doi: 10.1128/JVI.01839-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Echavarría M. Adenoviruses in immunocompromised hosts. Clin Microbiol Rev. 2008;21(4):704–715. doi: 10.1128/CMR.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidemann K, et al. Monitoring of adenovirus infection in pediatric transplant recipients by quantitative PCR: Report of six cases and review of the literature. Am J Transplant. 2004;4(12):2102–2108. doi: 10.1111/j.1600-6143.2004.00631.x. [DOI] [PubMed] [Google Scholar]
- 27. Razonable RR (2013) Human herpesviruses 6, 7, and 8 in solid organ transplant recipients. Am J Transplant 13(Suppl 3):67–77; quiz 77–78. [DOI] [PubMed]
- 28.Larroche C, et al. Epstein-Barr virus and human herpesvirus 8 coinfection and concomitant extranodal nasal-type NK/T cell lymphoma and Castleman disease: Case report. Clin Infect Dis. 2003;36(9):e107–e110. doi: 10.1086/374663. [DOI] [PubMed] [Google Scholar]
- 29.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9(8):811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dascomb K, Frazer T, Clark RA, Kissinger P, Didier E. Microsporidiosis and HIV. J Acquir Immune Defic Syndr. 2000;24(3):290–292. doi: 10.1097/00126334-200007010-00018. [DOI] [PubMed] [Google Scholar]
- 32.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 34.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105(42):16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson MR, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barzon L, et al. Next-generation sequencing technologies in diagnostic virology. J Clin Virol. 2013;58(2):346–350. doi: 10.1016/j.jcv.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magoc T, Salzberg S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia LC, Cram JA, Chen T, Fuhrman JA, Sun F. Accurate genome relative abundance estimation based on shotgun metagenomic reads. PLoS ONE. 2011;6(12):e27992. doi: 10.1371/journal.pone.0027992. [DOI] [PMC free article] [PubMed] [Google Scholar]