Abstract
The accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b, 9b and ORF14), predicted unknown proteins (PUPs) encoded by the genes, are considered to be unique to the severe acute respiratory syndrome coronavirus (SARS-CoV) genome. These proteins play important roles in various biological processes mediated by interactions with their partners. However, very little is known about the interactions among these accessory proteins. Here, a EYFP (enhanced yellow fluorescent protein) bimolecular fluorescence complementation (BiFC) assay was used to detect the interactions among accessory proteins. 33 out of 81 interactions were identified by BiFC, much more than that identified by the yeast two-hybrid (Y2H) system. This is the first report describing direct visualization of interactions among accessory proteins of SARS-CoV. These findings attest to the general applicability of the BiFC system for the verification of protein-protein interactions.
Abbreviations: aa, amino acids; AD, activation domain; BD, binding domain; BiFC, bimolecular fluorescence complementation; Co-IP, co-immunoprecipitation; E, envelope; EYFP, enhanced yellow fluorescent protein; M, membrane; N, nucleocapsid; NLS, nuclear localization signal; ORFs, open reading frames; PCR, polymerase chain reaction; PPIs, protein-protein interactions; PUPs, predicted unknown proteins; S, spike; SARS-CoV, severe acute respiratory syndrome coronavirus; Y2H, yeast two-hybrid
KEY WORDS: SARS-CoV, Accessory proteins, Y2H, Bimolecular fluorescence complementation assay
Graphical abstract
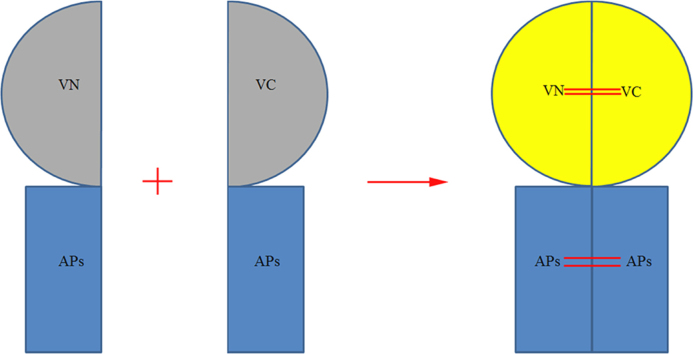
Two non-fluorescent fragments (VN and VC) derived from a fluorescent protein EYFP were separately attached to two accessory proteins of SARS-CoV (APs) and the resulting fusion proteins were co-expressed in Saccharomyces cerevisiae. When the two accessory proteins interact, the split EYFP fragments can be functionally reconstituted, allowing a fluorescent readout. Thereby, the BiFC method can be used for direct visualization of accessory protein interactions in living cells.
1. Introduction
The severe acute respiratory syndrome coronavirus (SARS-CoV) is an enveloped, positive-stranded RNA virus with a very large genomic size (about 30 kb) and constitutes 14 open reading frames (ORFs)1, 2. The two large overlapping (ORF1a and ORF1b) are translated to produce polyproteins that are involved in viral RNA replication and transcription. The downstream ORFs encode the structural proteins in the following order: spike (S), envelope (E), membrane (M) and nucleocapsid (N). In addition to these two groups of proteins, there is another group including accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b, 9b and ORF14) varying in length from 39 to 274 amino acids (aa) and with no significant sequence homology to proteins in other coronaviruses1, 2. The third group of proteins, named as predicted unknown proteins (PUPs), is unique to SARS-CoV1, 2.
Although the precise functions of these accessory proteins are still obscure, it seems clear that many of them play important roles in various biological processes mediated by interactions with their partners. For example, the 3a protein can interact with structural proteins like S, M and N proteins, and another accessory protein 7a in SARS-CoV infected cells; the resulting multiple complexes may be inferred to be important for viral assembly3. Additionally, the interaction of SARS-CoA protein 6 and another accessory protein 9b was also confirmed to be present in SARS-CoV infected cells4. Thus, the identification of the occurrence and components of protein-protein interactions (PPIs) among accessory proteins provided invaluable insights into the cellular functions of these proteins. However, very little is known about other interactions among these accessory proteins5. Given that several accessory proteins have been shown to exhibit functions in virus-host interactions during SARS-CoV infection in vivo, it is necessary to understand the putative interactions among accessory proteins6, 7.
A wide range of methods, including the yeast two-hybrid (Y2H) analysis8, co-immunoprecipitation (Co-IP) assay9 and bimolecular fluorescence complementation (BiFC) system10, 11, have been developed for the identification and analysis of PPIs in vitro and in vivo. Each method has its own advantages and limitations. Co-IP is one of the most commonly used methods for examining PPIs. This technique often requires the preparation of specific antibodies against each of the analyzed proteins, making it an expensive and time-consuming process12. Also, Co-IP can be performed using lysates of the cells, which generally prevents the determination of the exact subcellular localization of the interacting proteins13. Y2H is another means of assessing whether two single proteins interact. Although used extensively, Y2H suffers from several drawbacks, such as high occurrence of false-positives and the requirement that the interacting proteins accumulate in the cell nucleus8. Among these methods, the fluorescent protein–based BiFC is an effective and straightforward tool to study PPIs, enabling direct visualization of the occurrence and subcellular localization of PPIs with simple equipment. Moreover, this approach can avoid the possibility of non-physiological protein interactions caused by cell lysis and mixing the contents of different cellular compartments. Due to its stronger signal and direct readout, this assay has become widely accepted over the past decade10, 11, 14, 15.
In the present investigation, characterization of the interactions among accessory proteins of SARS-CoV was performed using BiFC and Y2H. Biochemical evidence revealed the interactions existed in accessory proteins.
2. Materials and methods
2.1. Strains and plasmids
Escherichia coli (E. coli) TG1 was used as a host strain for subcloning. The strain was grown in Luria–Bertani medium (10 g/L Bacto–Tryptone, 5 g/L Bacto-yeast extract, 10 g/L NaCl) supplemented with ampicillin (100 μg/mL) when required for selection.
BiFC vectors pFA6a-VN-KanMX6 and pFA6a-VC-KanMX6 were kindly supplied by Prof. Won-Ki Huh (Seoul National University, Republic of Korea)16. The haploid Saccharomyces cerevisiae (S. cerevisiae) strains AH109 (MATa, his3-200, leu2-3, 112, trp1-901, ura3-52, gal4Δ, gal80Δ, LYS2:: GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3:: MEL1UAS-MEL1TATA-lacZ, MEL1)17 and W303-1B (MATa, ura3-1, trp1-1, ade2-1, leu2-3, 112, his3-11, 15)18 were used to determine PPIs. The GAL4 DNA binding domain (BD) vector pGBKT7 and pGBT9, and the activation domain (AD) vector pGADT7 were from Clontech Laboratories, Inc. (Mountain View, USA) and were used throughout. The yeast expression vector pYeDP60 was kindly provided by Prof. Werck-Reichhart (France). pYeDP60 is a 2 µm plasmid with GAL10-CYC1 promoter, URA3 and ADE2 marker19, 20. All yeast strains were grown either in the non-selective YPD medium (10 g/L yeast extract, 20 g/L bactopeptone and 20 g/L glucose) or in the selective SD medium (0.7% yeast nitrogen base without amino acids, 0.1% casamino acids and 2% glucose) with appropriate amino acids dropped out at 30 °C. The detailed plasmids and strains used in the work are listed in Table S1 in Supporting information.
2.2. Enzymes and chemicals
In-Fusion® HD Cloning Kit, Restriction enzymes and X-α-gal were purchased from Takara Shuzo Co., Ltd. (Kyoto, Japan). KOD Plus Taq DNA polymerase was purchased from Toyobo Co., Ltd. (Osaka, Japan). All other fine chemicals were of analytical grade and commercial available.
2.3. Cloning of SARS-CoV accessory genes
ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, ORF9b and ORF14 genes were chemically synthesized respectively, according to the genome sequence of SARS-CoV Tor2 isolate (GenBank accession number AY274119.3)2. After the sequence confirmation, the genes were subcloned into the vector pMD™18-T to acquire nine vectors named pMD3a, pMD3b, pMD6, pMD7a, pMD7b, pMD8a, pMD8b, pMD9b and pMDORF14, which were used as template to construct Y2H expression vectors and BiFC plasmids by the In-Fusion method.
2.4. Plasmids construction for Y2H assay
Specific PCR primers (Table S2 in Supporting information) were designed according to the sequences of SARS-CoV accessory genes and used to amplify each accessory gene by using of KOD Plus Taq DNA polymerase. The PCR products were ligated into the NdeI and EcoRI double digested vectors pGADT7 and pGBKT7, respectively, using the In-Fusion method. The resulting constructs pGAD-APs and pGBK-APs (APs refer to ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, ORF9b and ORF14 genes) (Figs. S1 and S2 in Supporting information) containing SARS-CoV accessory genes were verified by custom sequencing.
2.5. Y2H assay
The resulting yeast two-hybrid vectors pGAD-APs and pGBK-APs were then co-transformed into S. cerevisiae AH109 by use of the LiAc transformation method21. The transformed yeasts were then grown on SD-Leu-Trp medium (leucine and tryptophan are omitted from the formulation) for 3–7 days at 30 °C. Colonies with 2–3 mm in diameter were transferred to solid SD-Ade-His-Leu-Trp medium and grown for 3–5 days at 30 °C.
For testing APs interactions, combinations of APs-bait and APs-prey constructs co-expressed in yeast were subjected to α- and β-galactosidase assay according to the manufacturer׳s instructions (Yeast Protocols Handbook, PT3024-1, Clontech Laboratories, Inc.). Each experiment was separately repeated in triplicate.
2.6. Construction of BiFC plasmid vectors
pFA6a-pGAL10CYC1-VN-KanMX6 and pFA6a-GAL10CYC1-VC-KanMX6: the DNA sequence coding for the GAL10-CYC1 promoter was amplified by polymerase chain reaction (PCR) from pYeDP60 using primer pair FGAL10CYC1 and RGAL10CYC1 (Table S2 in Supporting information). The amplified 800-bp PCR fragment was directionally cloned into the linearized vectors pFA6a-VN-KanMX6 and pFA6a-VC-KanMX6 with restriction digestion of SalI, respectively. The resulting recombinant vectors pFA6a-GAL10CYC1-VN-KanMX6 and pFA6a-GAL10CYC1-VC-KanMX6 were verified by sequencing.
pGAL10CYC1-VN and pGAL10CYC1-VC: the fusion fragment of GAL10-CYC1 promoter and ORF encoding N-terminus (aa 1−173, VN173) or C-terminus (aa 155−238, VC155) of Venus, a variant of yellow fluorescent protein, were amplified by PCR, using primers FGBT9GAL10CYC1 and RGBT9GAL10CYC 1 (VN173) or FGADT7GAL10CYC1 and RGADT7GAL10CYC1 (VC155). The resulting PCR products of GAL10CYC-VN173 and GAL10CYC-VC155 were inserted into pGBT9 linearized by SphI and pGADT7 double digested by SphI/NcoI to construct the yeast BiFC expression vector pGAL10CYC1-VN and pGAL10CYC1-VC, respectively.
pAPs-VN and pAPs-VC: the encoding sequences of SARS-CoV accessory proteins were amplified by individual primer and inserted into the SalI-digested vectors pGAL10CYC1-VN and pGAL10CYC1-VC to produce BiFC vectors pAPs-VN and pAPs-VC (Figs. S3 and S4 in Supporting information).
2.7. Microscopy and image analysis
The BiFC plasmid pAPs-VN and pAPs-VC were co-introduced into the S.cerevisiae strain W303-1B by the LiOAc/SS carrier DNA/PEG method according to Gietz et al.21 to give the yeast strain W303B[pAPs-VN+pAPs-VC]. Transformants were selected using a SD-Leu-Trp drop-out medium (SD medium without leucine and tryptophan). Verification of positive clones was done by extraction of yeast plasmids and further colony PCR. Yeast cultures were initially grown in 10 mL SD-Leu-Trp liquid medium at 30 °C to an OD600 of 2–3. The cells of 1 mL were centrifuged and washed three times by ddH2O. The resultant cell was resuspended in 50 mL induction YPD medium containing 2% galactose and grown at 30 °C for 16–24 h. 1 mL of S. cerevisiae cells was harvested by centrifugation at 13,000 rpm for 5 min. The cell pellets were resuspended in concanavalin A (1 mg/mL) buffer. 100 μL cells were removed and plated on the center of microscope slides covered with coverslips. Microscopic analysis was performed by a Nikon Eclipse 80i epifluorescence microscope equipped with YFP-, RFP- and UV-specific filters, a 40×/0.75 objective and a CCD camera applying differential interference contrast. The images were acquired using NIS-Elements F software and processed using Adobe Photoshop 7.0.1. YFP signal was detected between 515 and 565 nm after excitation by 450–499 nm laser.
3. Results
3.1. Detection of SARS-CoV accessory protein interactions by yeast two-hybrid assay
AH109 contains distinct ADE2, HIS3, lacZ and MEL1 reporter constructs that are under the control of distinct GAL4-responsive promoters and only expressed in the presence of GAL4-based protein interactions. To reduce false positive results, all of these reporter genes were used as part of the confirmation step of Y2H assay. SD-Ade-His-Leu-Trp medium was first used to examine the interaction. Cells harboring pGAD-APs and pGBK-APs are able to grow SD-Leu-Trp dropout medium because the plasmids encode tryptophan and leucine biosynthesis genes, respectively. When two proteins encoded by accessory genes of SARS-CoV interact, GAL4-responsive HIS3 and ADE2 expression is activated, allowing these cells to grow on SD-Ade-His-Leu-Trp minimal medium. The strains AH109[pGBKORF9b+pGADORF9b], AH109[pGBKORF8a+pGADORF9b] and AH109[pGBKORF14+pGADORF14] survived from SD-Ade-His-Leu-Trp medium after 3–5 d at 30 °C, implying the interactions of 8a-9b, 9b-9b and ORF14-ORF14 (Table 1). Yeast colonies turned blue when the strains mentioned above were plated on SD-Ade-His-Leu-Trp medium for 3–5 d at 30 °C in the presence of the chromagenic substrate X-α-Gal (an indicator of LacZ expression regulated by a GAL4-responsive promoter). This result further supported by the 8a-9b, 9b-9b and ORF14-ORF14 interactions.
Table 1.
Protein interactions identified by Y2H.
| pGBK3a | pGBK3b | pGBK6 | pGBK7a | pGBK7b | pGBK8a | pGBK8b | pGBK9b | pGBK ORF14 | |
|---|---|---|---|---|---|---|---|---|---|
| pGAD3a | − | − | − | − | − | − | − | − | − |
| pGAD3b | − | − | − | − | − | − | − | − | − |
| pGAD6 | − | − | − | − | − | − | − | − | − |
| pGAD7a | − | − | − | − | − | − | − | − | − |
| pGAD7b | − | − | − | − | − | − | − | − | − |
| pGAD8a | − | − | − | − | − | − | − | − | − |
| pGAD8b | − | − | − | − | − | − | − | − | − |
| pGAD9b | − | − | − | − | − | ++ | − | ++ | − |
| pGAD ORF14 | − | − | − | − | − | − | − | − | ++ |
“++” represents strong interaction; “−” represents no interaction.
The LacZ gene encodes β-galactosidase whereas MEL-1 encodes α-galactosidase, an enzyme occurring naturally in many yeast strains. As a result of two-hybrid interactions, both β-galactosidase and α-galactosidase are expressed and α-galactosidase is secreted by the yeast cells. The quantitative assays of β-galactosidase and α-galactosidase were performed and the detailed results are listed in Table 2.
Table 2.
α-Galactosidase and β-galactosidase assay of protein interactions of 9b-9b, 8a-9b and ORF14-ORF14 (n=3).
| Strain | α-Galactosidase | β-Galactosidase |
|---|---|---|
| AH109[pGBKORF9b+pGADORF9b] | 2.52±0.18 | 8.80±1.99 |
| AH109[pGBKORF8a+pGADORF9b] | 1.17±0.03 | 6.91±2.03 |
| AH109[pGBKORF14+pGADORF14] | 1.84±0.43 | 6.03±1.31 |
3.2. Detection of SARS-CoV accessory protein interactions by BiFC
In order to validate the interactions between SARS-CoV accessory proteins, we first constructed pFA6a-based plasmids, pAPs-VN and pAPs-VC (APs refer to SARS-CoV accessory genes) used for BiFC in S.cerevisiae. Fusion genes of the GAL10-CYC1 promoter and ORFs encoding the N- or C-terminal domains of the engineered YFP protein Venus were amplified by PCR and cloned separately into plasmids pGBT9 and pGADT7 to obtain BiFC plasmid pGAL10CYC1-VN and pGAL10CYC1-VC. The SARS-CoV accessory genes were inserted into the polylinker between the GAL10-CYC1 promoter and the sequence encoding N- or C-terminal domains of Venus. The BiFC plasmids carrying accessory genes were co-transformed into the single yeast cell to identify interactions. Figs. S3 and S4 in Supporting information show the schematic structure of BiFC plasmids pAPs-VN and pAPs-VC.
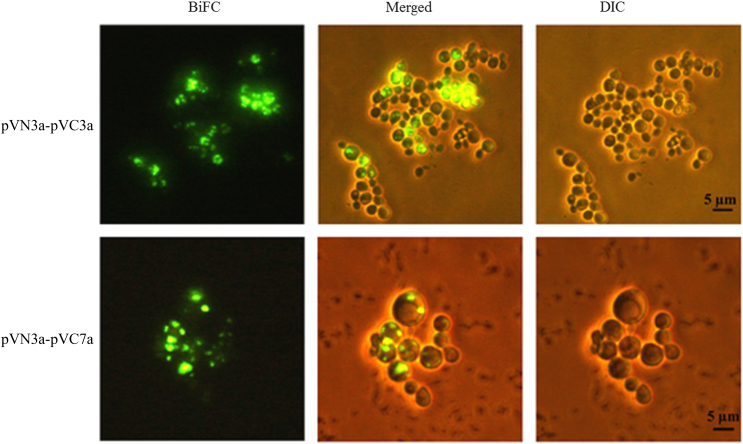
Following plasmid construction, pAPs-VN and pAPs-VC were co-transformed into yeast cell W303-1B in an effort to generate the 81 possible VN173-VC155 combinations for BiFC analysis. Galactose was added into the engineered yeast to induce the expression of SARS-CoV accessory protein labeled with Venus-VN173 and Venus-VC155, respectively. To visualize BiFC signals, exponentially-growing cells were examined under the fluorescence microscope. As illustrated in Fig. 1, strong fluorescence could be detected within 24 protein partners, including pVN3a-pVC3a and pVN3a-pVC7a (Table 3). The fluorescence was localized to discrete regions in the cell. Moreover, weak fluorescence signal was observed in the following 9 cells containing protein pairs: [pVN3b-pVC3a], [pVN3b-pVC7b], [pVN3b-pVC8a], [pVN6-pVC8a], [pVN7b-pVC8a], [pVN7b-pVCORF14], [pVN9b-pVC7a], [pVN9b-pVC7b] and [pVNORF14-pVC6] (Table 3). No BiFC signals were detected in the remaining 48 cells, suggesting that the N- and C-terminal fragments of Venus are insufficiently close to allow YFP complex formation (Table 3). Protein 8b did not interact with any other accessory protein. Results also found 5 self-interacting SARS-CoV accessory proteins 3a, 6, 7a, 7b and 9b, indicating that these proteins can form dimer or multimers in vivo. Eight pairs of interactions were detected in both directions that are 3a-6, 3a-7a, 3a-7b, 6-7a, 6-7b, 6-8a, 7a-7b and 7b-8a.
Figure 1.
Visualization of interactions of pVN3a-pVC3a (top panel) and pVN3a-pVC7a (bottom panel) using a split-EYFP based bimolecular fluorescence complementation assay. The rest of 31 interactions among accessory proteins were also visualized under the same conditions (data not shown). BiFC, bimolecular fluorescence complementation assay images; DIC, differential interference contrast images; Merged, merged images of DIC and BiFC.
Table 3.
Protein interactions identified by BiFC.
| pVC3a | pVC3b | pVC6 | pVC7a | pVC7b | pVC8a | pVC8b | pVC9b | pVCORF14 | |
|---|---|---|---|---|---|---|---|---|---|
| pVN3a | ++ | − | ++ | ++ | ++ | − | − | − | − |
| pVN3b | + | − | − | − | + | + | − | − | − |
| pVN6 | ++ | − | ++ | ++ | ++ | + | − | − | − |
| pVN7a | ++ | − | ++ | ++ | ++ | − | − | − | − |
| pVN7b | ++ | − | ++ | ++ | ++ | + | − | − | + |
| pVN8a | ++ | − | ++ | ++ | ++ | − | − | − | − |
| pVN8b | − | − | − | − | − | − | − | − | − |
| pVN9b | ++ | − | ++ | + | + | − | − | ++ | − |
| pVNORF14 | ++ | − | + | − | − | − | − | − | − |
“++” represents strong interaction; “+” represents weak interaction; “−” represents no interaction.
4. Discussion
Although the SARS epidemic has not reoccurred since the first outbreak in 2003, substantial research has continued in an effort to increase the understanding of SARS-CoV, and to search for effective anti-SARS-CoV therapeutic agents.
The group-specific accessory proteins of SARS-CoV have been shown to be dispensable for viral replication, but they can contribute to viral stability and pathogenesis in the natural hosts. Therefore, investigations on the interactions between accessory proteins will facilitate an understanding of the role of these genes in the pathogenesis and propagation of SARS-CoV.
In the present investigation, interactions of SARS-CoV accessory proteins were detected by the Y2H method and BiFC assay, respectively. The Y2H assay is a widely used method to study PPIs, based on the fact that many eukaryotic transcription factors, such as the yeast enhancer GAL4, are composed of two functionally distinct domains that mediate transcriptional activation domain (AD) and DNA binding domain (BD) respectively. As illustrated in Table 3, of 81 interactions detected by Y2H, only 3 interactions were indicated to be positive, viz., self-interactions of 9b and ORF14, and interaction of 8a-9b in one direction. Self-interactions were observed with 9b and ORF14, indicating the formation of dimeric or multimeric complexes in the nucleus, similar to the findings of von Brunn et al.5. PPIs of 8a-9b occurred in one direction only when 8a and 9b proteins were separately fused to pGBKT7 and pGADT7, but the opposite was reported by von Brunn et al.5. Moreover, the number of protein interactions is inconsistent with the study by von Brunn et al.5, who identified 11 interactions among accessory proteins of SARS-CoV. This discrepancy resulted from the selection method used. The interactions were identified mainly using the selective plates, which can result in false positive results. Moreover, of 13 interactions detected by Y2H, only 4 were corroborated by Co-IP, which further implied the detection of false positive interactions by Y2H. In the present investigation, more comprehensive selection methods were applied to characterize the interactions. Besides selective plates, colony PCR, quantitative β-galactosidase and α-galactosidase assays were used to validate the interaction among accessory proteins, which improve the reliability of Y2H.
Although Y2H results are major contributor to PPIs databases, these methods present several limitations. For example, the original Y2H cannot detect interactions between proteins targeting compartments other than the nucleus. Therefore, in order to avoid missing interactions in other compartments, BiFC was used to further verify the interacting partners after Y2H identification in the present investigation.
BiFC is based on the formation of a fluorescent complex through the association of two fragments of a fluorescent protein when they are brought together by an interaction between proteins fused to the fragments14, 22, 23. In principle, the BiFC assay can be used to detect interactions in any subcellular compartment in any aerobically growing organism or cell that can be genetically modified to express the fusion proteins24. BiFC, hence, can detect interactions not observed with Y2H. In the present study, 33 out of 81 interactions were visualized by BiFC. Only one Y2H interaction, namely self-interaction of 9b, was confirmed by BiFC. The self-interaction of 9b was also reported by von Brunn et al.5. The findings of the self-interaction of 9b identified by BiFC, coupled with the absence of nuclear localization signal (NLS) of 9b imply that the self-interactions also occur in the extra-nuclear domain. The conclusion that the 9b protein is retained in both the intra- and extra-nuclear domains is consistent with the paper by Sharma et al.25. In this report, the authors demonstrated that 9b protein can enter the nucleus by passive transport, though 9b was known to localize in the extra-nuclear region25.
The other two Y2H interactions, the self-interaction of ORF14 and interaction between 8a-9b, cannot be seen by BiFC. Additional comprehensive studies are needed to explain this discrepancy.
5. Conclusions
The present report has used multiple, overlapping, comprehensive methods to confirm the existence of well-characterized accessory protein interactions, but also to discover new interactions among these proteins. These interactions provide important clues for a more comprehensive understanding of the SARS-CoV accessory proteins.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81072673).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.apsb.2015.05.002.
Appendix A. Supplementary materials
Supplementary Material
References
- 1.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 2.Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 3.Tan YJ, Teng E, Shen S, Tan TH, Goh PY, Fielding BC. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo E, DeDiego ML, García P, López JA, Pérez-Breña P, Falcón A. Severe acute respiratory syndrome coronavirus accessory proteins 6 and 9b interact in vivo. Virus Res. 2012;169:282–288. doi: 10.1016/j.virusres.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Brunn A, Teepe C, Simpson JC, Pepperkok R, Friedel CC, Zimmer R. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu DX, Fung TS, Chong KK, Shukla A, Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBride R, Fielding BC. The role of severe acute respiratory syndrome (SARS)—coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva JV, Freitas MJ, Felgueiras J, Fardilha M. The power of the yeast two-hybrid system in the identification of novel drug targets: building and modulating PPP1 interactomes. Expert Rev Proteomics. 2015;12:147–158. doi: 10.1586/14789450.2015.1024226. [DOI] [PubMed] [Google Scholar]
- 9.Weis C, Pfeilmeier S, Glawischnig E, Isono E, Pachl F, Hahne H. Co-immunoprecipitation-based identification of putative BAX INHIBITOR-1-interacting proteins involved in cell death regulation and plant-powdery mildew interactions. Mol Plant Pathol. 2013;14:791–802. doi: 10.1111/mpp.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber-Boyvat M, Li S, Skarp KP, Olkkonen VM, Yan D, Jäntti J. Bimolecular fluorescence complementation (BiFC) technique in yeast Saccharomyces cerevisiae and mammalian cells. Methods Mol Biol. 2015;1270:277–288. doi: 10.1007/978-1-4939-2309-0_20. [DOI] [PubMed] [Google Scholar]
- 11.Miller KE, Kim Y, Huh WK, Park HO. Bimolecular fluorescence complementation (BiFC) analysis: advances and recent applications for genome-wide interaction studies. J Mol Biol. 2015;427:2039–2055. doi: 10.1016/j.jmb.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang JW, Fu JX, Li J, Cheng XL, Li F, Dong JF. A novel co-immunoprecipitation protocol based on protoplast transient gene expression for studying protein-protein interactions in rice. Plant Mol Biol Rep. 2014;32:153–161. [Google Scholar]
- 13.Masters SC. Co-immunoprecipitation from transfected cells. Methods Mol Biol. 2004;261:337–350. doi: 10.1385/1-59259-762-9:337. [DOI] [PubMed] [Google Scholar]
- 14.Pazos M, Natale P, Margolin W, Vicente M. Interactions among the early Escherichia coli divisome proteins revealed by bimolecular fluorescence complementation. Environ Microbiol. 2013;15:3282–3291. doi: 10.1111/1462-2920.12225. [DOI] [PubMed] [Google Scholar]
- 15.Kodama Y, Hu CD. Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. Biotechniques. 2012;53:285–298. doi: 10.2144/000113943. [DOI] [PubMed] [Google Scholar]
- 16.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 17.Waisberg M, Cerqueira GC, Yager SB, Francischetti IM, Lu J, Gera N. Plasmodium falciparum merozoite surface protein 1 blocks the proinflammatory protein S100P. Proc Natl Acad Sci U S A. 2012;109:5429–5434. doi: 10.1073/pnas.1202689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell. 2009;33:215–226. doi: 10.1016/j.molcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endo A, Kimura M, Kawakami N, Nambara E. Functional analysis of abscisic acid 8′-hydroxylase. Methods Mol Biol. 2011;773:135–147. doi: 10.1007/978-1-61779-231-1_9. [DOI] [PubMed] [Google Scholar]
- 20.Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 21.Gietz RD, Schiestl RH. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:35–37. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- 22.Sung MK, Huh WK. In vivo quantification of protein-protein interactions in Saccharomyces cerevisiae using bimolecular fluorescence complementation assay. J Microbiol Methods. 2010;83:194–201. doi: 10.1016/j.mimet.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma K, Akerström S, Sharma AK, Chow VT, Teow S, Abrenica B. SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS One. 2011;6:e19436. doi: 10.1371/journal.pone.0019436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material