Abstract
Influenza A virus is the major cause of seasonal or pandemic flu worldwide. Two main treatment strategies–vaccination and small molecule anti-influenza drugs are currently available. As an effective vaccine usually takes at least 6 months to develop, anti-influenza small molecule drugs are more effective for the first line of protection against the virus during an epidemic outbreak, especially in the early stage. Two major classes of anti-influenza drugs currently available are admantane-based M2 protein blockers (amantadine and rimantadine) and neuraminidase (NA) inhibitors (oseltamivir, zanamivir, and peramivir). However, the continuous evolvement of influenza A virus and the rapid emergence of resistance to current drugs, particularly to amantadine, rimantadine, and oseltamivir, have raised an urgent need for developing new anti-influenza drugs against resistant forms of influenza A virus. In this review, we first give a brief introduction of the molecular mechanisms behind resistance, and then discuss new strategies in small-molecule drug development to overcome influenza A virus resistance targeting mutant M2 proteins and neuraminidases, and other viral proteins not associated with current drugs.
KEY WORDS: Influenza A virus, Drug discovery, Resistance, M2 ion channel, Neuraminidase
Graphical abstract
Influenza A virus is the major cause of seasonal or pandemic flu worldwide. The continuous evolvement of influenza A virus and the rapid emergence of resistance to current drugs, particularly to amantadine, rimantadine, and oseltamivir, have raised an urgent need for developing new anti-influenza drugs against resistant forms of influenza A virus. In this review, we first give a brief introduction of the molecular mechanisms behind resistance, and then discuss new strategies in small-molecule drug development to overcome influenza A virus resistance targeting mutant M2 proteins and neuraminidases, and other viral proteins not associated with current drugs.
1. Introduction
Influenza A virus belongs to a family of RNA viruses termed the orthomyxoviridae. It is the main cause of seasonal or pandemic flu, an infectious disease characterized by high morbidity and significant mortality. Since influenza A virus is under continuous evolvement due to antigenic mutation, adaptation, and reassortment, highly virulent strains may appear unexpectedly, resulting in epidemics locally or pandemics worldwide such as the 1918 H1N1 (Spanish flu), 1957 H2N2 (Asian flu), 1968 H3N2 (Hong Kong flu), 2005 H5N1 (bird flu), 2009 H1N1 (swine flu), and more recently H7N9 (bird flu) of 2013. Influenza A virus pandemic may cause significant social health crisis and loss of life. For example, the pandemic of “Spanish flu” (H1N1) in 1918–1919 alone caused at least 20 million deaths. Besides, seasonal flu affects about 20% world population and causes 250,000 to 500,000 deaths per year based on a recent study1.
The potential of devasting pandemic influenza outbreaks has attracted a great amount of resources and efforts in the search for possible prevention and effective treatment methods of influenza A infections. Currently, two main strategies against the virus are available, which are vaccination and small molecule anti-influenza drugs. Anti-influenza small molecule drugs present the first line of protection against the virus during an epidemic outbreak, especially in the early stages, as an effective vaccine usually takes at least 6 months to develop for the circulating strains. Furthermore, vaccination has limited effectiveness in treatment of immunocompromised patients. Moreover, anti-influenza drugs have demonstrated benefit in clinical practices in terms of shortening the disease duration and reducing the risk of influenza-caused serious complications and death if patients are treated in a timely fashion. For all of these reasons, anti-influenza drugs are necessary for the control of influenza A virus pandemics. Currently, two major classes of drugs are approved by FDA for anti-influenza A virus treatment: admantane-based M2 ion channel protein blockers (amantadine and rimantadine) and neuraminidase inhibitors (oseltamivir, zanamivir, and peramivir). However, the continuous evolvement of influenza A virus and the rapid emergence of resistance to current drugs, particularly to admantanes2 and oseltamivir3, 4, has raised great concern for a possible pandemic flu, highlighting an urgent need for developing new anti-influenza drugs against resistant influenza A virus. In this review, we discuss recent progress made in small-molecule drug development to overcome influenza A virus resistance with a focus on novel drug design strategies targeting the mutant M2 ion channel proteins and neuraminidases, as well as other viral proteins not associated with current drugs.
2. Life cycle of influenza A virus: a brief introduction
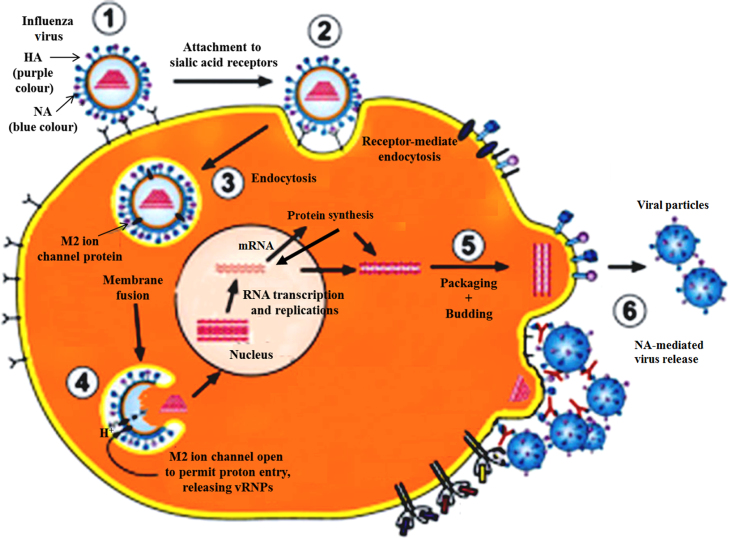
Influenza A virus life cycle is divided into several sequential steps (see Fig. 1). The virus first attaches to host cell surface sialic acid (SA) receptor via the viral surface glycoprotein hemagglutinin. The influenza virus then enters into the cell via receptor-mediated endocytosis, followed by low-pH-induced membrane fusion of the viral envelope with the endosomal membrane of the cell. In this step, the viral M2 protein transports protons from the late endosome into interior of the virus. The resulting acidification induces the conformation change of viral hemagglutinin, which leads to hemagglutinin-mediated membrane fusion followed by the dissociation of viral M1 matrix protein from the viral ribonucleoprotein complexes (vRNPs), resulting in the release of vRNPs into cytoplasm. The vRNPs containing viral genome are then transported into the nucleus to start transcription; mRNAs formed in the transcription process are transported to cytoplasm and are translated into proteins necessary for viral particle replication. Newly synthesized viral genome segments and proteins are assembled to form new vRNPs in the nucleus, which are then transported from nucleus back into the cytoplasm for final packaging. The exportation of vRNPs from the nucleus requires viral nucleoprotein (NP). New virions are then assembled in the cell membrane in a process called budding. During the process, part of the cell membrane is wrapped around virions to form lipid viral envelopes. Finally, neuraminidase (NA) on the surface of new budding viruses cleaves terminal sialic acid (SA) residues from hemagglutinin (HA) and new viruses are released to start a new cycle of infection and replication. All of these steps in the life cycle of influenza A virus are essential for its virulence, replication, and transmission. Development of small molecule based inhibitors that block any of these steps can generate potential efficient strategies to treat or prevent influenza A infections. In the following sections, we will go through new strategies currently being used or proposed for overcoming the resistance of influenza A virus to current M2 ion channel blocker drugs (amantadine and rimantadine) and NA inhibitor drugs (e.g. oseltamivir).
Figure 1.
A schematic presentation of influenza A virus life cycle (adapted with permission from Ref. 5 life cycle Copyright 2012 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim).
3. Drug development targeting mutant M2 ion channel protein
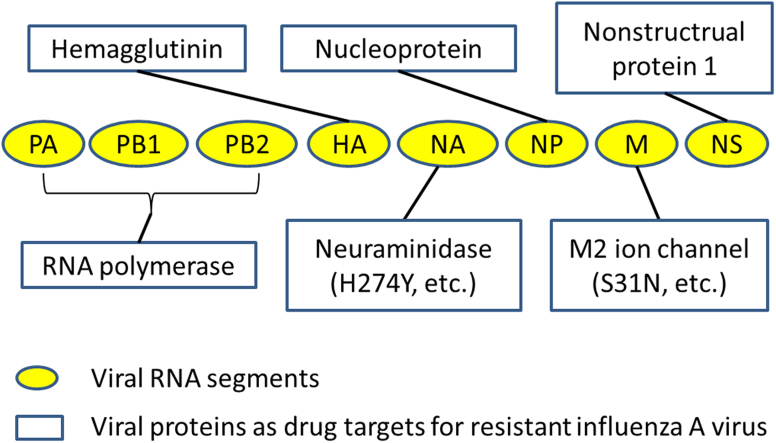
M2 ion channel protein is a tetrameric transmembrane protein forming proton ion channels across the viral membrane6. It functions as a proton transporter which delivers protons from the late endosome into virus interior. The resulting acidification induces the hemagglutinin conformation change, leading to hemagglutinin-mediated membrane fusion and subsequent release virus RNA into cytoplasm of the host cell to initiate subsequent virus replication steps. M2 ion channel blockers bind to the interior of the ion channel, block the influx of protons, and prevent hemagglutinin-mediated membrane fusion. The proton blocking effects may result from an electrostatic repulsion potential from the charged amino group7. Currently, two M2 ion channel blockers approved by the FDA are amantadine (SymmetrelTM, Endo Pharmaceuticals) and rimantadine (FlumadineTM, Forest Pharmaceuticals). But the emergence of drug-resistant mutations of the M2 protein renders these two drugs ineffective against most circulating strains7. Three major mutations identified in transmissible strains are L26F, V27A, and S31N2, 8, all of which are located on the transmembrane domain (residues 24–46) of the M2 protein. Among them, S31N mutation is the prevailing alteration presented in more than 95% of resistant viruses9, 10.
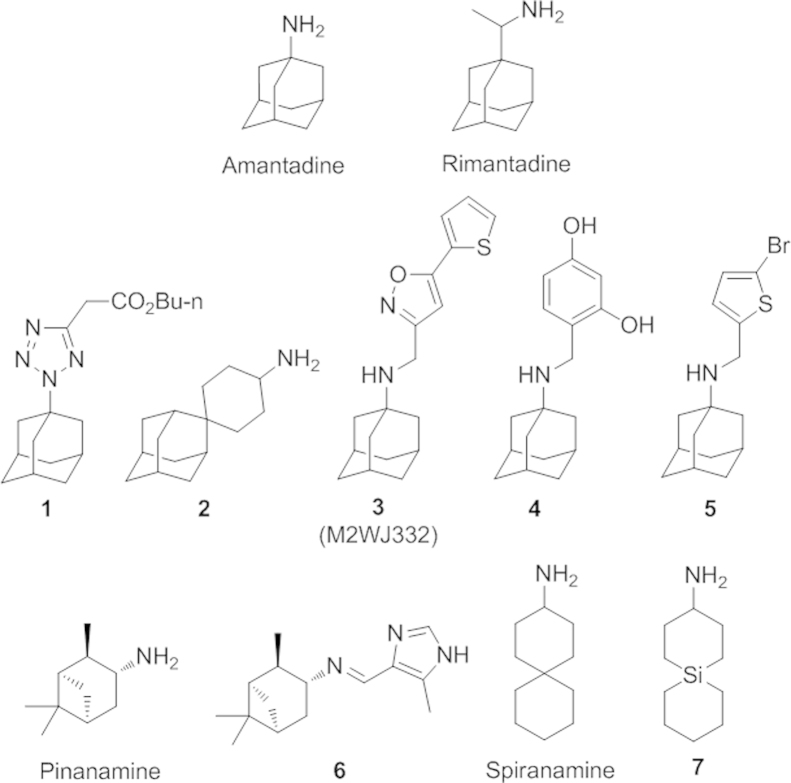
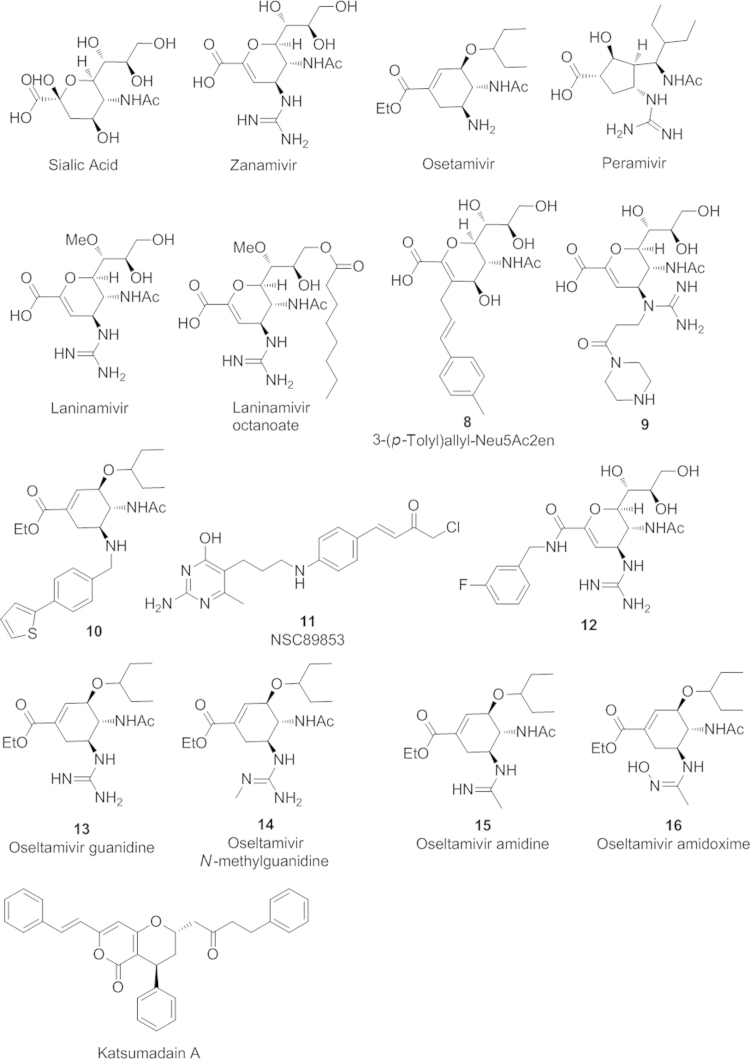
Many efforts have been undertaken to make derivatives of amantadine in order to find new M2 inhibitors against these mutant M2 proteins Fig. 2). Replacement of the amine group with pyrrolidine, azetidine, and aziridine rings did not lead to compounds with better antiviral activities11, 12, 13. In 2010, Zarubaev, et al.14, reported a tetrazole derivative of adamantane (1) which exhibits a highly potent antiviral activity against a rimantadine-resistant influenza strain containing a S31N mutation. More recently, DeGrado and coworkers15 rationally designed a spirane-adamantane derivative (2) with potent efficacy against both the V27A and L26P mutant strains of influenza virus with an activity comparable to amantadine to the wide-type strain. They also further developed the arylmethyl substituted amantadine derivative M2WJ332 (3) and the benzyl-substituted derivative (4). These two compounds showed both inhibitory activity against S31N and wild type M2 proteins. Notably, M2WJ332 has shown an even higher potency against the resistant S31N M2 protein than that of amantadine against wild type M2 protein. Evidences from NMR and molecular dynamic studies supported the drug bound inside the channel between the side chains of Asn31. Very recently, a new dual M2 inhibitor (5) was discovered by the same group to have novel flip-flop dual binding modes16. The drug binds to M2 proteins in different orientations depending upon whether it is a wide type S31 or mutant S31N M2 ion channel protein. Dual-binding inhibitors based upon the similar design strategy may provide new M2 blockers against amantadine or rimantadine resistant influenza virus A strains. Other scaffolds such as pinanamine derivatives17, 18, 19, 20 and spiranamine derivatives15, 21, 22 have also been explored as new M2 ion blockers. As exemplified by compound 620, pinanamine derivatives have shown low inhibiting effects towards resistan S31 mutant virus, while several spiranamine analogs, such as the spiranamine and a silico analog 7, have shown activity against V27A and L26P mutant virus.
Figure 2.
Structures of M2 ion channel blockers.
At present, no M2 blocker is active against both wild type and all circulating M2-mutant strains, likely due to difficulties in tuning the structure of small molecule-based inhibitors to fit each of the individual mutant M2 ion channel proteins. Recently, the allosteric binding site outside of the ion channel of M2 protein was proposed to be a possible target for developing inhibitors against resistant M2 proteins23, though no small molecule has been identified as of yet.
4. Drugs development targeting neuraminidase (NA)
4.1. NA and current NA inhibitor drugs
NA is one of the two major glycoproteins on the surface of viral lipid envelope. It functions as a sialidase to cleave off the terminal sialic acid (SA) on the budding virus, disrupting interactions between SA receptor and virus hemagglutinin24, 25. The SA binding site of NA is highly conserved among different influenza A strains26. Therefore, NA inhibitors which mimic SA structure in its transition state during NA catalyzed hydrolysis, represent a class of anti-influenza drugs with broad activities against different influenza virus strains.
At present, three NA inhibitors have been approved by the FDA, which are zanamivir (RelenzaTM, GlaxoSmithKline), oseltamivir (TamifluTM, Roche), and peramivir (RapivabTM, BioCryst). Zanamivir, a 4-deoxy-4-guanidino derivative of SA27, was the first approved NA inhibitor. It is administered through inhalation due to its poor oral bioavailability. This route of administration is often not possible for young children or some patients with respiratory disease. Thus, it is not as widely used as oseltamivir, the only orally administered drug among the above three approved agents. Oseltamivir is a prodrug, which quickly converts to the active form, oseltamivir carboxylate (GS4071), by an esterase-catalyzed hydrolysis reaction inside the body28. The replacement of the glycerol moiety with a hydrophobic 3-pentyloxy as well as the replacement of 4-guanidino group with an amine group make oseltamivir more lipophilic than zanamivir, and thus achieve a higher oral bioavailability than zanamivir (80% vs. 20%). The third drug paramivir, administered intravenously29, was recently approved by the FDA in December 2014 to treat adult influenza A infections. Structurally, paramivir is distinct from zanamivir and oseltamivir since it contains a unique five-membered ring. On the other hand, it contains structural features of both zanamivir and oseltamivir, the hydrophilic pharmacorphore of 4-guanidino group of zanamivir and the hydrophobic 3-pentanyloxy moiety of oseltamivir. Laninamivir30, another drug which is structurally similar to zanamivir, has been approved in Japan as an octanoyl prodrug (InavirTM, Daiichi Sankyo. Biota). It is currently involved in clinical trials in the United States.
4.2. An overview of resistance to NA inhibitors
Although the SA binding site of the viral NA is highly conserved, a number of mutations arise as a result of drug-induced selective pressure31, leading to the decreased sensitivity to NA inhibitors. The frequency of resistant mutations induced by NA inhibitors is relatively low compared with amantadine and rimantadine32, as the mutation often significantly reduces viral fitness in infection and replication. However, emergence of compensatory mutations may restore viral infectivity and ability to replicate in the host cell33. Understanding the mutation type and molecular mechanism behind the observed resistance helps to design new NA inhibitors against resistant strains.
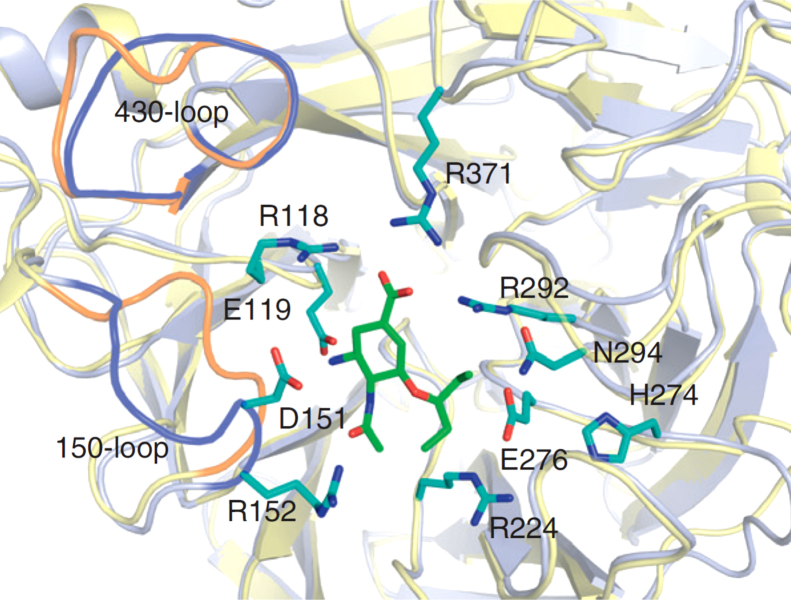
Residues surrounding the active site provide direct interactions with SA and SA-mimicing drugs (Fig. 3). To understand the cause of resistance, the details of the drug binding mode must also be understood. In the case of oseltamivir, the predominating resistant mutation is H274Y. Notably, it does not cause zanamivir resistance. The hydrophobic 3-pentyloxy group in oseltamivir induces a reposition of the Glu276 side chain to achieve optimal hydrophobic interactions. The H274Y mutation blocks these side chain conformational changes and thus reduces the drug binding efficiency of oseltamivir35. While the hydrophilic glycerol moiety in zanamivir interacts with Glu276 through hydrogen bonding in the same way as the natural substrate SA, no conformational rearrangement is required. Therefore, H274 mutation does not cause zanamivir resistance. As paramivir shares the similar hydrophobic interaction with Glu276 as does oseltamivir, H274 mutation may also reduce paramivir sensitivity31. Compared to oseltamivir, drug resistance to zanamivir and laninamivir are rare in clinic to date31, likely due to two reasons. First, their structures are more similar to the natural substrate SA. Secondly, these two drugs are less frequently administered. Therefore, the drug-induced selectivity pressure is low. However, resistance to these drugs is still possible, as a recombinant H3N2 virus containing the E276D mutation has shown resistance to both zanamivir and oseltamivir in one study26. In addition, compensatory mutation residues outside the binding pocket may also contribute to resistance fitness. For example, the compensatory mutations R222Q and V234M associated with H274Y significantly enhance the influenza virus infectivity and its ability to replicate in the host cells, likely due to the restoration of the flexibility of the Glu276 residue previously restricted by the mutation H274Y33.
Figure 3.
Residues around the SA active site of NA (N1) with oseltamivir and the relative positions of 150 and 430 loop of N1 versus N9 (N1: light blue, PDB 2HU0, N9: yellow, PDB 2C4A) (adapted with permission from Ref. 34, Copyright 2012 Elsevier Ltd.).
4.3. Drug development targeting mutant NA
Currently, NA-based drug development against resistant influenza A virus aims to search for novel compounds effective to treat predominant H274 mutant strains. Although zanamivir and laninamivir are still effective against H274 mutation, they are also associated with unfavorable pharmacokinetics and must be administered through inhalation or intravenously. New generations of NA inhibitors should have both excellent activity against resistant strains and improved oral bioavailability. Several strategies are employed to achieve this goal.
4.3.1. Structure-based rational drug design
Structure-based drug design is centered upon an understanding of the dynamic process of NA binding with a substrate and provides new opportunities to design new NA inhibitors. Crystal structures of N1 and N8 NA when each immerged with oseltamivir for a short period time revealed the presence of a transient “150-cavity” near the substrate binding pocket36. The initial binding of SA or NA inhibitors requires the adaptive opening of a 150-loop, and thus generates the 150-cavity. Several C-3 or C-4 modified Neu5Ac2en derivatives (e.g. 8, Fig. 4)37, 38, zanamivir derivatives (e.g. 9)39, and oseltamivir derivatives (e.g. 10)40 were designed and synthesized. The extended side chain at the 3 or 4 positions was believed to bind with the 150-cavity. These compounds demonstrated antiviral activity against both wide-type and oseltamivir-resistant influenza A virus, suggesting the use of the 150-cavity as a novel drug target in NA inhibitor design against resistant strains. However, there is a limitation for 150-cavity targeting NA inhibitors since the 150 cavity is likely be seen only in group 1 influenza A virus (N1, N4, N5, N8), but not in group 2 (N2, N9). Recently, the flexible “430-loop” generated cavity near the active SA binding site was proposed to be a new potential target to design NA inhibitors against both group-1 and group-2 NAs41, 42. This cavity is much larger than SA binding site and thus could possibly accommodate a larger molecular scaffold than current NA inhibiting drugs. An and coworkers reported that the small molecule NA inhibitor NSC89853 (11), which is structurally different from other SA mimics, could possibly bind to the pocket formed by the 430-loop based on molecular modeling43. Feng, et al.44 , recently designed and synthesized a series of zanamivir analogs with an extended side chain at 1-position (e.g., 12) projected towards the 430-loop. The most potent compound 12 shows an IC50 in low nmol/L range against both group-1 and group-2 NAs, demonstrating the potential of “430-cavity” as a powerful strategy in the design of new NA inhibitors.
Figure 4.
Sialic acid and neuraminidase (NA) inhibitors.
4.3.2. Bioisosteric design
Bioisosteric replacement of substituents in current NA inhibitors remains a powerful strategy in the design of new NA inhibitors. Fang and coworkers45 replaced the 2-carboxylic acid group with a phosphonic acid and the obtained phosphonate congeners exhibiting strong antiviral activities against both wide and H274 mutants. Continuous efforts in structural modification of oseltamivir, particularly the replacement of the 5-amino group with other bioisosteric groups such as guanidine (13)46, N-methylguanidine (14)47, amidine (15)48, amidoxime (16)48, etc., led to a number of potent NA inhibitors. Many of them were effective against the H274 mutation, but most of them were not able to retain the superior oral bioavailability of oseltamivir. Very recently, Schade and coworkers48 reported a 5-amidoxime substituted oseltamivir analog 16 which not only has a superior antiviral activity, but also has an oral bioavailability comparable to oseltamivir.
4.3.3. Screening of natural products
Natural products including extracts from traditional medicinal plants continue to serve as a source of new anti-influenza drug leads. Grienke and coworkers49 screened the seed extracts of Alpinia katsumadain, and identified the extract compound katsumadain A with a NA-inhibiting activity at low mM ranges against several strains of H1N1 swine influenza virus including one oseltamivir-resistant strain. Due to the large molecular volume, the binding site of katsumadain A is likely extended into the cavities formed by 430-loop or 245-loop based on docking analysis. Katsumadian A could serve as a leading compound for rational design of new NA inhibitors with novel binding modes. Other studies on tropical medicinal plants50, flavonoids51, 52, and chalcones53 have also been reported. These natural products may help in the development of safer and less toxic NA inhibitors for resistant influenza virus A strains.
5. Drug development targeting other viral proteins
Influenza A virus has eight segmented RNA genes and encodes twelve known peptides: PA, PB1, PB2, PB1-F2, N40, HA, NA, NP, M2 ion channel, M1 matrix proteins, NS1, and NS2. Besides the M2 ion channel protein and NA discussed previously, other viral proteins may also have potentials as drug targets. Many of them have already been under investigation for drug development against resistant influenza A virus.
5.1. Hemagglutinin based anti-influenza drug development
Hemagglutinin (HA) is a membrane-binding glycoprotein on the surface of an influenza virus. It is responsible for virus attachment to sialic acid receptors on the cell membrane in the initial stage of virus infection. It also facilitates the fusion of viral and cell membranes after the virus enters the cell via receptor-mediated endocytosis for subsequent release of viral nucleocapsids into cell cytoplasm54. Accordingly, two strategies have been adopted in anti-virus drug development.
The first strategy is to interfere with hemagglutinin binding to sialic acid receptors. One approach is the addition of SA-containing receptor-mimics as competing inhibitors. Such inhibitors include sialic acid containing natural compounds55, 56 and synthetic multivalent SA-containing inhibitors57. Multivalent SA-containing inhibitors offer better results than monovalent ones in inhibiting virus attachment58. However, multivalent SA-containing compounds often suffer from poor solubility, immunoreactivity, and toxicity issues59. One solution is to use liposome-based drug delivery system to encapsulate inhibitors as exemplified by the sialylneolacto-N-tetraose c (LSTc)-bearing liposomes developed by Hendricks, et al.57.
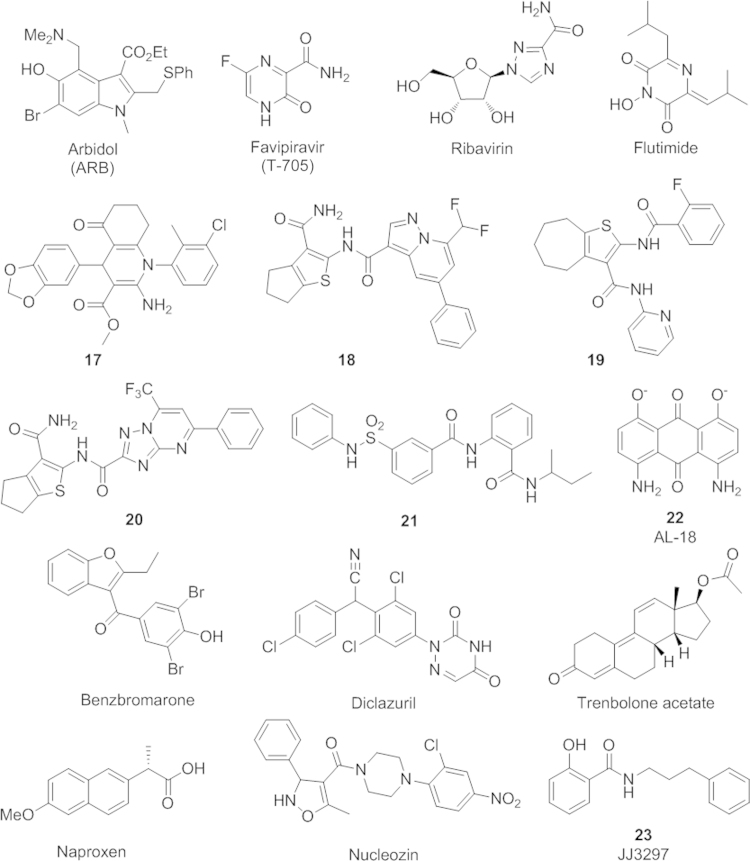
The second strategy involves prevention the formation of a low-pH induced conformation of hemagglutinin, a key step leading to fusion of the host endosomal membrane and the viral lipid envelop. The antiviral activity of this group of compounds is derived from their capability of stabilizing the nonfusogenic conformation of the hemagglutinin. The first compound in this class is tert-butylhydroquinone, which was discovered in 199360. Evidence from NMR study has shown that it binds to the stem loop region in H7 HA and stabilizes the neutral pH conformation61. Several anti-influenza A virus compounds with a similar mechanism were found later. Most of the compounds in this class were subtype-dependent with a low resistance barrier, and underwent no further investigation60, 62, 63, 64, 65. One exception is arbidol (ARB), currently an approved drug in Russia and China to treat influenza virus infection (Fig. 5)66, 67. Aribidol was shown to stabilize the HA by a 0.2 pH unit reduction at the transition phase66. Recent studies pointed out that both the binding of the ARB to the polar head groups of phospholipids in the cell membrane and the presence of the aromatic residues of the viral surface proteins could contribute to its antivirus activity67. A better understanding the details of the drug binding mode and resulting effects on hemagglutinin and lipid interaction would help to design new ARB-like molecules.
Figure 5.
Anti-influenza drugs targeting other viral proteins besides M2 ion channel and NA.
5.2. RNA polymerase as the drug target
Influenza A׳s RNA polymerase is a heterotrimeric protein containing three subunits PB1, PB2, and PA. It is responsible for transcription and replication of the virus genome. All three subunits are important for its functions. PB1 has polymerase activity, and PB2 is involved in cap-binding of host cell pre-mRNAs, whereas PA possesses the endonuclease domain which cleaves capped host pre-mRNAs and initiates transcription. Influenza virus polymerase is highly conserved among different strains68 and contains multiple sites for potential anti-virus drug development. Currently, three major classes of drugs are under development: RNA synthesis inhibitors, cap-snatching inhibitors, and inhibitors targeting protein–protein interactions between the three polymerase subunits (e.g. PA–PB1, PB1–PB2).
RNA synthesis inhibitors are mainly nucleoside mimics such as favipiravir (T-705)69 and ribavirin (Fig. 5)70. Inside the cells, T-705 is metabolized to its active form T-705 ribofuransosyl 5-triphosphate and functions as a purine mimic to inhibit viral RNA-dependent RNA polymerase. Notably, favipiravir does not interfere with mammalian cell RNA or DNA synthesis, so it is nontoxic to the host cell. Moreover, the drug has been shown to be effective against a broad range of influenza strains including amantadine and oseltamivir resistant strains. It demonstrated anti-virus activity against the H5N1 avian flu, recently emerged 2009 H1N1 swine flu, and 2013 H7N9 bird flu71. Additionally, favipiravir was reported to have a low resistance rate69. Favipiravir was approved in Japan in 2014 as a stockpiling drug (AviganTM) during periods of a possible influenza pandemic. Currently, favipiravir is under clinical trials in the United States. Ribavirin, another compound in this class, is converted to ribavirin monophosphate and functions as an inosine 5′-monophosphate (IMP) dehydrogenase in the guanosine 5′-triphosphate sodium salt hydrate (GTP) synthesis pathway, leading to the inhibition of viral RNA synthesis70.
Cap-snatching is a key process in virus RNA transcription. During the cap-snatching process, PB2 first binds to the 5′-methyl cap of host pre-mRNA which is then cleaved by PA׳s endonuclease site to produce a capped primer for transcription initiation72. Both the PB2 cap-binding site and PA′s endonuclease site appear to be potential drug targets. From their calcuations, Lv, et al.73, found two small molecules RO and PPT28, which have a higher affinity with the viral PB 2 cap-binding domain than a known cap analog, m7GTP. RO and PPT28 could serve as potential anti-influenza drugs targeting PB2 cap-binding domain. PA׳s endonuclease active site resides in the N-terminal region of PA and is highly conserved. X-ray crystal structures reveal a conserved cleft in the active site74, 75. A number of cap-snatching inhibitors discovered targeting the endonuclease site have been reported including 4-substituted 2,4-dioxobutanoic acid derivatives76, flutimide (Fig. 5)77, and N-hydroxamic acid/N-hydroxyimide78. Most of these compounds did not undergo further development, partially due to their unknown binding mode to allow for further optimization of their structures. Recently, the X-ray crystal structures of the PA endonuclease domain bound with several inhibitors in this class were reported by two different groups79, 80. These structures provide a better understanding of the binding of the inhibitors at the PA endonuclease active site and may help in designing new PA inhibitors with improved activities.
The interactions between the polymerase′s subunits have recently been proposed to be attractive targets81, as their proper assembly is crucial for the normal activities of the polymerase. Evidence from crystallography data show that the protein–protein interactions between subunits PA and PB1 involve relatively few residues82, 83, suggesting great promise in designing small-molecule based inhibitors. Muratore, et al.84, screened a library of 3 million compounds in silico based on one of the reported crystal structures82, and identified several compounds (17–19) as effective inhibitors against PA–PB1 binding in an ELISA-based assay and in cells (Fig. 5). Particularly, compound 17 was demonstrated to be active against an oseltamivir-resistant strain84. These compounds were identified as lead compounds for further antivirus drug development85. Studies with compound 19 and its analogs have shown that cycloheptathiophene-3-carboxamide scaffold is promising for new anti-influenza virus drug development85. Very recently, continuous efforts along this line led to the discovery of new RNA-dependent RNA polymerase (RdRP) inhibitors (20, 21) through dissection of the thiophene-3-carboxamide molecule via a molecular interaction field (MIF)-based scaffold-hopping method86. Serendipitously, the same group also discovered that the compound AL18 (22), previously reported as a potent inhibitor of human cytomegalovirus DNA polymerase87, blocks the PA/PB1 interaction and inhibits influenza A virus replication88. In another study, Fukuoka and coworkers89 performed a docking simulation-based screening of ~4000 drugs. Benzbromarone, diclazuril, and trenbolone acetate, were identified as three successful hit compounds with strong anti-influenza activities (Fig. 5). Two compounds, benzbromarone and diclazuril, were confirmed to be bound to PA and decreased the activity of the viral RNA polymerase89. More recently, Kessler, et al., discovered benzofurazan-based novel inhibitors targeting PA–PB1 binding interfaces using an ELISA-based high-throughput screening method. However, this serie of compounds has unfavorable toxicity profiles for in vivo use. Overall, the PA–PB1 binding interface has emerged to be a potential fruitful target for a new generation of anti-influenza A drug development.
Similar anti-viral opportunities are also seen in PB1–PB2 interactions, since the crystal structure of the binding interface of PB1–PB2 shows that only a small section of PB1 (residues 678–757) and of PB2 (residues 1–37) are involved in the interface90. To date, several synthetic-peptide-based inhibitors have been reported to be able to interfere with PB1–PB2 interactions and disrupt normal function of the polymerase91, 92. However, nonpeptide small-molecule based inhibitors remain to be discovered. One particular challenge is to design small-molecules fitting in the flat PB1–PB2 interface.
5.3. Nucleoprotein (NP) as the drug target
The influenza virus nucleoprotein (NP) is one of the most abundant viral proteins produced in the virus replication process inside the host cell. During the viral life cycle, NP binds with influenza viral RNA and polymerase subunits (PB1, PB2, and PA) to form the viral ribonucleoprotein complexes (vRNPs). It also involved in vRNPs nuclear import, replication, and export93, 94. NP has emerged as a novel target for anti-virus drug development because of its important structural and regulatory roles in virus replication. Recent large-scale sequence analysis of influenza A virus NP has identified several already known functional region as well as new highly conserved sites as potential drug-targets95.
Currently, NP-based antiviral drug development has formed two major classes of inhibitors that target on NP interaction with virus RNA96 and block formation of vRNPs by the mechanism of NP oligomerization97, 98, 99. Naproxen, an approved nonsteroidal anti-inflammatory drug that inhibits inducible cyclooxygenase-2 (COX-2), was the first inhibitor discovered through in silico virtual screening to inhibit the NP-RNA interactions (Fig. 5)96. It is effective against H1N1 and H3N2 strains both in vitro and in vivo. Single-point mutation experiments indicated that naproxen targets the RNA binding groove of NP and prevents its interaction with virus RNA. Naproxen serves as a lead compound for further development, particularly as a dual-functional drug with both antiviral and anti-inflammatory benefits. Nucleozin (Fig. 5) and its derivatives represent the other class of small molecules targeting NP oligomerization97, 98, 99. Nucleozin was first identified in 2010 by two independent groups97, 100. It appears to exert inhibitory effects at both early and late stages in the influenza virus life cycle101. Nucleozin not only inhibits viral RNA and protein synthesis, but also stabilizes NP self-oligomerization and promotes nonfunctional aggregates of NP. Moreover, it blocks the exportation of the vRNPs from the nucleus and facilitates the formation of large perinuclear aggregates of vRNPs along with the cellular protein Rab11101.
5.4. Nonstructrual protein I (NS1) as the drug target
The NS1 protein of influenza A virus has multiple functions102. It suppresses the host responses to the virus infection, in particular the interferon(IFN)-mediated response103, through binding with viral dsRNA104, CPSF30105, the ubiquitin-ligase TRIM25106, and phosphoinositide 3-kinase (PI3K)107. The distinct binding sites of NS1 with these biomolecules provide potential opportunities for drug discovery108. Currently, drug development targeting the NS1 protein is still in the early stages.
Serving as a proof of concept that NS1 could be a feasible target, a few lead compounds that inhibit NS1 functions were identified by either a yeast-based assay109 or virtual screening110. Engel and coworkers109 tested 2000 compounds in yeast model expressing NS1. Four compounds were found to reverse the growth inhibiting effect of NS1. These compounds potentially exhibited antivirus activity by blocking NS1-mediated IFN response. Further derivatization of these compounds leads to the compound JJ3297 (23) (Fig. 5)111, which facilitated the restoration of an IFN-like antiviral state and inhibited virus replication and spread in a RNase L dependent manner. Very recently, Ai et al.110 screened a library of more than 30,000 compounds from traditional Chinese medicine using a structural-based docking method, and identified two compounds 32056 and 31674 that show antiviral activities by stably binding to the CPSF30-binding site of NS1. In addition, several compounds interfering with NS1/viral RNA binding were identified either through virtual112 or high-throughput screening methods113, 114.
6. Combination therapy
Combination therapy uses several antivirus drugs with different mechanisms of action in one regime to achieve greater efficacy than one drug dose alone based on drug synergistic effect. It allows for lower doses of each component, resulting in reduced side effects. In addition, it may significantly increase the selection barrier for resistance. Thus, it presents a promising strategy for the treatment of influenza A virus, particularly for severe and immunocompromised patients115. A combination of amantadine+oseltamivir116 as well as favirpiravir+oseltamivir117 have demonstrated enhanced drug efficacy in H5N1 infected mice. Recently, combination therapy with three antiviral drugs (amantadine, ribavirin, and oseltamivir) in patients with severe A/H1N1 2009 (pH1N1) influenza in Korea has reduced the 14-day mortality118. In another study, triple-combination antiviral drug therapy showed similar pharmacokinetics to mono-drug therapy and demonstrated increased safety in immunocompromised patients118, 119. Currently, several combination therapies based on approved and/or new drugs are under investigation.
7. Conclusions
The rapid emergence of influenza virus A resistance to current anti-virus drugs and the unpredictable nature of a potential influenza pandemic outbreak has impelled scientists and researchers to search for new antiviral strategies to overcome resistance and provide for better prophylactic and therapeutic outcomes than what current drugs can offer us. Recent progress in basic molecular biology, biophysics, and virology has provided important information regarding the molecular and structural basis behind the resistant influenza A virus and has helped to guide new drug development strategies for antiviral treatments. These new strategies target the mutant M2 proteins, neuraminidases, or other viral proteins. Ideally, the next generation of antivirus drug should have strong potency against a broad range of current resistant strains with a much higher resistance barrier. A number of small-molecule based drugs have been identified through structure-based drug design and in vitro or in vivo screening. A few have advanced into different phases of clinical trials. They may provide new options for drug treatment and prevention of influenza virus A infections in the near future, either alone or in combination with other drugs, to keep flu pandemics under check.
Acknowledgments
The work was supported by the Fundamental Research Funds for the Central Universities (Kaiyan Lou) and East China University of Science and Technology (start-up funds to Wei Wang). We also would like to thank William Hanafin at the Univeristy of Illinois for his editing suggestions.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Contributor Information
Kaiyan Lou, Email: kylou@ecust.edu.cn.
Wei Wang, Email: wwang@unm.edu.
References
- 1.World Health Organization. Influenza (seasonal ) fact sheet N˚211. 2014. Available from: 〈http://www.who.int/mediacentre/factsheets/fs211/en/〉.
- 2.Deyde VM, Xu XY, Bright RA, Shaw M, Smith CB, Zhang Y. Surveillance of resistance to adamantanes among influenza A (H3N2) and A (H1N1) viruses isolated worldwide. J Infect Dis. 2007;196:249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- 3.Moscona A. Oseltamivir resistance-disabling our influenza defenses. N Engl J Med. 2005;353:2633–2636. doi: 10.1056/NEJMp058291. [DOI] [PubMed] [Google Scholar]
- 4.Sheu TG, Fry AM, Garten RJ, Deyde VM, Shwe T, Bullion L. Dual resistance to adamantanes and oseltamivir among seasonal influenza A(H1N1) viruses: 2008–2010. J Infect Dis. 2011;203:13–17. doi: 10.1093/infdis/jiq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng EG, Ye DJ, Li J, Zhang DY, Wang JF, Zhao F. Recent advances in neuraminidase inhibitor development as anti-influenza drugs. ChemMedChem. 2012;7:1527–1536. doi: 10.1002/cmdc.201200155. [DOI] [PubMed] [Google Scholar]
- 6.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 7.Leonov H, Astrahan P, Krugliak M, Arkin IT. How do aminoadamantanes block the influenza M2 channel, and how does resistance develop? J Am Chem Soc. 2011;133:9903–9911. doi: 10.1021/ja202288m. [DOI] [PubMed] [Google Scholar]
- 8.Furuse Y, Suzuki A, Oshitani H. Large-scale sequence analysis of M gene of influenza A viruses from different species: mechanisms for emergence and spread of amantadine resistance. Antimicrob Agents Chemother. 2009;53:4457–4463. doi: 10.1128/AAC.00650-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krumbholz A, Schmidtke M, Bergmann S, Motzke S, Bauer K, Stech J. High prevalence of amantadine resistance among circulating European porcine influenza A viruses. J Gen Virol. 2009;90:900–908. doi: 10.1099/vir.2008.007260-0. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Wu YB, Ma CL, Fiorin G, Wang JZ, Pinto LH. Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc Natl Acad Sci U S A. 2013;110:1315–1320. doi: 10.1073/pnas.1216526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolocouris N, Kolocouris A, Foscolos GB, Fytas G, Neyts J, Padalko E. Synthesis and antiviral activity evaluation of some new aminoadamantane derivatives. 2. J Med Chem. 1996;39:3307–3318. doi: 10.1021/jm950891z. [DOI] [PubMed] [Google Scholar]
- 12.Stamatiou G, Kolocouris A, Kolocouris N, Fytas G, Foscolos GB, Neyts J. Novel 3-(2-adamantyl)pyrrolidines with potent activity against influenza A virus-identification of aminoadamantane derivatives bearing two pharmacophoric amine groups. Bioorg Med Chem Lett. 2001;11:2137–2142. doi: 10.1016/s0960-894x(01)00388-2. [DOI] [PubMed] [Google Scholar]
- 13.Zoidis G, Fytas C, Papanastasiou I, Foscolos GB, Fytas G, Padalko E. Heterocyclic rimantadine analogues with antiviral activity. Bioorg Med Chem. 2006;14:3341–3348. doi: 10.1016/j.bmc.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 14.Zarubaev VV, Golod EL, Anfimov PM, Shtro AA, Saraev VV, Gavrilov AS. Synthesis and anti-viral activity of azolo-adamantanes against influenza A virus. Bioorg Med Chem. 2010;18:839–848. doi: 10.1016/j.bmc.2009.11.047. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Ma CL, Wu YB, Lamb RA, Pinto LH, DeGrado WF. Exploring organosilane amines as potent inhibitors and structural probes of influenza A virus M2 proton channel. J Am Chem Soc. 2011;133:13844–13847. doi: 10.1021/ja2050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YB, Canturk B, Jo H, Ma CL, Gianti E, Klein ML. Flipping in the pore: discovery of dual inhibitors that bind in different orientations to the wild-type versus the amantadine-resistant 531N mutant of the influenza A virus M2 proton channel. J Am Chem Soc. 2014;136:17987–17995. doi: 10.1021/ja508461m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu WH, Zeng SG, Li CF, Jie YL, Li ZY, Chen L. Identification of hits as matrix-2 protein inhibitors through the focused screening of a small primary amine library. J Med Chem. 2010;53:3831–3834. doi: 10.1021/jm901664a. [DOI] [PubMed] [Google Scholar]
- 18.Hu FH, Luo WB, Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Li CF, Zeng SG, Hu WH. Discovery of highly potent agents against influenza A virus. Eur J Med Chem. 2011;46:52–57. doi: 10.1016/j.ejmech.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X, Jie YL, Rosenberg MR, Wan JT, Zeng SG, Cui W. Design and synthesis of pinanamine derivatives as anti-influenza A M2 ion channel inhibitors. Antivir Res. 2012;96:91–99. doi: 10.1016/j.antiviral.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Balannik V, Wang J, Ohigashi Y, Jing XH, Magavern E, Lamb RA. Design and pharmacological characterization of inhibitors of amantadine-resistant mutants of the M2 ion channel of influenza A virus. Biochemistry. 2009;48:11872–11882. doi: 10.1021/bi9014488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz S, Luo GX, Hahnenberger KM, Brooks C, Gecha O, Ingalls K. Growth impairment resulting from expression of influenza virus M2 protein in saccharomyces cerevisiae: identification of a novel inhibitor of influenza virus. Antimicrob Agents Chemother. 1995;39:2204–2209. doi: 10.1128/aac.39.10.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk H-D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T, Takahashi T, Guo CT, Hidari KI-PJ, Miyamoto D, Goto H. Sialidase activity of influenza A virus in an endocytic pathway enhances viral replication. J Virol. 2005;79:11705–11715. doi: 10.1128/JVI.79.18.11705-11715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yen HL, Hoffmann E, Taylor G, Scholtissek C, Monto AS, Webster RG. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J Virol. 2006;80:8787–8795. doi: 10.1128/JVI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 28.Davies BE. Pharmacokinetics of oseltamivir: an oral antiviral for the treatment and prophylaxis of influenza in diverse populations. J Antimicrob Chemother. 2010;65:II5–II10. doi: 10.1093/jac/dkq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohno S, Kida H, Mizuguchi M, Hirotsu N, Ishida T, Kadota J. Intravenous peramivir for treatment of influenza A and B virus infection in high-risk patients. Antimicrob Agents Chemother. 2011;55:2803–2812. doi: 10.1128/AAC.01718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koyama K, Takahashi M, Oitate M, Nakai N, Takakusa H, Miura S. CS-8958, a prodrug of the novel neuraminidase inhibitor R-125489, demonstrates a favorable long-retention profile in the mouse respiratory tract. Antimicrob Agents Chemother. 2009;53:4845–4851. doi: 10.1128/AAC.00731-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samson M, Pizzorno A, Abed Y, Boivin G. Influenza virus resistance to neuraminidase inhibitors. Antivir Res. 2013;98:174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Pizzorno A, Bouhy X, Abed Y, Boivin G. Generation and characterization of recombinant pandemic influenza A (H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis. 2011;203:25–31. doi: 10.1093/infdis/jiq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J, Cross TA, Zhou HX. Recent progress in structure-based anti-influenza drug design. Drug Disc Today. 2012;17:1111–1120. doi: 10.1016/j.drudis.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins PJ, Haire LF, Lin YP, Liu JF, Russell RJ, Walker PA. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature. 2008;453:1258–1261. doi: 10.1038/nature06956. [DOI] [PubMed] [Google Scholar]
- 36.Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 37.Rudrawar S, Dyason JC, Rameix-Welti M-A, Rose FJ, Kerry PS, Russell RJM. Novel sialic acid derivatives lock open the 150-loop of an influenza A virus group-1 sialidase. Nat Commun. 2010;1:113. doi: 10.1038/ncomms1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudrawar S, Kerry PS, Rameix-Welti M-A, Maggioni A, Dyason JC, Rose FJ. Synthesis and evaluation of novel 3-C-alkylated-Neu5Ac2en derivatives as probes of influenza virus sialidase 150-loop flexibility. Org Biomol Chem. 2012;10:8628–8639. doi: 10.1039/c2ob25627d. [DOI] [PubMed] [Google Scholar]
- 39.Wen WH, Wang SY, Tsai KC, Cheng YSE, Yang AS, Fang JM. Analogs of zanamivir with modified C4-substituents as the inhibitors against the group-1 neuraminidases of influenza viruses. Bioorg Med Chem. 2010;18:4074–4084. doi: 10.1016/j.bmc.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Xie YC, Xu DQ, Huang B, Ma XL, Qi WB, Shi FY. Discovery of N-substituted oseltamivir derivatives as potent and selective inhibitors of H5N1 influenza neuraminidase. J Med Chem. 2014;57:8445–8458. doi: 10.1021/jm500892k. [DOI] [PubMed] [Google Scholar]
- 41.Amaro RE, Minh DDL, Cheng LS, Lindstrom WM, Jr, Olson AJ, Lin JH. Remarkable loop flexibility in avian influenza N1 and its implications for antiviral drug design. J Am Chem Soc. 2007;129:7764–7765. doi: 10.1021/ja0723535. [DOI] [PubMed] [Google Scholar]
- 42.Kirchmair J, Rollinger JM, Liedl KR, Seidel N, Krumbholz A, Schmidtke M. Novel neuraminidase inhibitors: identification, biological evaluation and investigations of the binding mode. Future Med Chem. 2011;3:437–450. doi: 10.4155/fmc.10.292. [DOI] [PubMed] [Google Scholar]
- 43.An JH, Lee DCW, Law AHY, Yang CLH, Poon LLM, Lau ASY. A novel small-molecule inhibitor of the avian influenza H5N1 virus determined through computational screening against the neuraminidase. J Med Chem. 2009;52:2667–2672. doi: 10.1021/jm800455g. [DOI] [PubMed] [Google Scholar]
- 44.Feng EG, Shin W-J, Zhu XL, Li J, Ye DJ, Wang J. Structure-based design and synthesis of C-1- and C-4-modified analogs of zanamivir as neuraminidase inhibitors. J Med Chem. 2013;56:671–684. doi: 10.1021/jm3009713. [DOI] [PubMed] [Google Scholar]
- 45.Shie JJ, Fang JM, Wang SY, Tsai KC, Cheng YS, Yang AS. Synthesis of tamiflu and its phosphonate congeners possessing potent anti-influenza activity. J Am Chem Soc. 2007;129:11892–11893. doi: 10.1021/ja073992i. [DOI] [PubMed] [Google Scholar]
- 46.Kim CU, Lew W, Williams MA, Wu HW, Zhang LJ, Chen XW. Structure-activity relationship studies of novel carbocyclic influenza neuraminidase inhibitors. J Med Chem. 1998;41:2451–2460. doi: 10.1021/jm980162u. [DOI] [PubMed] [Google Scholar]
- 47.Mooney CA, Johnson SA, ′t Hart P, Quarles Van Ufford L, De Haan CAM, Moret EE. Oseltamivir analogues bearing N-substituted guanidines as potent neuraminidase inhibitors. J Med Chem. 2014;57:3154–3160. doi: 10.1021/jm401977j. [DOI] [PubMed] [Google Scholar]
- 48.Schade D, Kotthaus J, Riebling L, Kotthaus J, Müller-Fielitz H, Raasch W. Development of novel potent orally bioavailable oseltamivir derivatives active against resistant influenza A. J Med Chem. 2014;57:759–769. doi: 10.1021/jm401492x. [DOI] [PubMed] [Google Scholar]
- 49.Grienke U, Schmidtke M, Kirchmair J, Pfarr K, Wutzler P, Dürrwald R. Antiviral potential and molecular insight into neuraminidase inhibiting diarylheptanoids from Alpinia katsumadai. J Med Chem. 2010;53:778–786. doi: 10.1021/jm901440f. [DOI] [PubMed] [Google Scholar]
- 50.Rajasekaran D, Palombo EA, Yeo TC, Ley DLS, Tu CL, Malherbe F. Identification of traditional medicinal plant extracts with novel anti-influenza activity. PLoS One. 2013;8:e79293. doi: 10.1371/journal.pone.0079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dao T-T, Tung B-T, Nguyen P-H, Thuong P-T, Yoo S-S, Kim E-H. C-methylated flavonoids from Cleistocalyx operculatus and their inhibitory effects on novel influenza A (H1N1) neuraminidase. J Nat Prod. 2010;73:1636–1642. doi: 10.1021/np1002753. [DOI] [PubMed] [Google Scholar]
- 52.Liu AL, Wang HD, Lee SM, Wang YT, Du GH. Structure-activity relationship of flavonoids as influenza virus neuraminidase inhibitors and their in vitro anti-viral activities. Bioorg Med Chem. 2008;16:7141–7147. doi: 10.1016/j.bmc.2008.06.049. [DOI] [PubMed] [Google Scholar]
- 53.Dao TT, Nguyen PH, Lee HS, Kim E, Park J, Lim SI. Chalcones as novel influenza A (H1N1) neuraminidase inhibitors from Glycyrrhiza inflata. Bioorg Med Chem Lett. 2011;21:294–298. doi: 10.1016/j.bmcl.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 54.Hamilton BS, Whittaker GR, Daniel S. Influenza virus-mediated membrane fusion: determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses. 2012;4:1144–1168. doi: 10.3390/v4071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terabayashi T, Morita M, Ueno M, Nakamura T, Urashima T. Inhibition of influenza-virus-induced cytopathy by sialylglycoconjugates. Carbohydr Res. 2006;341:2246–2253. doi: 10.1016/j.carres.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 56.Job ER, Bottazzi B, Gilbertson B, Edenborough KM, Brown LE, Mantovani A. Serum amyloid P is a sialylated glycoprotein inhibitor of influenza A viruses. PLoS One. 2013;8:e59623. doi: 10.1371/journal.pone.0059623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hendricks GL, Weirich KL, Viswanathan K, Li J, Shriver ZH, Ashour J. Sialylneolacto-N-tetraose c (LSTc)-bearing liposomal decoys capture influenza A virus. J Biol Chem. 2013;288:8061–8073. doi: 10.1074/jbc.M112.437202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun XL. Recent anti-influenza strategies in multivalent sialyloligosaccharides and sialylmimetics approaches. Curr Med Chem. 2007;14:2304–2313. doi: 10.2174/092986707781696582. [DOI] [PubMed] [Google Scholar]
- 59.Gambaryan AS, Tuzikov AB, Chinarev AA, Juneja LR, Bovin NV, Matrosovich MN. Polymeric inhibitor of influenza virus attachment protects mice from experimental influenza infection. Antivir Res. 2002;55:201–205. doi: 10.1016/s0166-3542(02)00020-7. [DOI] [PubMed] [Google Scholar]
- 60.Bodian DL, Yamasaki RB, Buswell RL, Stearns JF, White JM, Kuntz ID. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry. 1993;32:2967–2978. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- 61.Antanasijevic A, Cheng H, Wardrop DJ, Rong LJ, Caffrey M. Inhibition of influenza H7 hemagglutinin-mediated entry. PLoS One. 2013;8:e76363. doi: 10.1371/journal.pone.0076363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plotch SJ, O׳ Hara B, Morin J, Palant O, LaRocque J, Bloom JD. Inhibition of influenza A virus replication by compounds interfering with the fusogenic function of the viral hemagglutinin. J Virol. 1999;73:140–151. doi: 10.1128/jvi.73.1.140-151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshimoto J, Kakui M, Iwasaki H, Fujiwara T, Sugimoto H, Hattori N. Identification of a novel HA conformational change inhibitor of human influenza virus. Arch Virol. 1999;144:865–878. doi: 10.1007/s007050050552. [DOI] [PubMed] [Google Scholar]
- 64.Vanderlinden E, Göktas F, Cesur Z, Froeyen M, Reed ML, Russell CJ. Novel inhibitors of influenza virus fusion: structure-activity relationship and interaction with the viral hemagglutinin. J Virol. 2010;84:4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo G, Torri A, Harte WE, Danetz S, Cianci C, Tiley L. Molecular mechanism underlying the action of a novel fusion inhibitor of influenza A virus. J Virol. 1997;71:4062–4070. doi: 10.1128/jvi.71.5.4062-4070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leneva IA, Russell RJ, Boriskin YS, Hay AJ. Characteristics of arbidol-resistant mutants of influenza virus: implications for the mechanism of anti-influenza action of arbidol. Antivir Res. 2009;81:132–140. doi: 10.1016/j.antiviral.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Teissier E, Zandomeneghi G, Loquet A, Lavillette D, Lavergne J-P, Montserret R. Mechanism of inhibition of enveloped virus membrane fusion by the antiviral drug arbidol. PLoS One. 2011;6:e15874. doi: 10.1371/journal.pone.0015874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Babar MM, Zaidi NU, Tahir M. Global geno-proteomic analysis reveals cross-continental sequence conservation and druggable sites among influenza virus polymerases. Antivir Res. 2014;112:120–131. doi: 10.1016/j.antiviral.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 69.Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antivir Res. 2009;82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sidwell RW, Bailey KW, Wong MH, Barnard DL, Smee DF. In vitro and in vivo influenza virus-inhibitory effects of viramidine. Antivir Res. 2005;68:10–17. doi: 10.1016/j.antiviral.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 71.Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fodor E. The RNA polymerase of influenza A virus: mechanisms of viral transcription and replication. Acta Virol. 2013;57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 73.Lv HM, Guo XL, Gu RX, Wei DQ. Free energy calculations and binding analysis of two potential anti-influenza drugs with polymerase basic protein-2 (PB2) Protein Peptide Lett. 2011;18:1002–1009. doi: 10.2174/092986611796378675. [DOI] [PubMed] [Google Scholar]
- 74.Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 75.Yuan PW, Bartlam M, Lou ZY, Chen SD, Zhou J, He XJ. Crystal structure of an avian influenza polymerase PAN reveals an endonuclease active site. Nature. 2009;458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 76.Tomassini J, Selnick H, Davies ME, Armstrong ME, Baldwin J, Bourgeois M. Inhibition of cap (m-7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2, 4-dioxobutanoic acid compounds. Antimicrob Agents Chemother. 1994;38:2827–2837. doi: 10.1128/aac.38.12.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tomassini JE, Davies ME, Hastings JC, Lingham R, Mojena M, Raghoobar SL. A novel antiviral agent which inhibits the endonuclease of influenza viruses. Antimicrob Agents Chemother. 1996;40:1189–1193. doi: 10.1128/aac.40.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parkes KEB, Ermert P, Fässler J, Ives J, Martin JA, Merrett JH. Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J Med Chem. 2003;46:1153–1164. doi: 10.1021/jm020334u. [DOI] [PubMed] [Google Scholar]
- 79.DuBois RM, Slavish PJ, Baughman BM, Yun MK, Bao J, Webby RJ. Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 2012;8:e1002830. doi: 10.1371/journal.ppat.1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kowalinski E, Zubieta C, Wolkerstorfer A, Szolar OHJ, Ruigrok RWH, Cusack S. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 2012;8:e1002831. doi: 10.1371/journal.ppat.1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palu G, Loregian A. Inhibition of herpesvirus and influenza virus replication by blocking polymerase subunit interactions. Antivir Res. 2013;99:318–327. doi: 10.1016/j.antiviral.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 82.He XJ, Zhou J, Bartlam M, Zhang RG, Ma JY, Lou ZY. Crystal structure of the polymerase PAC–PB1N complex from an avian influenza H5N1 virus. Nature. 2008;454:1123–1126. doi: 10.1038/nature07120. [DOI] [PubMed] [Google Scholar]
- 83.Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature. 2008;454:1127–1131. doi: 10.1038/nature07225. [DOI] [PubMed] [Google Scholar]
- 84.Muratore G, Goracci L, Mercorelli B, Foeglein A, Digard P, Cruciani G. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc Natl Acad Sci U S A. 2012;109:6247–6252. doi: 10.1073/pnas.1119817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Massari S, Nannetti G, Goracci L, Sancineto L, Muratore G, Sabatini S. Structural investigation of cycloheptathiophene-3-carboxamide derivatives targeting influenza virus polymerase assembly. J Med Chem. 2013;56:10118–10131. doi: 10.1021/jm401560v. [DOI] [PubMed] [Google Scholar]
- 86.Lepri S, Nannetti G, Muratore G, Cruciani G, Ruzziconi R, Mercorelli B. Optimization of small-molecule inhibitors of influenza virus polymerase: from thiophene-3-carboxamide to polyamido scaffolds. J Med Chem. 2014;57:4337–4350. doi: 10.1021/jm500300r. [DOI] [PubMed] [Google Scholar]
- 87.Loregian A, Coen DM. Selective anti-cytomegalovirus compounds discovered by screening for inhibitors of subunit interactions of the viral polymerase. Chem Biol. 2006;13:191–200. doi: 10.1016/j.chembiol.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 88.Muratore G, Mercorelli B, Goracci L, Cruciani G, Digard P, Palu G. Human cytomegalovirus inhibitor AL18 also possesses activity against influenza A and B viruses. Antimicrob Agents Chemother. 2012;56:6009–6013. doi: 10.1128/AAC.01219-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fukuoka M, Minakuchi M, Kawaguchi A, Nagata K, Kamatari YO, Kuwata K. Structure-based discovery of anti-influenza virus A compounds among medicines. Biochim Biophys Acta – Gen Subj. 2012;1820:90–95. doi: 10.1016/j.bbagen.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 90.Sugiyama K, Obayashi E, Kawaguchi A, Suzuki Y, Tame JRH, Nagata K. Structural insight into the essential PB1–PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 2009;28:1803–1811. doi: 10.1038/emboj.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chase G, Wunderlich K, Reuther P, Schwemmle M. Identification of influenza virus inhibitors which disrupt of viral polymerase protein–protein interactions. Methods. 2011;55:188–191. doi: 10.1016/j.ymeth.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 92.Li CF, Ba Q, Wu AP, Zhang H, Deng T, Jiang TJ. A peptide derived from the C-terminus of PB1 inhibits influenza virus replication by interfering with viral polymerase assembly. FEBS J. 2013;280:1139–1149. doi: 10.1111/febs.12107. [DOI] [PubMed] [Google Scholar]
- 93.Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 94.Amorim M-J, Read EK, Dalton RM, Medcalf L, Digard P. Nuclear export of influenza A virus mRNAs requires ongoing RNA polymerase II activity. Traffic. 2007;8:1–11. doi: 10.1111/j.1600-0854.2006.00507.x. [DOI] [PubMed] [Google Scholar]
- 95.Kukol A, Hughes DJ. Large-scale analysis of influenza A virus nucleoprotein sequence conservation reveals potential drug-target sites. Virology. 2014;454–455:40–47. doi: 10.1016/j.virol.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 96.Lejal N, Tarus B, Bouguyon E, Chenavas S, Bertho N, Delmas B. Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob Agents Chemother. 2013;57:2231–2242. doi: 10.1128/AAC.02335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kao RY, Yang D, Lau L-S, Tsui WH, Hu LH, Dai J. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol. 2010;28:600–605. doi: 10.1038/nbt.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gerritz SW, Cianci C, Kim S, Pearce BC, Deminie C, Discotto L. Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers. Proc Natl Acad Sci U S A. 2011;108:15366–15371. doi: 10.1073/pnas.1107906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng HM, Wan JT, Lin M-I, Liu YX, Lu XY, Liu JS. Design, synthesis, and in vitro biological evaluation of 1H-1,2,3-triazole-4-carboxamide derivatives as new anti-influenza a agents targeting virus nucleoprotein. J Med Chem. 2012;55:2144–2153. doi: 10.1021/jm2013503. [DOI] [PubMed] [Google Scholar]
- 100.Su C-Y, Cheng T-JR, Lin M-I, Wang SY, Huang W-I, Lin-Chu S-Y. High-throughput identification of compounds targeting influenza RNA-dependent RNA polymerase activity. Proc Natl Acad Sci U S A. 2010;107:19151–19156. doi: 10.1073/pnas.1013592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amorim MJ, Kao RY, Digard P. Nucleozin targets cytoplasmic trafficking of viral ribonucleoprotein-rab11 complexes in influenza A virus infection. J Virol. 2013;87:4694–4703. doi: 10.1128/JVI.03123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ayllon J, García-Sastre A. The NS1 protein: a multitasking virulence factor. In: Oldstone MBA, Compans RW, editors. Springer International Publishing; Switzerland: 2015. pp. 73–107. [Google Scholar]
- 103.Ayllon J, Hale BG, García-Sastre A. Strain-specific contribution of NS1-activated phosphoinositide 3-kinase signaling to influenza A virus replication and virulence. J Virol. 2012;86:5366–5370. doi: 10.1128/JVI.06722-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marc D, Barbachou S, Soubieux D. The RNA-binding domain of influenzavirus non-structural protein-1 cooperatively binds to virus-specific RNA sequences in a structure-dependent manner. Nucl Acids Res. 2013;41:434–449. doi: 10.1093/nar/gks979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Das K, Ma L-C, Xiao R, Radvansky B, Aramini J, Zhao L. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A. 2008;105:13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gack MU, Albrecht RA, Urano T, Inn K-S, Huang I-C, Carnero E. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hale BG, Kerry PS, Jackson D, Precious BL, Gray A, Killip MJ. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc Natl Acad Sci U S A. 2010;107:1954–1959. doi: 10.1073/pnas.0910715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Engel DA. The influenza virus NS1 protein as a therapeutic target. Antivir Res. 2013;99:409–416. doi: 10.1016/j.antiviral.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Basu D, Walkiewicz MP, Frieman M, Baric RS, Auble DT, Engel DA. Novel influenza virus NS1 antagonists block replication and restore innate immune function. J Virol. 2009;83:1881–1891. doi: 10.1128/JVI.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ai HX, Zhang L, Chang AK, Wei HY, Che YC, Liu HS. Virtual screening of potential inhibitors from TCM for the CPSF30 binding site on the NS1A protein of influenza A virus. J Mol Model. 2014;20:2142. doi: 10.1007/s00894-014-2142-7. [DOI] [PubMed] [Google Scholar]
- 111.Walkiewicz MP, Basu D, Jablonski JJ, Geysen HM, Engel DA. Novel inhibitor of influenza non-structural protein 1 blocks multi-cycle replication in an RNase L-dependent manner. J Gen Virol. 2011;92:60–70. doi: 10.1099/vir.0.025015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ai HX, Zheng FL, Zhu CY, Sun TT, Zhang L, Liu X. Discovery of novel influenza inhibitors targeting the interaction of dsRNA with the NS1 protein by structure-based virtual screening. Int J Bioinform Res Appl. 2010;6:449–460. doi: 10.1504/IJBRA.2010.037985. [DOI] [PubMed] [Google Scholar]
- 113.Cho EJ, Xia SL, Ma LC, Robertus J, Krug RM, Anslyn EV. Identification of influenza virus inhibitors targeting NS1A utilizing fluorescence polarization-based high-throughput assay. J Biomol Screen. 2012;17:448–459. doi: 10.1177/1087057111431488. [DOI] [PubMed] [Google Scholar]
- 114.Maroto M, Fernandez Y, Ortin J, Pelaez F, Cabello MA. Development of an HTS assay for the search of anti-influenza agents targeting the interaction of viral RNA with the NS1 protein. J Biomol Screen. 2008;13:581–590. doi: 10.1177/1087057108318754. [DOI] [PubMed] [Google Scholar]
- 115.Dunning J, Baillie JK, Cao B, Hayden FG. International Severe Acute Respiratory and Emerging Infection Consortium. Antiviral combinations for severe influenza. Lancet Infect Dis. 2014;14:1259–1270. doi: 10.1016/S1473-3099(14)70821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ilyushina NA, Hoffmann E, Salomon R, Webster RG, Govorkova EA. Amantadine-oseltamivir combination therapy for H5N1 influenza virus infection in mice. Antivir Ther. 2007;12:363–370. [PubMed] [Google Scholar]
- 117.Smee DF, Hurst BL, Wong M-H, Bailey KW, Tarbet EB, Morrey JD. Effects of the combination of favipiravir (T-705) and oseltamivir on influenza A virus infections in mice. Antimicrob Agents Chemother. 2010;54:126–133. doi: 10.1128/AAC.00933-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim W-Y, Suh GY, Huh JW, Kim S-H, Kim M, Kim YS. Triple-combination antiviral drug for pandemic H1N1 influenza virus infection in critically ill patients on mechanical ventilation. Antimicrob Agents Chemother. 2011;55:5703–5709. doi: 10.1128/AAC.05529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seo S, Englund JA, Nguyen JT, Pukrittayakamee S, Lindegardh N, Tarning J. Combination therapy with amantadine, oseltamivir and ribavirin for influenza A infection: safety and pharmacokinetics. Antivir Ther. 2013;18:377–386. doi: 10.3851/IMP2475. [DOI] [PMC free article] [PubMed] [Google Scholar]