Abstract
The increasing interest in studying cells using more in vivo-like three-dimensional (3D) microenvironments has created a need for advanced 3D screening platforms with enhanced functionalities and increased throughput. 3D screening platforms that better mimic in vivo microenvironments with enhanced throughput would provide more in-depth understanding of the complexity and heterogeneity of microenvironments. The platforms would also better predict the toxicity and efficacy of potential drugs in physiologically relevant conditions. Traditional 3D culture models (e.g., spinner flasks, gyratory rotation devices, non-adhesive surfaces, polymers) were developed to create 3D multicellular structures. However, these traditional systems require large volumes of reagents and cells, and are not compatible with high-throughput screening (HTS) systems. Microscale technology offers the miniaturization of 3D cultures and allows efficient screening of various conditions. This review will discuss the development, most influential works, and current advantages and challenges of microscale culture systems for screening cells in 3D microenvironments.
Keywords: Microarray, Extracellular matrix, Microwells, Technology, Collagen, Hydrogels, Microfluidics
Introduction
Tissues and organs reside within a three-dimensional (3D) microenvironment that provides cells with the physical structure and various external stimuli that are necessary for various biological processes such as tissue regeneration, wound healing, and cancer initiation and progression [1]. However, for decades, cell culture, drug screening, and cytotoxicity studies have been done mostly using traditional two-dimensional (2D) culture systems, in which cells are cultured on a flat surface treated to promote cellular attachment. Although 2D culture systems are useful to study cell–cell interactions and cellular responses to biochemical properties of extracellular matrix (ECM) proteins, there are myriad interactions in tissues that are not completely recapitulated in 2D culture systems, such as cell–ECM interactions and cellular responses to physical properties (i.e., density and stiffness) of the ECM. For example, luminal mammary epithelial cells show polarized acini structures similar to those seen in vivo, and express milk proteins in response to lactogenic hormones when they are cultured in 3D laminin-rich gels [2]. In addition, inhibition of β1 integrins in 3D cultures of breast cancer cells changes the abnormal morphology of the cancer cells and their patterns of growth to a normal phenotype, whereas no effects have been observed in 2D cultures [3]. These examples demonstrate the dramatic difference between 2D and 3D culture systems, highlighting the importance of the 3D microenvironment. Even though the relevance of the 3D system has become more recognized, traditional 3D systems are not well suited to investigate the complexity and heterogeneity of 3D microenvironments. Accordingly, there has been an increase in the design, development, and utilization of more in vivo-like 3D culture systems with better functions, lower cost and labor, and enhanced throughput.
This review discusses the pros and cons of traditional and emerging microscale 3D in vitro systems and the practical applications of microscale 3D systems. The first section reviews traditional 3D culture systems, especially designed to overcome the limitations of traditional 2D in vitro systems. The second section reviews existing microscale in vitro 3D cell culture screening systems developed to increase the throughput capacity of the traditional 3D in vitro cell culture systems. The advantages and limitations of microscale 3D cell culture systems are also discussed in detail. Lastly, an analysis of current challenges for screening in 3D cell cultures is given. By providing a broad overview of current microscale technologies for 3D cultures, this review aims at highlighting the influence and impact of 3D cell cultures in biology and medical research, providing insight for future directions.
Macroscale 3D culture systems
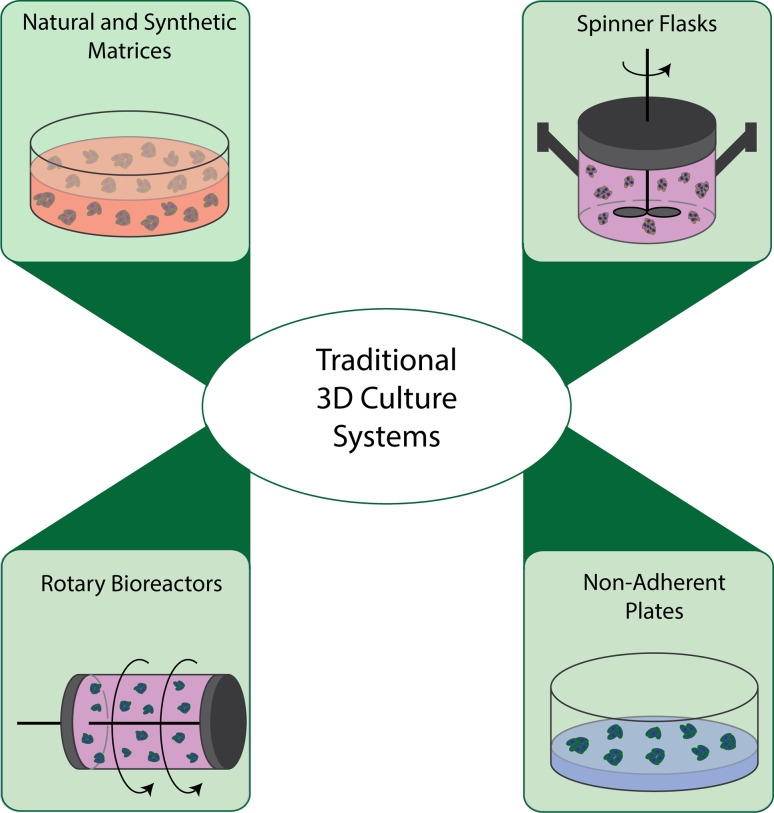
Macroscale 3D culture systems were originally developed to culture large amounts of cells using more in vivo-like cell–cell and cell–matrix interactions. Macroscale assays typically utilize milliliters to liters of reagents in enclosed bioreactors or hundreds of microliters in multi-well plate formats. The 3D multicellular spheroid (MCS) model is a 3D macroscale culture system that has been developed to offer a higher similarity to real tissues than 2D cultures. For example, MCSs have shown growth kinetics similar to those found in tumors in vivo by decreasing the proportion of proliferating cells and increasing cell death and necrosis in the interior of the spheroid [4]. In addition, MCSs show enhanced differentiation similar to tumors found in vivo [4]. Some of the most commonly used 3D macroscale culture systems for creating 3D MCSs include: spinner flasks, rotating cell bioreactors, plates with non-adherent surfaces, and natural and synthetic matrices (Fig. 1).
Fig. 1.
The most commonly used 3D culture systems fall within four categories: natural and synthetic matrices, spinner flasks, rotary reactors, and non-adherent plates
Spinner flasks were first developed by Sutherland in the 1970s for culturing large amounts of 3D MCSs [5]. By continuously stirring the cell suspension with an impeller, cells in spinner flasks are allowed to grow and form MCSs, while preventing cell attachment to the surface [6, 7]. However, a drawback of the spinner flask culture is that cells are subjected to sheer stress by the spinning impeller, which may disrupt the integrity of the spheroids and alter cell physiology. To address this limitation, in 1992 the NASA developed a rotating cell bioreactor to mimic microgravity and exert low sheer stress on the cells [8, 9]. Low sheer stress was achieved by rotating the chamber containing the cell culture, instead of stirring the culture suspension. Although large numbers of 3D spheroids can be obtained using spinner flasks and rotating cell bioreactors, major limitations of both systems are the variety of number of cells per spheroid, meaning the size of the spheroids cannot be controlled, and the slow rate of the MCS formation (typically hours or days). The variance in the number of cells per spheroid can be critical for drug screening applications where cell proliferation or cell death is monitored. Moreover, MCSs of different sizes may exhibit different drug resistances due to differences in the penetration of the drug, and thus affect the outcome of the screening.
The control of the size and formation rate of MCSs was first achieved by loading approximately the same number of cells into each well of multi-well plates, which have been treated to have non-adherent surfaces to prevent the attachment of cells to the surface while promoting cell-to-cell attachment to form MCSs. For example, Ivascu et al. [10] formed 3D MCSs by coating 96-well plates with a non-adherent polymer (0.5 % poly-HEMA), and adding a specific number of cells to each well. Next, the 96-well plate was centrifuged to stimulate the aggregation of all the cells at the bottom of the well. Using this method, tumor spheroids of homogenous sizes and morphologies were obtained within 24 h, thus providing a faster alternative to spinner flasks and rotating cell bioreactors. Natural polymers such as 1–1.5 % agarose, instead of poly-HEMA, are also used to coat plates and create MCSs aggregates because cells typically do not adhere to agarose [11–13].
Unlike previous methods in which MSCs are generated in liquid environments and have minimized interactions with any substrates, MCSs are also generated within ECM gels (3D embedded) or on top of the ECM gels (3D on-top). For the 3D embedded method, cells are mixed with ECM solutions followed by the polymerization of the ECM solution. For the 3D on-top method, cells are placed on top of the polymerized ECM gels [14, 15]. Both methods allow enhanced interactions with ECM gels; thus, cells embedded in natural matrices such as collagen I and Matrigel® have shown different morphologies and behaviors depending on the composition and mechanical properties of matrices [16]. For example, mouse mammary epithelial cells cultured on collagen floating gels (i.e., softer) with added hormones showed an increased production of milk proteins, similarly to the epithelial cells found in vivo. This result was not obtainable from the attached collagen gels (i.e., stiffer) of same density [17]. In addition to the mammary epithelial cells, it has been reported that chondrocytes differentiate into cartilaginous nodules, endothelial cells into capillary-like structures, and salivary glands into acinar-like structures when cultured in 3D Matrigel® [18].
Although natural matrices help recreate the in vivo environment better by providing cells with the geometry, chemistry, and signaling cues found in vivo, batch-to-batch variation and limited modification of chemical and mechanical properties are considered major limitations for obtaining consistent and reproducible outcomes [19]. Synthetic matrices offer more defined properties than natural matrices by controlling adhesive moieties, proteolytic sites, and the mechanics of the material more precisely. For example, by increasing the pore size and decreasing the mechanical strength of chitosan hydrogels, the exchange of nutrients and waste is enhanced, and cell proliferation is improved [20]. Another method is to functionalize polyethylene glycol diacrylate (PEGDA) gels with RGDS (Arg-Gly-Asp-Ser) adhesion peptide moieties to culture human Mesenchymal Stem Cells (hMSCs) and to examine the effect of the RGDS adhesion peptides on hMSC viability and chondrogenic differentiation [21]. Other common synthetic matrices utilized for 3D cell culture include polyethylene glycol (PEG), polylactic acid (PLA), polyglycolic acid (PGA), and the synthetic matrix peptide PuraMatrix [22–24].
The need for microscale 3D culture screening systems
Although traditional 3D culture systems have helped to bridge the gap between 2D culture systems and more biologically relevant tissue samples, the high costs associated with the reagents needed for traditional 3D culture have hindered the widespread use of these systems. Microscale 3D culture systems provide a more cost-effective alternative to traditional 3D culture systems by significantly reducing the amount of materials, reagents, and cells needed for an assay. For example, one data point in a microscale system requires approximately 2–15 μL of sample, whereas a traditional 3D culture cellular assay (96-well plate) requires a minimum of 50–100 μL. Therefore, a 7- to 25-fold increase in saving reagents can be obtained using 3D microscale systems compared to traditional 3D culture systems. This significant decrease in the volumes of reagents and the number of cells required per assay makes microscale 3D culture systems attractive and convenient for drug screening applications and for the testing and identification of hundreds of compounds in more in vivo-like conditions. In particular, the ability of microscale 3D culture systems to reduce total costs while increasing experimental conditions is particularly useful for analysis of rare cells such as primary cells isolated from patient biopsies. Patient samples are also limited and difficult to examine using traditional culture systems when only a small amount of cells are available. Some microscale 3D culture systems can provide better spatial and temporal control over traditional 3D cell cultures by patterning cells and ECM gels within a single channel to create more in vivo-like tissue structures. Therefore, the power of microscale 3D screening systems to examine more experimental conditions with fewer cells shows promise to provide a more comprehensive understanding about the complexity and heterogeneity of microenvironment.
Microscale 3D culture screening systems
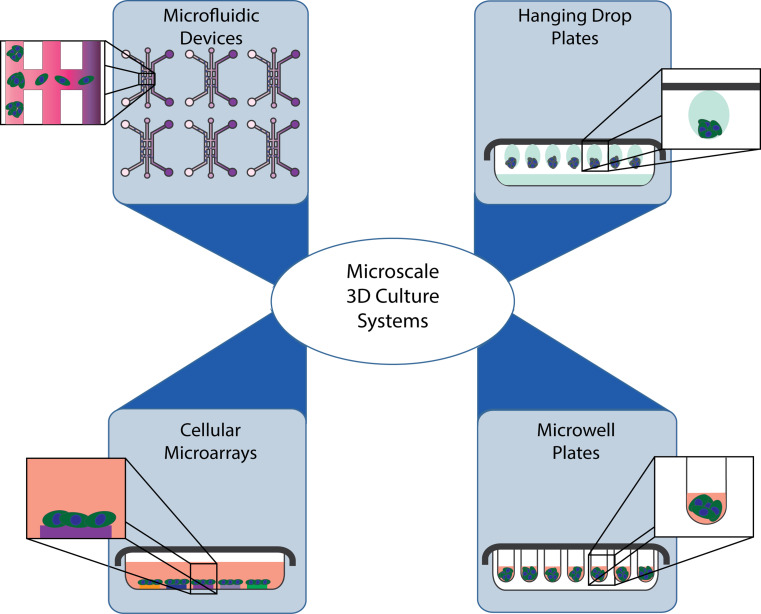
Current microscale 3D screening systems can be classified into four main platforms: hanging drop plates, microwell plates, cellular microarrays, and microfluidic devices (Fig. 2). First, the hanging drop plate was the first microscale 3D culture system developed, and is still widely applied to generate hundreds of 3D embryonic bodies, tumor spheroids, and cultures of bacteria in confined liquid environments. Second, microwell plates are an adaptation of the traditional well plate formats, with the added advantage of reduced well size to manipulate 3D spheroid size and shape. Third, cellular microarrays are based on spotting technology developed for DNA and protein microarrays. Cellular microarray technology allows the deposition of hundreds, sometimes thousands, of materials on a flat surface, and the examination of cells under multiple culture conditions. Last, microfluidic devices allow a more controllable culture by manipulating the flow of liquids and the precise placement of cells within geometrically confined microchannels.
Fig. 2.
Microscale 3D culture systems can be classified into four main platforms: microfluidic devices, hanging drop plates, cellular microarrays, and microwell plates. Inserts show a closer view of individual compartments for each microscale system
Each of these microscale 3D culture systems is designed to address specific limitations of traditional 3D culture systems. The next section includes a detailed discussion of these microscale 3D culture systems including their advantages and limitations, which are also summarized in Table 1.
Table 1.
Benefits and limitations of microscale 3D culture systems
| Microscale 3D culture system | Benefits | Limitations |
|---|---|---|
| Hanging drop plates |
Relatively reproducible Offer control over spheroid size Can culture multiple cell types |
Short-term culture (few days) Media replacement is difficult Low stability of the system can detach droplets from plate Formation of small spheroids No ECM molecules Need to transfer spheroids for analysis Risk of evaporation |
| Microwell plates |
Longer culture time (weeks–month) Offer control over spheroid size and shape Can culture multiple cell lines Can be used for screening applications |
Little work done with ECM molecules Requires fabrication of microwells Often requires specialized equipment compatible with microwell infrastructure No spatial control over multiple cell lines |
| Cellular microarrays |
A variety of biomaterials and ECM molecules can be added High content screening of cell–cell, cell–biomaterials interactions |
Most are limited to 3D on-top cultures Soluble formulations are shared across the patterned surface Quantitative analysis is often limited to antibody staining and microscopy |
| Microfluidic devices |
Offer control of fluids and cell locations to specific regions Can culture multiple cell lines Can generate chemical gradients Tubeless microfluidics are suitable for drug screening applications Tubeless microfluidics do not need external connectors, pump, valves |
Cells are difficult to isolate from culture regions Quantitative analysis is often limited to antibody staining and microscopy |
Hanging drop plate
The hanging droplet plate was developed over a century ago by Harrison to create 3D MCSs [25]. This simple method consists of depositing drops of a cell suspension onto a well plate, inverting the plate, and allowing the cell-containing drops to hang in place via surface tension. The cells inside the hanging drop aggregate at the tip of the drop and form tight cell–cell adhesions that lead to the formation of 3D cell spheroids. In this method, spheroids must be hand-picked or pulled through nylon mesh sieves to obtain uniformly sized spheroids [26]. Also, the 3D spheroids could not be cultured over a long period of time (less than several days) because of the challenges associated with replenishing the media and the nutrients [6]. The process was laborious but often yielded useful results.
Recently, hanging drop systems have become more efficient. Tung et al. [27] have developed a 384-well plate with access holes on the top surface of the plate that allow the removal and addition of fresh media to the hanging drops. Using this platform, media are exchanged and spheroids are able to proliferate over 7 days of culture [27]. Moreover, the exchange of fluids using this platform allowed the addition of drugs to screen for drug toxicity and anti-proliferative effects on cells [27]. Another limitation of the hanging drop method is the low stability of the droplets towards mechanical shocks and plate movement. To address this limitation, micro-ring structures were added to the 384-well plate to stabilize the droplets against mechanical perturbations and extend spheroid culture time for up to 22 days [28].
There are two other limitations that still remain unaddressed. First, hanging drop plates can only support the formation of small 3D spheroids inside droplets less than 50 μL in size due to the instability of heavier and larger drops. Second, the lack of ECM molecules in the hanging drops does not provide cells with additional biochemical cues and cell–ECM interactions that naturally occur in vivo, thus decreasing the biological relevance of the assay. Recently, in attempts to address this limitation 3D spheroids were collected from hanging drop plates and transferred to plates with ECM proteins. However, the transfer of spheroids can be time consuming, and their manipulation can damage their integrity [11]. Alternatively, Truong et al. [29] introduced a microinjection method for culturing 3D spheroids inside collagen gels to circumvent the transferring step.
Microwell plates
Microwell plates are commonly fabricated using micromolding techniques on non-adhesive, inert materials such as PEG, chitosan, polydimethylsiloxane (PDMS), polyacrylamide, or agarose [6, 30, 31]. After microwells are fabricated, a cell suspension is added to the wells, and cells are allowed to settle down on non-adhesive wells via gravity. Cells that remain outside of the microwells are gently washed away, and only cells that docked in the wells remain for culture to form MSCs. For example, a microfabricated chip with multicavities coated with PEG was developed to culture thousands of HEPG2 spheroids of uniform diameter for over a month [32]. This system allowed the collection of spheroids and their culture media to further quantify the secretory activities of three proteins necessary for proper liver function: alpha-fetoprotein, albumin, and fibrinogen [32]. More recently, newer designs have been used to manipulate the shape of 3D cell constructs. For example, agarose and chitosan microwells of different shapes were used to generate cellular constructs in the shape of rods, tori, and honeycombs [33, 34]. Controlling the 3D geometry of cell constructs showed to be particularly advantageous for increasing the size of the construct while maintaining an adequate diffusion of nutrients along the structure.
The high stability and compatibility of microwell plates with traditional well plate formats and microscopes make them more attractive for high-throughput screening (HTS) applications than the hanging drop platform. One example of a screening application utilized a hexagonal honeycomb microwell plate previously fabricated from a Polymer Live Cell Array (PLCA) to culture 3D spheroids of MCF7 breast cancer cells and to screen for antitumor drugs [35, 36]. In addition to providing separate wells where the 3D spheroids could be cultured, the hexagonal microwell plate allowed for the treatment of the 3D spheroids with two anticancer drugs. More importantly, the multiwell plate was equipped with a glass bottom surface that permitted the continuous monitoring of spheroid growth and the cytotoxic effect of the drugs without removing the spheroids or their media from the culture wells [36].
Although both microwell plates and hanging droplets are capable of controlling the size and shape of MSCs, added advantages of microwell plates over the hanging drop method are that microwell plates enable a longer culture, an easy collection of the spheroids and their culture media for post-culture analysis, and are more compatible with imaging systems for the continuous monitoring of cell culture and spheroid formation.
Cellular microarrays
Cellular microarrays are fabricated using contact and non-contact printing techniques previously utilized in DNA and protein microarrays [37–39]. The advancement in robotic spotting and microprinting technologies introduced cellular microarrays as attractive screening platforms for incorporating ECM molecules in cell cultures and for identifying key cell–biomaterials and cell–ECM interactions. Using these technologies, thousands of adhesive patterns of natural or synthetic polymers can be deposited on non-adhesive surfaces (i.e., glass, PEG, PDMS) to conduct 3D on-top or 3D cell-embedded cultures [6, 40].
To achieve the 3D on-top culture in cellular microarrays, a cell suspension is added to the ECM patterned surface, and cells are allowed to interact with that surface. After the cells have adhered to the patterns, the microarray is washed and non-attached cells are washed away. A variety of synthetic and natural materials have been used to pattern cellular microarrays for 3D on-top cultures. Previously, glass substrates coated with PEG gels were used to create a 100 × 100 cellular microarray of hepatocyte cells co-cultured with endothelial cells [41]. Using this PEG microarray, hepatocytes retained their liver-specific functions and were able to secrete albumin for up to a month, thus demonstrating the utility of the microarray for modulating cellular behavior [41]. Additionally, blends of biodegradable polymers such as PLGA/PCL and PLLA/PDLLA were used to present smooth muscle cells and osteoblasts with gradients of different surface roughness and to screen for differences in cell proliferation [42, 43].
Natural materials have also been used to study cell–ECM interactions in 3D on-top cultures. For instance, human liver cells and epithelial cells were previously cultured on collagen type I and Matrigel® microarrays, respectively [44, 45]. In 2005, Flaim et al. [46] utilized a combinatory approach to study the effect of 32 combinations of five ECM molecules (collagen I, collagen III, collagen IV, laminin and fibronectin) on mouse embryonic stem cell differentiation. The effect of growth factors on stem cell differentiation was later examined with the addition of gasket well structures around the cellular microarray platform [47]. More recently, a screening with 192 combinations of five natural ECM molecules such as collagen I, collagen IV, laminin, RGD, and Matrigel® has suggested that progenitor cell fate decisions are influenced by their microenvironment [48].
3D cell-embedded cultures in cellular microarrays are prepared by mixing cells with a gel of interest (i.e., alginate, collagen, Matrigel®), and depositing the cell–gel mixture on surfaces. One particular advantage of alginate over other matrices is that alginate remains as a liquid in the absence of multivalent cations (e.g., Ba2+, Ca2+), and a three-dimensional lattice of ionically cross-linked alginate can be formed upon being deposited on Ca2+ or Ba2+ surfaces. This is particularly useful during microarray fabrication due to the constant pipetting, spotting, and mixing steps that require a liquid cell suspension. Fernandes et al. [40] developed a 3D cell-embedded microarray by spotting a solution of BaCl2 on a glass slide, and depositing human pancreatic tumor cells mixed with alginate on the BaCl2 spots. However, some materials such as collagen spots degrade more quickly in cellular microarrays perhaps due to matrix metalloproteinases produced by cells in culture, and often result in cells leaching of the collagen spots [49].
Cellular microarray technologies are continuously evolving, and promise to be an influential platform for high content screening and identification of molecules and drug candidates. For example, a 3D on-top cellular microarray of 768 combinations of ECM molecules has been reported recently for the identification of ECM–integrin interactions in tumor-derived cells [50]. Also, a 3D cell-embedded alginate microarray was used to screen for the anti-proliferative effects of four drugs on human hepatocellular carcinoma [51]. However, one remaining limitation of cellular microarrays is that, in general, only one type of soluble formulation (i.e., medium, drug formulation) can be added to the patterned surface. Previous work has utilized wells to separate cells within a group of spots to treat different cells with different growth factors [47]. However, ECM spots within that group still shared the same soluble formulation. Therefore, cellular microarrays can be particularly unfavorable for cell signaling studies, as cross talk between samples can occur.
Microfluidic devices
The introduction of microfluidics to the biological sciences has provided additional microscale screening platforms to enable the individualized treatment of cells with different soluble and drug formulations within a single array. For example, a microfluidic 3D hepatocyte chip (3D HepTox Chip) with multiplexed channels was utilized to allow the simultaneous administration of multiple drugs and dosages to hepatocyte cells [52]. Using this microfluidic platform it has been reported that cells in 2D and 3D conditions show a significant different response to certain drug treatments, highlighting the value of 3D microfluidic systems for drug screening applications [53]. Other microfluidic systems have been developed to simplify their operation and make the loading and analysis process more compatible with existing HTS infrastructures. For instance, tubeless microfluidics using passive pumping offers a simplified 3D screening system. Using surface tension-driven passive pumping, samples are loaded into each microchannel with a pipette that touches off drops on the microchannel’s port surfaces without the need of any connectors, pumps, or cables [54]. A 3D screening system operated by passive pumping has been fully automated and applied to various 3D screening experiments. Previous works have demonstrated that tubeless microfluidic devices can be used to culture human mammary fibroblasts (HMF) and T47D carcinoma cells with collagen gels, and to screen for the effect of stromal cells and seven combinations of three different ECM molecules on T47D cell proliferation [55, 56]. The automated microfluidic platform was also utilized to perform small molecule screening and to identify the influence of different paracrine mediators on cells isolated from different patients [55, 57]. These microfluidic systems demonstrate increased capabilities for cell culture and treatment than other microscale systems such as hanging drop plates, microwell plates, or cellular microarrays. Their ability to culture cells with different ECM molecules and treat cells with different drug formulations per assay makes microfluidic devices more attractive for HTS of many compounds and drugs.
Microfluidic devices also provide enhanced spatial control by creating distinct compartments within a single channel to enable the recapitulation of in vivo-like tissue organization. Sudo et al. [58] presented a compartmentalized system to co-culture primary hepatocytes and endothelial cells in a microchannel. In this system, the compartments containing hepatocytes and endothelial cells are separated by a blank collagen gel, allowing the physical separation of both cell lines and the exchange of secreted proteins across the collagen gel. Using this microfluidic device it was found that endothelial cells on one side of the microfluidic device are able to form 3D capillary-like structures that extended through the collagen gel towards the hepatocytes [58]. Compartmentalized co-cultures in microfluidic devices can also be used to investigate the effect of stromal cells during the invasive transition of ductal carcinoma in situ (DCIS) in breast cancer. For instance, a compartmentalized culture in which cancer cells and stromal fibroblasts are loaded in separate but adjacent compartments have interestingly revealed that the transition is dependent on the distance between the two cell types, and also that the physical contacts between the two cell types are necessary to fully facilitate the transition [16]. These results demonstrate that the spatial control offered by microfluidic devices during cell culture can be particularly beneficial for studying paracrine signaling and the influence of physical contacts between different cell types.
Microfabrication allows the integration of geometrical confinement inside channels to control fluids and trap cell suspensions in specific regions. For example, an array of micropillars within a microfluidic device has been fabricated to immobilize cells to a specific region of the microchannel and to increase cell–cell interactions [59]. A similar microfluidic device is being utilized to culture hepatocytes in 3D without natural or synthetic matrices by trapping the cells between micropillars, and continuously flowing media through the microchannel [60]. Others have designed microfluidic devices to create vascular networks in 3D gels by confining cells in specific regions within a 3D gel. One example is a vascular network developed by Tuong et al. where cells are embedded in a collagen matrix and confined to specific regions with perpendicular fluid flows that mimic in vivo interstitial flows near vascular vessels [61, 62]. Another example utilizes carbohydrate glass pillars embedded in 3D ECM gels containing cells that dissolve via the addition of cell culture media. Lumen-like networks are formed within the 3D gel after network dissolution, and endothelial cells are injected into the vascular structure to line up and create vessel-like structures. The resulting multicellular vascular network can be perfused with a culture medium under high pressure through the vascular structure to mimic vascular vessels with active flow [63]. By controlling fluids and location of cells, these microfluidic devices are great tools for creating well-defined compartments for each cell type within a microfluidic channel that can ultimately be used to resemble more complex in vivo-like organizations helping increase the relevance of 3D cultures systems. The microfluidic systems with micropillars have also simplified the operation process, enabling compatibility with existing HTS infrastructure.
Microfluidic devices offer greater control over the generation of chemical gradients in 3D cultures. The diffusion of molecules becomes more controllable in microsystems due to the scale of the system and the ability to control the shape, length, and material of channels, allowing accurate control of the gradient [64]. Several microfluidic systems have been developed to create gradients of epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), and netrin-1 in collagen gels to examine brain cancer cell migration [65], angiogenesis [66–68], and neurite cell guidance [69], respectively; and to investigate the synergetic effect of multiple growth factors on sprouting angiogenesis [68, 70]. Taken together, these microfluidic systems have shown to be useful for generating chemical gradients and controlling cellular processes such as cell migration and proliferation. More importantly, the generation of gradients with microfluidic devices shows promise for investigating specific mechanisms involved in wound healing, cancer progression, and angiogenesis.
Other microscale 3D culture systems
Recently developed 3D microscale culture systems continue to introduce new materials and integrate more control and function to investigate broader topics in biology using even more practical approaches. For example, Derda et al. [71] introduced a new microscale 3D screening platform made from stacks of paper. This paper-based system called “cells-in-gel-in-paper” (CiGiP) allows stacking and destacking of layers of papers to better analyze characteristics of generated 3D spheroids. Using this paper-based 3D culture platform, Derda et al. [71] found that proliferating cells are located in the outer layer, whereas growth-arrested or necrotic cells are concentrated in the core, which correlates with patient tumor biopsies. The paper-based 3D culture platform also allows the study of 3D cell migration of cells labeled with different fluorescent markers [72]. A 96-zone holder has recently been added to this platform to allow each tissue to be separated from neighboring wells, and to treat cells with a soluble toxic compound [73].
Magnetic cell levitation has also been applied to place cells labeled with magnetic nanoparticles into a spheroid-like configuration in collagen gels using a magnet [74]. This method is very simply operated and provides good control over cell density and cell–cell interactions, and enhanced monitoring of cell morphology. This method is also used to co-culture cancer cells with stromal fibroblasts, by creating MSCs containing both cell types and demonstrates that coexistence with stromal fibroblasts affects cancer cell invasion [75]. Souza et al. [76] also utilized a magnetic force system based on the cellular uptake of bacteriophage and magnetic iron oxide (MIO) nanoparticles to levitate cells in culture. Using this method, they showed that magnetically levitated glioblastoma cells produce similar protein expression patterns to xenografts.
Additional examples of microscale 3D culture platforms include microinjection of cells into 3D ECM scaffolds, microencapsulation of cells in alginate-poly-l-lysine-alginate (APA) capsules, and generation of PDMS microbubbles. The microinjection method consists of an automated needle that injects cell-polymer suspensions into polymerized collagen gels, thus allowing the cells to assemble into spheroids inside the gels. In addition to providing an automated platform for higher throughput screening, this method has showed benefits in terms of providing a faster generation of 3D spheroids, producing spheroids of homogeneous size, and controlling the spatial distribution of the spheroids [29]. Microencapsulation methods are also being explored for 3D cell culture applications. For example, Zhang et al. [77] utilized microcapsule generation technology to produce microcapsules made of alginate-poly-l-lysine-alginate (APA) for growing tumor spheroids inside the microcapsules. This technique allowed the screening of several chemotherapy drugs, and demonstrated the potential to be a rapid screening tool for 3D cultures [77]. The microfabrication of PDMS microbubbles via low surface energy Gas Expansion Molding (GEM) also showed promises as a screening tool for 3D cultures. Previously described for cell sorting applications, and later on applied to the generation of 3D colon tumor spheroids, PDMS microbubbles provided the means for growing 3D spheroids under physiological flow conditions [78, 79]. Moreover, a toxicity test showed that 3D colon tumor spheroids are more resistant to drugs when cultured inside PDMS microbubbles than spheroids cultured under static conditions [79]. These results suggested that the microbubble and flow conditions might be a better tumor model than static culture models.
Challenges of microscale 3D screening platforms
As we have discussed, 3D cell cultures have proved to be better in vitro models for recapitulating the in vivo environment compared to 2D cell culture systems. They have also increased our understanding of multiple autocrine and paracrine signals, and cell–ECM interactions found in vivo. In addition, microscale 3D screening platforms have advanced the capabilities of traditional 3D screening platforms by adding more functions while reducing the amount of reagents and cells significantly. However, as the biological relevance of 3D culture platforms keeps increasing with the addition of components such as stromal cells, growth factors, chemokines, hormones, and ECM molecules, the complexity of the system increases and presents several challenges that need to be considered.
First, as we strive to recapitulate the cellular microenvironment better, more specialized equipment may be needed. For example, better control over fluid flow, mechanics, and 3D morphology may require additional compartments, pumps, and connectors. These components are often cumbersome and require trained personnel for operation. Moreover, the addition of more components increases the volume of the system and makes their incorporation into higher throughput screening systems difficult. Therefore, special consideration must be taken before designing 3D culture methods to maintain a simple system operation, and to ensure their seamless incorporation into traditional high-throughput screening (HTS) infrastructures. Along this line, Tung et al. and Montanez-Sauri et al. designed a hanging drop plate and a tubeless microfluidic platform for 3D cell cultures that were compatible with existing liquid handling robots and fluorescent microplate readers to demonstrate the feasibility of their systems as HTS platforms [27, 55, 56].
Second, generation and collection of data from 3D microscale screening platforms can be challenging. Most imaging systems with enhanced throughput such as automated imaging systems and scanners were originally designed for 2D cell cultures that can be imaged on a single focal plane. This can be troublesome for imaging cells seeded on non-adherent surfaces or embedded within 3D gels because cells usually lie on multiple focal planes. In addition, spheroids generated with the hanging drop method usually need to be transferred to a different plate for their analysis, which is laborious and not favorable for screening. Microfluidic devices have partly addressed these limitations by allowing the culture and imaging of cells within the device, and providing smaller compartments where cells can remain within a short distance from each other. Also, because the thickness of gels in microfluidic channels is much thinner, there are fewer variations along the z-axis. In addition, image analysis algorithms such as JeX (http://jexperiment.wikidot.com), CurvePrep, and CurveAlign (http://loci.wisc.edu/software/curvelet-based-alignment-analysis) have been developed and are currently available for data processing and automated quantitative analysis of 3D culture data. The use of this software will make the microscale 3D screening platform more compatible with the HTS approach by reducing time and effort required for post-culture analysis.
Third, common methods for analyzing cellular response to certain treatment or condition in 3D cultures are generally restricted to the immunocytochemistry (ICC) assay. Although the ICC assay can provide information regarding the localization, concentration, and activation of biomolecules, the quantification of fluorescent signals is often interrupted by background signals and non-specificity of primary or secondary antibodies. Therefore, conventional biochemical assays (i.e., ELISA, western blots, qPCR) need to accompany the ICC assay analysis. Recently, Berry et al. developed an efficient sample isolation method, the Immiscible Phase Filtration Assisted by Surface Tension (IFAST) technology, for gene expression analysis [80]. The IFAST technology relies on immiscible phase filtration to reduce the time and effort required to purify DNA. IFAST replaces the multiple wash and centrifugation steps required by traditional DNA sample preparation methods with a single step and has been successfully integrated with a compartmentalized co-culture microfluidic system. Using the integrated system, the mRNA from co-cultures of MCF-7 and HS-5 cell co-cultures are extracted and purified, and the expression of estrogen receptor alpha (ERα) can be quantified [81]. Also, microfluidic devices have been coupled with mass spectrometers to enable small molecule [82], proteomic [83] and glycomic studies [84]. However, all these quantitative analysis methods have been applied to 2D cell cultures, and still need to be validated for 3D cell cultures.
Conclusions and Future remarks
In the last century, 2D cell cultures proved to be useful for culturing cells in vitro and for the development and testing of new drugs. However, not all drugs identified in 2D culture drug screenings are fully developed after clinical tests [85]. This might be due to the differences in drug resistance between 2D cultures used in the drug screening systems, and the 3D nature of cells in vivo. In fact, studies have demonstrated that cells grown in 3D cultures exhibit greater resistance to drugs compared to cells cultured in 2D possibly due to different levels of oxygenation in 3D cell clusters and changes in integrin-based signaling in 3D conditions [86, 87]. In addition, some microfluidic 3D systems have shown in vivo-like cellular arrangement as well as functions by utilizing channel geometry and the physical attributes of microsystems. It has been shown that microfluidic systems allowed the regeneration of in vivo-like endothelial barrier functions such as permeability [88]. Therefore, several 3D cell culture systems have evolved during the last decade as alternatives to 2D culture systems to provide cells with increased cell–cell and cell–ECM interactions that are more similar to those found in tissues.
Increasing interest in performing 3D cell culture and drug screening in 3D systems has stimulated the commercialization of 3D culture platforms. One hanging drop plate called Perfecta3D® is commercially available from 3D Biomatrix, for the culture of 3D MCSs in 96- and 384-well plate formats. Platypus Technologies has also developed a 3D Invasion Assay known as Oris-Pro™ to quantify the migration of cells cultured in 3D collagen gels. Other companies such as Scivax, The Lab Depot, Inc., and Reinnervate are distributing well plates known as NanoCulture® Plate, CELLTREAT, and Alvetex®, respectively, which contain synthetic materials that stimulate the aggregation of cells into MCSs via non-adherent surfaces. Iuvo™ microfluidic devices that use passive pumping for loading cells and reagents into microchannel arrays are commercially available as well through BellBrook Labs, which use HTS to study the 3D migration of cells through collagen gels.
As more microscale 3D culture systems are developed, their potential for newer applications continues to emerge. The ultimate goal of creating more biologically relevant assays may be achieved using “organ-on-a-chip” systems that incorporate organ-level functions into 3D culture platform where cells from different tissues can interact with each other. Multiple microscale culture systems have been reported that mimic tissue functions such as liver [89–91], blood vessels [92, 93], lung [94, 95], heart [96, 97], muscle [98] kidney [99], and bone [100], among others. Another promising future direction for 3D microscale systems and organ-on-chips will be the inclusion of stem cells or inducible pluripotent stem (iPS) cells into the culture, and the ability to differentiate these cells along specific lineages within the 3D system. Although the utilization of iPS cells is still in its infancy, including them in 3D culture systems will increase the biological relevance of the system and the predictability of drug performance, while decreasing the use of the ethically questionable embryonic cells.
It is true that there are still many remaining questions that need to be addressed to fully utilize 3D microscale systems as HTS platforms in industry. However, 3D microscale systems do offer a powerful alternative to current 2D or 3D traditional macroscale culture systems by enhancing biological relevance and increasing throughput. In addition, using unique functions and capabilities offered by microtechnology, researchers are able to reveal new mechanisms that have been challenging to accomplish with traditional macroscale systems. As researchers aim to develop more complex in vivo-like systems, more questions are raised regarding whether we indeed need more in vivo-like screening platforms and how complex the system could be. It is clear that both simple and complex systems are useful depending on the end goal of screening. If the goal of screening is at the early stage of drug development focusing on the identification of drug targets, simple systems could be more useful to investigate cellular level interactions. However, for more advanced screening such as drug toxicity and efficiency, it may be critical to incorporate more in vivo-like structures and functions. The continued development and integration of microscale in vitro systems, 3D biology, and drug screening will facilitate the identification and validities of new drugs and therapeutics.
Acknowledgments
The authors would like to thank Dr. Lindsey Maccoux, and Won Hong for helpful discussions, and Josue Baeza for his help in figures design. This work was supported by University of Wisconsin Carbone Cancer Center Cancer Center Support Grant P30 CA014520, NIH grants (R01EB10039, 1R01CA155192-01A1, and R33CA137673-01), NIH Training Grant T32GM083499, NSF EFRI-1136903. David J. Beebe has an ownership interest in Bellbrook Labs LLC, and Salus Discovery, LLC, which have licensed technology reported in this publication.
Abbreviations
- 2D
Two-dimensional
- 3D
Three-dimensional
- APA
Alginate-poly-l-lysine-alginate
- ECM
Extracellular matrix
- EGF
Epidermal growth factor
- ERα
Estrogen receptor alpha
- HMF
Human mammary fibroblasts
- hMSC
Human mesenchymal stem cell
- HTS
High-throughput screening
- IFAST
Immiscible phase filtration assisted by surface tension
- iPS
Inducible pluripotent stem
- MCS
Multicellular spheroids
- PCL
Poly(ε-caprolactone)
- PDLLA
Poly(d,l-lactic acid)
- PDMS
Poly(dimethylsiloxane)
- PEG
Poly(ethylene glycol)
- PEGDA
Poly(ethylene glycol) diacrylate
- PGA
Poly(glycolic acid)
- PLA
Poly(lactic acid)
- PLCA
Polymer live cell array
- PLGA
Poly(d,l-lactic-co-glycolic acid)
- PLLA
Poly(l-lactic acid)
- Poly-HEMA
Poly(2-hydroxyethyl methacrylate)
- PS
Polystyrene
- RGDS
Arg-Gly-Asp-Ser
- VEGF
Vascular endothelial growth factor
References
- 1.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130(4):601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105(2):223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137(1):231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutherland RM. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science. 1988;240(4849):177–184. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland RM, Inch WR, McCredie JA, Kruuv J. A multicomponent radiation survival curve using an in vitro tumour model. Int J Radiat Biol Relat Stud Phys Chem Med. 1970;18:491–495. doi: 10.1080/09553007014551401. [DOI] [PubMed] [Google Scholar]
- 6.Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J. 2008;3(9–10):1172–1184. doi: 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- 7.Kim JB. Three-dimensional tissue culture models in cancer biology. Semin Cancer Biol. 2005;15(5):365–377. doi: 10.1016/j.semcancer.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discovery Today. 2013;18(5–6):240–249. doi: 10.1016/j.drudis.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin TJ, Prewett TL, Wolf DA, Spaulding GF. Reduced shear stress: a major component in the ability of mammalian tissues to form three-dimensional assemblies in simulated microgravity. J Cell Biochem. 1993;51(3):301–311. doi: 10.1002/jcb.240510309. [DOI] [PubMed] [Google Scholar]
- 10.Ivascu A, Kubbies M. Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J Biomol Screen. 2006;11(8):922–932. doi: 10.1177/1087057106292763. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat Protoc. 2009;4(3):309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Chen C, Kapadia A, Zhou Q, Harper MK, Schaack J, LaBarbera DV. 3D models of epithelial-mesenchymal transition in breast cancer metastasis: high-throughput screening assay development, validation, and pilot screen. J Biomol Screen. 2011;16(2):141–154. doi: 10.1177/1087057110392995. [DOI] [PubMed] [Google Scholar]
- 13.Fang C, Avis I, Salomon D, Cuttitta F. Novel phenotypic fluorescent three-dimensional platforms for high-throughput drug screening and personalized chemotherapy. J Cancer. 2013;4(5):402–415. doi: 10.7150/jca.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 15.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung KE, Yang N, Pehlke C, Keely PJ, Eliceiri KW, Friedl A, Beebe DJ. Transition to invasion in breast cancer: a microfluidic in vitro model enables examination of spatial and temporal effects. Integr Biol Quant Biosci Nano Macro. 2011;3(4):439–450. doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emerman JT, Enami J, Pitelka DR, Nandi S. Hormonal effects on intracellular and secreted casein in cultures of mouse mammary epithelial cells on floating collagen membranes. Proc Natl Acad Sci USA. 1977;74(10):4466–4470. doi: 10.1073/pnas.74.10.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15(5):378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Cuddihy MJ, Kotov NA. Three-dimensional cell culture matrices: state of the art. Tissue Eng Part B Rev. 2008;14(1):61–86. doi: 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- 20.Ji C, Khademhosseini A, Dehghani F. Enhancing cell penetration and proliferation in chitosan hydrogels for tissue engineering applications. Biomaterials. 2011;32(36):9719–9729. doi: 10.1016/j.biomaterials.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324(5923):59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci USA. 2003;100(9):5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 24.Takei J. 3-Dimensional cell culture scaffold for everyone: drug screening, tissue engineering and cancer biology. AATEX. 2006;11(3):170–176. [Google Scholar]
- 25.Harrison RG. Observations on the living developing nerve fiber. Proc Soc Exp Biol Med. 1907;4:140–143. doi: 10.3181/00379727-4-98. [DOI] [Google Scholar]
- 26.Kunz-Schughart LA, Kreutz M, Knuechel R. Multicellular spheroids: a three-dimensional in vitro culture system to study tumour biology. Int J Exp Pathol. 1998;79(1):1–23. doi: 10.1046/j.1365-2613.1998.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst. 2011;136(3):473–478. doi: 10.1039/c0an00609b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiao AY, Tung YC, Kuo CH, Mosadegh B, Bedenis R, Pienta KJ, Takayama S. Micro-ring structures stabilize microdroplets to enable long term spheroid culture in 384 hanging drop array plates. Biomed Microdevices. 2012;14(2):313–323. doi: 10.1007/s10544-011-9608-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truong HH, de Sonneville J, Ghotra VP, Xiong J, Price L, Hogendoorn PC, Spaink HH, van de Water B, Danen EH. Automated microinjection of cell-polymer suspensions in 3D ECM scaffolds for high-throughput quantitative cancer invasion screens. Biomaterials. 2012;33(1):181–188. doi: 10.1016/j.biomaterials.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 30.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci USA. 2006;103(8):2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Napolitano AP, Chai P, Dean DM, Morgan JR. Dynamics of the self-assembly of complex cellular aggregates on micromolded nonadhesive hydrogels. Tissue Eng. 2007;13(8):2087–2094. doi: 10.1089/ten.2006.0190. [DOI] [PubMed] [Google Scholar]
- 32.Sakai Y, Nakazawa K. Technique for the control of spheroid diameter using microfabricated chips. Acta Biomater. 2007;3(6):1033–1040. doi: 10.1016/j.actbio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Dean DM, Napolitano AP, Youssef J, Morgan JR. Rods, tori, and honeycombs: the directed self-assembly of microtissues with prescribed microscale geometries. FASEB Journal Off Publ Fed Am Soc Exp Biol. 2007;21(14):4005–4012. doi: 10.1096/fj.07-8710com. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda J, Khademhosseini A, Yeo Y, Yang X, Yeh J, Eng G, Blumling J, Wang CF, Kohane DS, Langer R. Micromolding of photocrosslinkable chitosan hydrogel for spheroid microarray and co-cultures. Biomaterials. 2006;27(30):5259–5267. doi: 10.1016/j.biomaterials.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 35.Zurgil N, Afrimzon E, Deutsch A, Namer Y, Shafran Y, Sobolev M, Tauber Y, Ravid-Hermesh O, Deutsch M. Polymer live-cell array for real-time kinetic imaging of immune cells. Biomaterials. 2010;31(18):5022–5029. doi: 10.1016/j.biomaterials.2010.02.035. [DOI] [PubMed] [Google Scholar]
- 36.Markovitz-Bishitz Y, Tauber Y, Afrimzon E, Zurgil N, Sobolev M, Shafran Y, Deutsch A, Howitz S, Deutsch M. A polymer microstructure array for the formation, culturing, and high throughput drug screening of breast cancer spheroids. Biomaterials. 2010;31(32):8436–8444. doi: 10.1016/j.biomaterials.2010.07.050. [DOI] [PubMed] [Google Scholar]
- 37.Barbulovic-Nad I, Lucente M, Sun Y, Zhang M, Wheeler AR, Bussmann M. Bio-microarray fabrication techniques—a review. Crit Rev Biotechnol. 2006;26(4):237–259. doi: 10.1080/07388550600978358. [DOI] [PubMed] [Google Scholar]
- 38.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 39.Hartmann M, Sjodahl J, Stjernstrom M, Redeby J, Joos T, Roeraade J. Non-contact protein microarray fabrication using a procedure based on liquid bridge formation. Anal Bioanal Chem. 2009;393(2):591–598. doi: 10.1007/s00216-008-2509-7. [DOI] [PubMed] [Google Scholar]
- 40.Fernandes TG, Kwon SJ, Lee MY, Clark DS, Cabral JM, Dordick JS. On-chip, cell-based microarray immunofluorescence assay for high-throughput analysis of target proteins. Anal Chem. 2008;80(17):6633–6639. doi: 10.1021/ac800848j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otsuka H, Hirano A, Nagasaki Y, Okano T, Horiike Y, Kataoka K. Two-dimensional multiarray formation of hepatocyte spheroids on a microfabricated PEG-brush surface. Chembiochem Eur J Chem Biol. 2004;5(6):850–855. doi: 10.1002/cbic.200300822. [DOI] [PubMed] [Google Scholar]
- 42.Sung FJ, Su J, Berglund JD, Russ BV, Meredith JC, Galis ZS. The use of temperature-composition combinatorial libraries to study the effects of biodegradable polymer blend surfaces on vascular cells. Biomaterials. 2005;26(22):4557–4567. doi: 10.1016/j.biomaterials.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 43.Simon CG, Jr, Eidelman N, Kennedy SB, Sehgal A, Khatri CA, Washburn NR. Combinatorial screening of cell proliferation on poly(l-lactic acid)/poly(d, l-lactic acid) blends. Biomaterials. 2005;26(34):6906–6915. doi: 10.1016/j.biomaterials.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 44.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26(1):120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 45.Sodunke TR, Turner KK, Caldwell SA, McBride KW, Reginato MJ, Noh HM. Micropatterns of Matrigel for three-dimensional epithelial cultures. Biomaterials. 2007;28(27):4006–4016. doi: 10.1016/j.biomaterials.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 47.Flaim CJ, Teng D, Chien S, Bhatia SN. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev. 2008;17(1):29–39. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 48.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, Stampfer MR, Petersen OW, Bissell MJ. Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integr Biol Quant Biosci Nano Macro. 2009;1(1):70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee MY, Kumar RA, Sukumaran SM, Hogg MG, Clark DS, Dordick JS. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc Natl Acad Sci USA. 2008;105(1):59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reticker-Flynn NE, Malta DFB, Winslow MM, Lamar JM, Xu MJ, Underhill GH, Hynes RO, Jacks TE, Bhatia SN. A combinatorial extracellular matrix platform identifies cell-extracellular matrix interactions that correlate with metastasis. Nat Commun. 2012 doi: 10.1038/ncomms2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meli L, Jordan ET, Clark DS, Linhardt RJ, Dordick JS. Influence of a three-dimensional, microarray environment on human cell culture in drug screening systems. Biomaterials. 2012;33(35):9087–9096. doi: 10.1016/j.biomaterials.2012.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toh YC, Lim TC, Tai D, Xiao G, van Noort D, Yu H. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip. 2009;9(14):2026–2035. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- 53.Aref AR, Huang RY-J, Yu W, Chua K-N, Sun W, Tu T-Y, Bai J, Sim W-J, Zervantonakis IK, Thiery JP, Kamm RD. Screening therapeutic EMT blocking agents in a three-dimensional microenvironment. Integr Biol. 2013;5(2):381–389. doi: 10.1039/c2ib20209c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker GM, Beebe DJ. A passive pumping method for microfluidic devices. Lab Chip. 2002;2(3):131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 55.Montanez-Sauri SI, Sung KE, Berthier E, Beebe DJ. Enabling screening in 3D microenvironments: probing matrix and stromal effects on the morphology and proliferation of T47D breast carcinoma cells. Integr Biol Quant Biosci Nano Macro. 2013;5(3):631–640. doi: 10.1039/c3ib20225a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montanez-Sauri SI, Sung KE, Puccinelli JP, Pehlke C, Beebe DJ. Automation of three-dimensional cell culture in arrayed microfluidic devices. Journal of laboratory automation. 2011;16(3):171–185. doi: 10.1016/j.jala.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su G, Sung KE, Beebe DJ, Friedl A. Functional screen of paracrine signals in breast carcinoma fibroblasts. PLoS One. 2012;7(10):e46685. doi: 10.1371/journal.pone.0046685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sudo R, Chung S, Zervantonakis IK, Vickerman V, Toshimitsu Y, Griffith LG, Kamm RD. Transport-mediated angiogenesis in 3D epithelial coculture. FASEB J Off Publ Fed Am Soc Exp Biol. 2009;23(7):2155–2164. doi: 10.1096/fj.08-122820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toh YC, Zhang C, Zhang J, Khong YM, Chang S, Samper VD, van Noort D, Hutmacher DW, Yu H. A novel 3D mammalian cell perfusion-culture system in microfluidic channels. Lab Chip. 2007;7(3):302–309. doi: 10.1039/b614872g. [DOI] [PubMed] [Google Scholar]
- 60.Goral VN, Hsieh YC, Petzold ON, Clark JS, Yuen PK, Faris RA. Perfusion-based microfluidic device for three-dimensional dynamic primary human hepatocyte cell culture in the absence of biological or synthetic matrices or coagulants. Lab Chip. 2010;10(24):3380–3386. doi: 10.1039/c0lc00135j. [DOI] [PubMed] [Google Scholar]
- 61.C-k Tung, Krupa O, Apaydin E, Liou J-J, Diaz-Santana A, Kim BJ, Wu M. A contact line pinning based microfluidic platform for modelling physiological flows. Lab Chip. 2013;13(19):3876–3885. doi: 10.1039/c3lc50489a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lopez JA, Stroock AD. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci USA. 2012;109(24):9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012;11(9):768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berthier E, Surfus J, Verbsky J, Huttenlocher A, Beebe D. An arrayed high-content chemotaxis assay for patient diagnosis. Integr Biol Quant Biosci Nano Macro. 2010;2(11–12):630–638. doi: 10.1039/c0ib00030b. [DOI] [PubMed] [Google Scholar]
- 65.Zervantonakis IK, Chung S, Sudo R, Zhang M, Charest JL, Kamm RD. Concentration gradients in microfluidic 3D matrix cell culture systems. Int J Micro Nano Scale Transp. 2010;1(1):27–36. doi: 10.1260/1759-3093.1.1.27. [DOI] [Google Scholar]
- 66.Jeong GS, Han S, Shin Y, Kwon GH, Kamm RD, Lee SH, Chung S. Sprouting angiogenesis under a chemical gradient regulated by interactions with an endothelial monolayer in a microfluidic platform. Anal Chem. 2011;83(22):8454–8459. doi: 10.1021/ac202170e. [DOI] [PubMed] [Google Scholar]
- 67.Song JW, Munn LL. Fluid forces control endothelial sprouting. Proc Natl Acad Sci USA. 2011;108(37):15342–15347. doi: 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim S, Lee H, Chung M, Jeon NL. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip. 2013;13(8):1489–1500. doi: 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- 69.Kothapalli CR, van Veen E, de Valence S, Chung S, Zervantonakis IK, Gertler FB, Kamm RD. A high-throughput microfluidic assay to study neurite response to growth factor gradients. Lab Chip. 2011;11(3):497–507. doi: 10.1039/c0lc00240b. [DOI] [PubMed] [Google Scholar]
- 70.Shin Y, Jeon JS, Han S, Jung GS, Shin S, Lee SH, Sudo R, Kamm RD, Chung S. In vitro 3D collective sprouting angiogenesis under orchestrated ANG-1 and VEGF gradients. Lab Chip. 2011;11(13):2175–2181. doi: 10.1039/c1lc20039a. [DOI] [PubMed] [Google Scholar]
- 71.Derda R, Laromaine A, Mammoto A, Tang SK, Mammoto T, Ingber DE, Whitesides GM. Paper-supported 3D cell culture for tissue-based bioassays. Proc Natl Acad Sci USA. 2009;106(44):18457–18462. doi: 10.1073/pnas.0910666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Derda R, Tang SK, Laromaine A, Mosadegh B, Hong E, Mwangi M, Mammoto A, Ingber DE, Whitesides GM. Multizone paper platform for 3D cell cultures. PLoS One. 2011;6(5):e18940. doi: 10.1371/journal.pone.0018940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deiss F, Mazzeo A, Hong E, Ingber DE, Derda R, Whitesides GM. Platform for high-throughput testing of the effect of soluble compounds on 3D cell cultures. Anal Chem. 2013;85(17):8085–8094. doi: 10.1021/ac400161j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okochi M, Takano S, Isaji Y, Senga T, Hamaguchi M, Honda H. Three-dimensional cell culture array using magnetic force-based cell patterning for analysis of invasive capacity of BALB/3T3/v-src. Lab Chip. 2009;9(23):3378–3384. doi: 10.1039/b909304d. [DOI] [PubMed] [Google Scholar]
- 75.Okochi M, Matsumura T, Honda H. Magnetic force-based cell patterning for evaluation of the effect of stromal fibroblasts on invasive capacity in 3D cultures. Biosens Bioelectron. 2013;42:300–307. doi: 10.1016/j.bios.2012.09.067. [DOI] [PubMed] [Google Scholar]
- 76.Souza GR, Molina JR, Raphael RM, Ozawa MG, Stark DJ, Levin CS, Bronk LF, Ananta JS, Mandelin J, Georgescu MM, Bankson JA, Gelovani JG, Killian TC, Arap W, Pasqualini R. Three-dimensional tissue culture based on magnetic cell levitation. Nat Nanotechnol. 2010;5(4):291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang XL, Wang W, Yu WT, Xie YB, Zhang XH, Zhang Y, Ma XJ. Development of an in vitro multicellular tumor spheroid model using microencapsulation and its application in anticancer drug screening and testing. Biotechnol Prog. 2005;21(4):1289–1296. doi: 10.1021/bp050003l. [DOI] [PubMed] [Google Scholar]
- 78.Giang U-BT, King MR, DeLouise LA. Microfabrication of bubbular cavities in PDMS for cell sorting and microcell culture applications. J Bionic Eng. 2008;5(4):308–316. doi: 10.1016/S1672-6529(08)60175-4. [DOI] [Google Scholar]
- 79.Agastin S, Giang UB, Geng Y, Delouise LA, King MR. Continuously perfused microbubble array for 3D tumor spheroid model. Biomicrofluidics. 2011;5(2):24110. doi: 10.1063/1.3596530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berry SM, Strotman LN, Kueck JD, Alarid ET, Beebe DJ. Purification of cell subpopulations via immiscible filtration assisted by surface tension (IFAST) Biomed Microdev. 2011;13(6):1033–1042. doi: 10.1007/s10544-011-9573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berry SM, Singh C, Lang JD, Strotman LN, Alarid ET, Beebe DJ. Streamlining gene expression analysis: integration of co-culture and mRNA purification. Integr Biol Quant Biosci Nano Macro. 2014;6(2):224–231. doi: 10.1039/c3ib40136g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin SL, Bai HY, Lin TY, Fuh MR. Microfluidic chip-based liquid chromatography coupled to mass spectrometry for determination of small molecules in bioanalytical applications. Electrophoresis. 2012;33(4):635–643. doi: 10.1002/elps.201100380. [DOI] [PubMed] [Google Scholar]
- 83.Lee J, Soper SA, Murray KK. Microfluidic chips for mass spectrometry-based proteomics. J Mass Spectrom JMS. 2009;44(5):579–593. doi: 10.1002/jms.1585. [DOI] [PubMed] [Google Scholar]
- 84.Bindila L, Peter-Katalinic J. Chip-mass spectrometry for glycomic studies. Mass Spectrom Rev. 2009;28(2):223–253. doi: 10.1002/mas.20197. [DOI] [PubMed] [Google Scholar]
- 85.Kunz-Schughart LA, Freyer JP, Hofstaedter F, Ebner R. The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. J Biomol Screen. 2004;9(4):273–285. doi: 10.1177/1087057104265040. [DOI] [PubMed] [Google Scholar]
- 86.David L, Dulong V, Le Cerf D, Cazin L, Lamacz M, Vannier JP. Hyaluronan hydrogel: an appropriate three-dimensional model for evaluation of anticancer drug sensitivity. Acta Biomater. 2008;4(2):256–263. doi: 10.1016/j.actbio.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 87.Li Q, Chow AB, Mattingly RR. Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. J Pharmacol Exp Ther. 2010;332(3):821–828. doi: 10.1124/jpet.109.160390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc Natl Acad Sci. 2012;109(34):13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leclerc E, Sakai Y, Fujii T. Microfluidic PDMS (polydimethylsiloxane) bioreactor for large-scale culture of hepatocytes. Biotechnol Prog. 2004;20(3):750–755. doi: 10.1021/bp0300568. [DOI] [PubMed] [Google Scholar]
- 90.Ho CT, Lin RZ, Chen RJ, Chin CK, Gong SE, Chang HY, Peng HL, Hsu L, Yew TR, Chang SF, Liu CH. Liver-cell patterning lab chip: mimicking the morphology of liver lobule tissue. Lab Chip. 2013;13(18):3578–3587. doi: 10.1039/c3lc50402f. [DOI] [PubMed] [Google Scholar]
- 91.Legendre A, Baudoin R, Alberto G, Paullier P, Naudot M, Bricks T, Brocheton J, Jacques S, Cotton J, Leclerc E. Metabolic characterization of primary rat hepatocytes cultivated in parallel microfluidic biochips. J Pharm Sci. 2013;102(9):3264–3276. doi: 10.1002/jps.23466. [DOI] [PubMed] [Google Scholar]
- 92.Booth R, Kim H. Characterization of a microfluidic in vitro model of the blood-brain barrier (mu BBB) Lab Chip. 2012;12(10):1784–1792. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 93.Shin M, Matsuda K, Ishii O, Terai H, Kaazempur-Mofrad M, Borenstein J, Detmar M, Vacanti JP. Endothelialized networks with a vascular geometry in microfabricated poly(dimethyl siloxane) Biomed Microdevices. 2004;6(4):269–278. doi: 10.1023/B:BMMD.0000048559.29932.27. [DOI] [PubMed] [Google Scholar]
- 94.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328(5986):1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. 2012 doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agarwal A, Goss JA, Cho A, McCain ML, Parker KK. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip. 2013;13(18):3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grosberg A, Alford PW, McCain ML, Parker KK. Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab Chip. 2011;11(24):4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grosberg A, Nesmith AP, Goss JA, Brigham MD, McCain ML, Parker KK. Muscle on a chip: in vitro contractility assays for smooth and striated muscle. J Pharmacol Toxicol Methods. 2012;65(3):126–135. doi: 10.1016/j.vascn.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jang KJ, Suh KY. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip. 2010;10(1):36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 100.Zhang Y, Gazit Z, Pelled G, Gazit D, Vunjak-Novakovic G. Patterning osteogenesis by inducible gene expression in microfluidic culture systems. Integr Biol. 2011;3(1):39–47. doi: 10.1039/c0ib00053a. [DOI] [PMC free article] [PubMed] [Google Scholar]