Abstract
Sparse systems are usually parameterized by a tuning parameter that determines the sparsity of the system. How to choose the right tuning parameter is a fundamental and difficult problem in learning the sparse system. In this paper, by treating the the tuning parameter as an additional dimension, persistent homological structures over the parameter space is introduced and explored. The structures are then further exploited in drastically speeding up the computation using the proposed soft-thresholding technique. The topological structures are further used as multivariate features in the tensor-based morphometry (TBM) in characterizing white matter alterations in children who have experienced severe early life stress and maltreatment. These analyses reveal that stress-exposed children exhibit more diffuse anatomical organization across the whole white matter region.
Index Terms: GLASSO, maltreated children, persistent homology, sparse brain networks, sparse correlations, tensor-based morphometry
I. Introduction
In the usual tensor-based morphometry (TBM), the spatial derivatives of deformation fields obtained during nonlinear image registration for warping individual magnetic resonance imaging (MRI) data to a template is used in quantifying neuroanatomical shape variations [3], [20], [70]. The Jacobian determinant of a deformation field is most frequently used in quantifying the brain tissue growth or atrophy at a voxel level. [20], [22], [25], [54], [71] used the Jacobian determinant of the deformation field as a measure of regional brain change. Subsequently, the statistical parametric maps are obtained by fitting the tensor maps as a response variable in a linear model at each voxel, which results in a massive number of univariate test statistics.
Recently, there have been attempts at explicitly modeling the structural variation of one region to another [11], [37], [38], [50], [62], [75], [76] using network approaches. This provides additional information that complements existing univariate approaches. In most of these multivariate approaches, anatomical measurements such as mesh coordinates, cortical thickness or Jacobian determinant across different voxels are correlated using models such as canonical correlations [4], [62], cross-correlations [11], [37], [38], [50], [75], [76], partial correlations, which are equivalent to the inverse of covariances [6], [8], [30], [40], [48]. However, these multivariate techniques suffer the small-n large-p problem [17], [31], [48], [66], [73]. Specifically, when the number of voxels are substantially larger than the number of images, it produces an under-determined linear model. The estimated covariance matrix is rank deficient and no longer positive definite. In turn, the resulting correlation matrix is not considered as a good approximation to the true correlation matrix.
The small-n large-p problem can be remedied by using sparse methods, which regularize the under-determined linear model with additional sparse penalties. There exist various sparse models: sparse correlation [17], [48], sparse partial correlation [8], [40], [48], sparse canonical correlation [4] and sparse log-likelihood [6], [7], [30], [41], [55], [74]. Sparse model 𝒜(λ) is usually parameterized by a tuning parameter λ that controls the sparsity of the representation. Increasing the sparse parameter makes the solution more sparse. So far, all previous sparse network approaches use a fixed parameter λ that may not be optimal. Depending on the choice of the sparse parameter, the final statistical results will be different. Instead of performing statistical inference at one fixed sparse parameter λ that may not be optimal, we introduce a new framework that performs statistical inferences over the whole parameter space using persistent homology [12], [17], [18], [27], [32], [46], [47], [67].
Persistent homology is a recently popular branch of computational topology with applications in protein structures [64], gene expression [24], brain cortical thickness [18], activity patterns in visual cortex [67], sensor networks [23], complex networks [39] and brain networks [46], [47]. However, as far as we are aware, it is yet to be applied to sparse models in any context. This is the first study that introduces persistent homology in sparse models. The proposed persistent homological framework is similar to the existing multi-thresholding framework that has been used in modeling connectivity matrices at many different thresholds [1], [37], [47], [69]. However, such an approach has not been applied in sparse networks before. In a sparse network, sparsity is controlled by the sparse parameter λ and the estimated sparse matrix entries. So it is unclear how the existing multi-thresholding framework can be applicable in this situation. In this paper, we prove that thresholding the sparse parameter is equivalent to thresholding correlations under some conditions. Thus, we resolve the unclarity of applying the existing multi-threshold method to the sparse networks.
The main methodological contributions of this paper are as follows. (i) We introduce a new sparse model based on Pearson correlation. Although various sparse models have been proposed for other correlations such as partial correlations [8], [40], [48] and canonical correlations [4], the sparse version of the Pearson correlation was not often studied.
(ii) We introduce persistent homology in the proposed sparse model for the first time. We explicitly show that persistent homological structures can be found in the sparse model. This paper differs substantially from our previous study [47], which studies the persistent homology in graphs and networks. Sparse models and sparse networks were never considered in [47].
(iii) We show that the identification of persistent homological structures can yield greater computational speed and efficiency in solving the proposed sparse correlation model without any numerical optimization. Note that most sparse models require numerical optimization for minimizing sparse penalty, which can be a computational bottleneck for solving large scale problems. There are few attempts at speeding up the computation for sparse models. By identifying block diagonal structures in the estimated (inverse) covariance matrix, it is possible to bypass the numerical optimization in the penalized log-likelihood method [55], [74]. LASSO (least absolute shrinkage and selection operator) can be done without numerical optimization if the design matrix is orthogonal [72]. The proposed method substantially differs from [55], [74] in that we do not need to assume the data to follow normality since there is no need to specify the likelihood function. Further the cost functions we are optimizing are different. The proposed method also differs from [72] in that our problem is not orthogonal.
As an application of the proposed method, we applied the techniques to the quantification of interregional white matter abnormality in stress-exposed children's magnetic resonance images (MRI). Early and severe childhood stress, such as experiences of abuse and neglect, have been associated with a range of cognitive deficits [52], [59], [65] and structural abnormalities [35], [36], [42]. However, little is known about the underlying biological mechanisms leading to cognitive problems in these children [60] due to the difficulties in the existing methods that do not have enough discriminating power. However, we demonstrate that the proposed method is very well suited to this problem.
II. Methods
A. Sparse Correlations
Correlations
Consider measurement vector xj on node j. If we center and rescale the measurement xj such that
the sample correlation between nodes i and j is given by . Since the data is normalized, the sample covariance matrix is reduced to the sample correlation matrix.
Consider the following linear regression between nodes j and k (k ≠ j):
| (1) |
We are correlating data at node j to data at node k. In this particular case, γjk is the usual Pearson correlation. The least squares estimation (LSE) of γjk is then given by
| (2) |
which is the sample correlation. For the normalized data, the estimated regression coefficient is exactly the sample correlation. For the normalized and centered data, the regression coefficient is the correlation. Equation (2) minimizes the sum of least squares over all nodes:
| (3) |
Note that we do not really care about correlating xj to itself since the correlation is then trivially γjj = 1.
1) Sparse Correlations
Let Γ = (γjk) be the correlation matrix. The sparse penalized version of (3) is given by
| (4) |
The sparse correlation is given by minimizing F(Γ). By increasing λ, the estimated correlation matrix Γ̂(λ) becomes more sparse. When λ = 0, the sparse correlation is simply given by the sample correlation, i.e., . As λ increases, the correlation matrix Γ shrinks to zero and becomes more sparse.
This sparse regression is not orthogonal, i.e., , the Dirac delta, so the existing soft-thresholding method for LASSO [72] is not applicable. The minimization of (4) can be done by the proposed soft-thresholding method analytically by exploiting the topological structure of the problem.
Theorem 1
For λ ≥ 0, the minimizer of (4) is given by the soft-thresholding
| (5) |
Proof
Write (4) as
| (6) |
where
Since f (γjk) is nonnegative and convex, F(Γ) is minimum if each component f (γjk) achieves minimum. So we only need to minimize each component f (γjk). This differentiates our sparse correlation formulation from the standard compressed sensing or LASSO that cannot be optimized in this component wise fashion. f (γjk) can be rewritten as
We used the fact .
For λ = 0, the minimum of f (γjk) is achieved when , which is the usual LSE. For λ > 0, Since f (γjk) is quadratic in γjk, the minimum is achieved when
| (7) |
The sign of λ depends on the sign of γjk. Thus, sparse correlation γ̂jk is given by a soft-thresholding of :
| (8) |
Theorem 1 is heuristically introduced in the conference paper [17]. This paper extends [17] with clearly spelled out soft-thresholding rule and the detailed proof. The estimated sparse correlation (8) basically thresholds the sample correlation that is larger or smaller than λ by the amount λ. Due to this simple expression, there is no need to optimize (4) numerically as often done using the coordinate descent learning or the active-set algorithm in compressed sensing or LASSO [30], [58]. Note Theorem 1 is only applicable in a separable compressed sensing or LASSO type problem.
Since different choices of sparsity parameter λ will produce different solutions in sparse model 𝒜(λ), we propose to use the collection of all the sparse solutions for many different values of λ for the subsequent statistical analysis. This avoids the problem of using an arbitrary threshold or identifying the optimal sparse parameter that may not be optimal in practice. The question is then how to use the collection of 𝒜(λ) in a coherent mathematical fashion. For this, we propose to apply persistent homology [26], [46], [47] and establish Theorem 2.
B. Persistent Homology in Graphs
Using persistent homology, topological features such as the connected components and cycles of a graph can be tabulated in terms of the Betti numbers. The Betti numbers β0 and β1, which are topological invariants, respectively denote the number of connected components and holes in the graph [27]. The network difference is then quantified using the Betti numbers of the graph [46], [47]. The graph filtration is a new graph simplification technique that iteratively builds a nested subgraphs of the original graph. The algorithm simplifies a complex graph by piecing together the patches of locally connected nearest nodes. The process of graph filtration is related to the single linkage hierarchical clustering and dendrogram construction [46], [47].
Consider a weighted graph with node set V = {1, …, p} and edge weights ρ = (ρjk), where ρjk is the weight between nodes j and k. Weighted graph X = (V, ρ) is formed by the pair of node set V and edge weights ρ. The edge weights in many brain imaging applications are usually given by some similarity measures such as correlation or covariance between nodes [46], [51], [56], [57], [68]. Given weighted network X = (V, ρ), we induce binary network 𝒢(λ) by thresholding the weighted network at λ. The adjacency matrix A = (ajk) of 𝒢(λ) is defined as
| (9) |
Any edge weight less than or equal to λ is made into zero while edge weight larger than λ is made into one. The binary network 𝒢(λ) is a simplicial complex consisting of 0-simplices (nodes) and 1-simplices (edges), a special case of the Rips complex [32]. Then it can be seen that 𝒢(λ1) ⊃ 𝒢(λ2) for λ1 < λ2 in a sense the vertex and edge sets of 𝒢(λ2) are contained in those of 𝒢(λ1). Just as in the case of Rips filtration, which is a collection of nested Rips complexes, we can construct the filtration on the collection of binary networks:
| (10) |
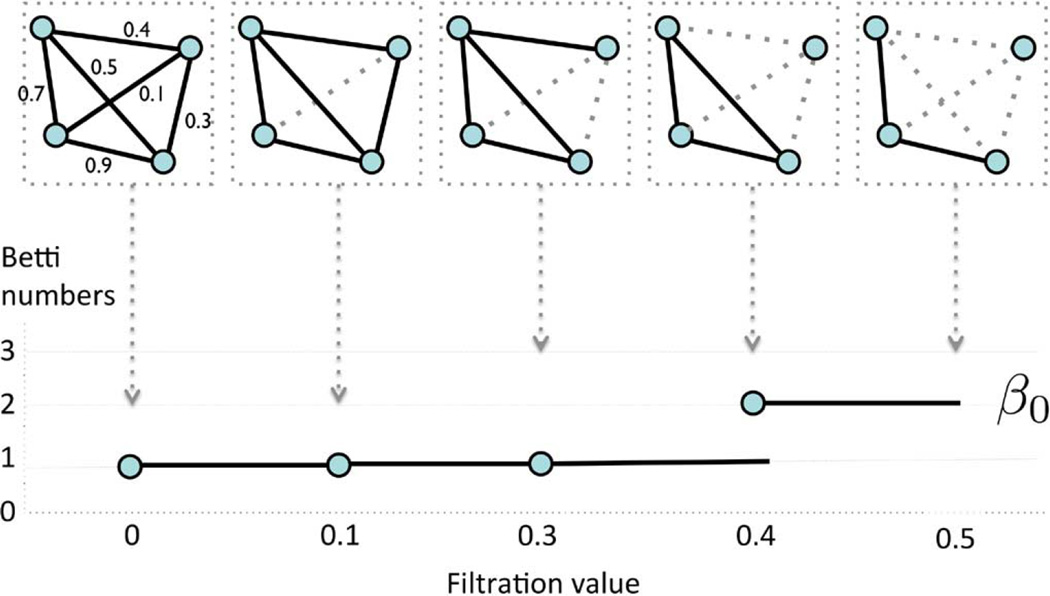
for 0 = λ0 < λ1 < λ2 < ⋯. Note that 𝒢(0) is the complete weighted graph while 𝒢(∞) is the node set V. By increasing the λ value, we are thresholding at higher correlation so more edges are removed. Such the nested sequence of the Rips complexes (10) is called a Rips filtration, the main object of interest in persistent homology [26]. The sequence of λ values are called the filtration values. Since we are dealing with a special case of Rips complexes restricted to graphs, we will call such structure graph filtration. Fig. 1 illustrates an example of a graph filtration with 4 nodes. Sequentially we are deleting edges based on the ordering of the edge weights. Since the graph filtration is a special case of the Rips filtration, it inherits all the topological properties of the Rips filtration. Given a weighted graph, there are infinitely many different filtrations. In Fig. 1 example, we have two filtrations 𝒢(0.0) ⊃ 𝒢(0.1) ⊃ 𝒢(0.3) ⊃ 𝒢(0.4) ⊃ 𝒢(0.5) and 𝒢(0.0) ⊃ 𝒢(0.2) ⊃ 𝒢(0.6) among many other possibilities. So a question naturally arises if there is a unique filtration that can be used in characterizing the graph. Let the level of a filtration be the number of nested unique sublevel sets in the given filtration.
Fig. 1.
Schematic of graph filtration. We start with a weighted graph (top left). We sort the edge weights in an increasing order. We threshold the graph at filtration value λ and obtain unweighted binary graph 𝒢(λ) based on rule (9). The thresholding is performed sequentially by increasing λ values. Then we obtain the sequence of nested graphs such as 𝒢(0.0) ⊃ 𝒢(0.1) ⊃ 𝒢(0.3) ⊃ ⋯. The collection of such nested graph is defined as a graph filtration. The dotted lines are thresholded edges. The first Betti number β0, which counts the number of connected components, is then plotted over the filtration.
Theorem 2
For graph X = (V, ρ) with q unique positive edge weights, the maximum level of a filtration on the graph is q + 1. Further, the filtration with q + 1 filtration level is unique.
Proof
For a graph with p nodes, the maximum number of edges is (p2 − p)/2, which is obtained in a complete graph. If we order the edge weights in the increasing order, we have the sorted edge weights:
where q ≤ (p2 − p)/2. The subscript () denotes the order statistic. For all λ < ρ(1), 𝒢(λ) = 𝒢(0) is the complete graph of V. For all ρ(r) ≤ λ < ρ(r + 1) (r = 1, …, q − 1), 𝒢(λ) = 𝒢(ρ(r)). For all ρ(q) ≤ λ, 𝒢(λ) = 𝒢(ρ(q)) = V, the vertex set. Hence, the filtration given by
| (11) |
is maximal in a sense that we cannot have any additional level of filtration.
The condition of having unique edge weights is not restrictive in practice. Assuming edge weights to follow some continuous distribution, the probability of any two edges being equal is zero. Among many possible filtrations, we will use the maximal filtration (11) in the study since it is uniquely given. The finiteness and uniqueness of the filtration levels over finite graphs are intuitively clear by themselves and are already applied in software packages such as javaPlex. [2]. However, we still need a rigorous statement to specify the type of filtration we are using.
C. Persistent Homology in Sparse Regression
We introduce a persistent homological structure in sparse correlations now as follows. Let A = (ajk(λ)) be the adjacency matrix obtained from sparse correlation (8):
Let 𝒢(λ) be the graph defined by the adjacency matrix A. Then we have the main result of this paper, which relies on the results of Theorem 1 and Theorem 2.
Theorem 3
For centered and normalized data xj (j = 1, …, p), ρ(1), ρ(2), …, ρ(q) be the order statistic of edge weights (1 ≤ j, k ≤ p, k ≠ j). Then graph 𝒢(λ) obtained from the sparse regression (4) forms the maximal graph filtration
| (12) |
Proof
The proof follows by simplifying the adjacency matrix A into a simpler but equivalent adjacency matrix B = (bjk). From (8), γ̂jk(0) ≠ 0 if and 0 otherwise. Thus, the adjacency matrix A is equivalent to the adjacency matrix B = (bjk):
| (13) |
Let ℋ(λ) be the graph defined by adjacency matrix B. Graph ℋ(λ) is formed by thresholding edge weights given by the absolute value of sample correlations . From Theorem 2, such graph must have maximal filtration:
| (14) |
Since A = B, graph 𝒢 also must have the identical maximal filtration.
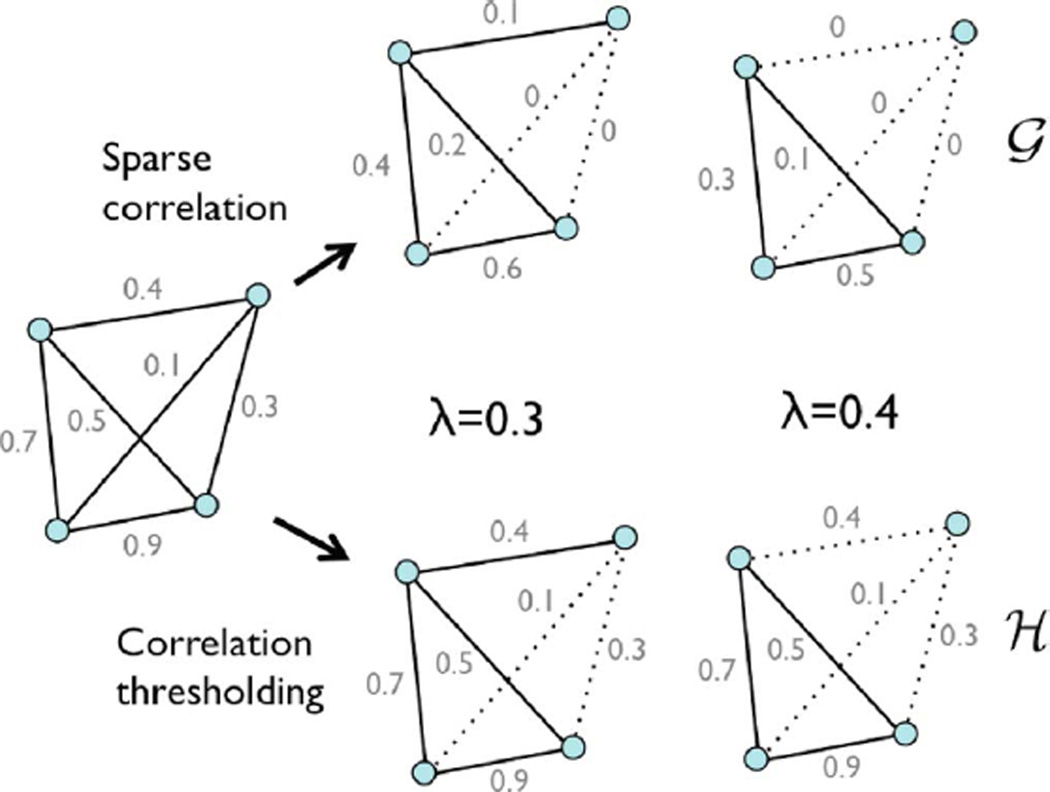
Theorem 3 is illustrated in Fig. 2 with a 4-nodes example. In this study, much larger p = 548 and p = 1856 nodes will be used. In obtaining the topological structure of a graph induced by sparse correlation, it is not necessary to solve the sparse regression by the direct optimization, which can be very time consuming. Identical topological information can be obtained by performing the soft-thresholding on the sample correlations.
Fig. 2.
Comparison between the sparse correlation estimation via numerical optimization (top) and the proposed soft-thresholding method in Theorem 3 (bottom). The direct numerical optimization makes the graph sparse by shrinking the edge weights to zero. Nonzero edges form binary graph 𝒢. The persistent homological approach thresholds the sample correlations at given filtration value and construct binary graph ℋ. The both methods produce the identical binary graphs, i.e., 𝒢 = ℋ. If the methods are applied at two different parameters λ = 0.3, 0.4, we obtain nested binary graphs 𝒢(0.3) ⊃ 𝒢(0.4) and ℋ(0.3) ⊃ ℋ(0.4). Theorem 3 generalizes this example.
The resulting maximal filtration can be quantified by plotting the change of Betti numbers over increasing filtration values [27], [32], [46]. The first Betti number β0(λ) counts the number of connected components of the given graph 𝒢(λ) at the filtration value λ [47]. Given graph filtration 𝒢(λ0) ⊃ 𝒢(λ1) ⊃ 𝒢(λ2) ⊃ ⋯, we plot the first Betti numbers β0(λ0) < β0(λ1) < β0(λ2) ⋯ over filtration values λ0 < λ1 < λ2 ⋯ (Fig. 1). The number of connected components increase as the filtration value increases. The pattern of increasing number of connected components visually show how the topology of the graph changes over different parameter values. The overall pattern of Betti (number) plots can be used as a summary measure of quantifying how the graph changes over increasing edge weights. The Betti number plots are related but different from barcodes in literature. The Betti number is equal to the number of bars in the barcodes at the specific filtration value. To construct Betti plots, it is not necessary to perform filtrations for infinitely many possible λ values. From Theorem 2, the maximum possible number of filtration level for plotting the Betti numbers is one plus the number of unique edge weights. For a tree, which is a graph with no cycle, we can come up with a much stronger statement than this.
Theorem 4
For a tree with p ≥ 2 nodes and unique positive edge weights ρ(1) < ρ(2) < ⋯ ρ(p−1), the plot for the first Betti number (β0) corresponding to the maximal graph filtration is given by the coordinates
Proof
For a tree with p nodes, there are total p − 1 edges. Then from Theorem 2, we have the maximal filtration
| (15) |
Since all the edge weights are above filtration value ρ(0) = 0, all the nodes are connected, i.e., β0(ρ(0)) = 1. Since no edge weight is above the threshold ρ(q−1), β0(ρ(p−1)) = p. At each time we threshold an edge, the number of components increases exactly by one in the tree. Thus, we have
For a general graph, it is not possible to analytically determine the coordinates for its Betti-plot. The best we can do is to compute the number of connected components β0 numerically using the single linkage dendrogram method (SLD) [47], the Dulmage-Mendelsohn decomposition [16], [61] or existing simplical complex approach [12], [23], [27]. For our study, we used the SLD method.
D. Statistical Inference on Betti Number Plots
The first Betti number will be used as features for characterizing network differences statistically. We assume there are n subjects and p nodes in Group 1. For subject i, we have measurement xij at node j. Denote data matrix as X = (xij), where xij is the measurement for subject i at node j. We then construct a sparse network and corresponding Betti number using X. Thus, is a function of X. Consider another Group 2 consists of m subjects. For Group 2, data matrix is denoted as Y = (yij), where yij is the measurement for subject i at node j. Group 2 will also generate single Betti number plot as a function of Y. We are then interested in testing if the shapes of Betti number plots are different between the groups. This can be done by comparing the areas under the Betti plots. So the null hypothesis of interest is
| (16) |
while the alternate hypothesis is
This inference avoids the use of multiple comparisons. The null hypothesis (16) is related to the following pointwise null hypothesis:
| (17) |
If the hypothesis (17) is true, the hypothesis (16) is also true (but inverse is not true). Thus, testing the area under the curve is related to testing the height of the curve at every point. The advantage of using the area under the curve is that we do not need to worry about multiple comparisons associated with testing (17). The area under the curve seems a reasonable approach to use for Betti-plots. A similar approach has been introduced in [14] in removing the multiple comparisons and produce a single summary test statistic.
There is no prior study on the statistical distribution on the Betti numbers so it is difficult to construct a parametric test procedure. Further, since there is only one Betti-plot per group, it is necessary to empirically construct the null distribution and determine the p-value by resampling techniques such as the permutation test and jackknife [15], [17], [28], [47]. For this study, we use the jackknife resampling.
For Group 1 with n subjects, one subject is removed at a time and the remaining n − 1 subjects are used in constructing a network and a Betti-plot. Let X−l be the data matrix, where the l-th row (subject) is removed from X. Then for each l-th subject removed, we compute , which is a function of λ and X−l. Repeating this process for each subject, we obtain n Betti-plots . For Group 2, the l-th row (subject) is removed from the original data matrix Y and obtain data matrix as Y−l. For each l-th subject removed, we compute , which is a function of λ and X−l. Repeating this process for each subject, we obtain m Betti-plots . There are 23 maltreated and 31 control children in the study, so we have 23 and 31 Jackknife resampled Betti-plots. Subsequently we compute the areas under the Betti-plots by discretizing the integral. The area differences between the groups are then tested using the Wilcoxon rank-sum test, which is a nonparametric test on median differences [33].
We did not use the permutation test. For the permutation test to converge for our data set, it requires tens of thousands permutations and it is really time consuming even with the proposed time-saving soft-thresholding method. The proposed method takes about a minute of computation in a desktop but ten-thousands permutations will take about seven days of computation. Hence, we used a much simpler Jackknife resampling technique. The procedure is validated using the simulation with the known ground truth. The MATLAB codes for constructing network filtration, barcodes and performing statical inference on are given in http://brainimaging.waisman.wisc.edu/~chung/barcodes with the post-processed Jacobian determinant and FA data that were used for this study.
1) Simulations
We performed two simulations. In each simulation, the sample sizes are n = 20 in Group 1 and m = 20 in Group 2. There are p = 100 nodes. In Group 1, data xij at node j for subject i is simulated as independent standard normal N (0, 1) for the both simulations.
2) Study1 (No Group Difference)
In Group 2, we simulated data yij at node j for subject i as yij = xij + N (0, 0.052). Small noise N (0, 0.052) is added to perturb Group 1 data a little bit. It is expected there is no group difference. Following the proposed framework, we constructed the sparse correlation networks and constructed a Betti-plot. Then performed the Jackknife resampling and constructed 20 Betti-plots in each group. The rank sum test was applied and obtained the p-value of 0.78. So we could not detect any group difference as expected.
3) Study 2 (Group Difference)
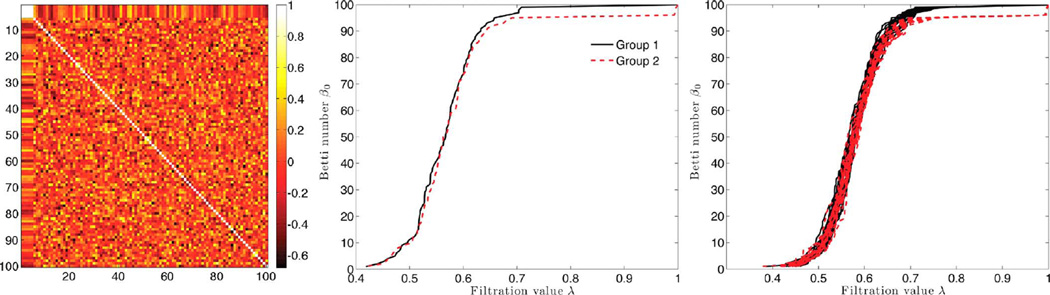
We first simulate data as yij = xij + N (0, 0.052) independently for all the nodes. Then for four nodes indexed by i = 2, 3, 4, 5, we introduce additional dependency: yij = 0.5xi1 + N (0, 0.052). We added small noise to perturb the node values further. This dependency gives any connection between 1 to 5 to have high correlation. Fig. 3 shows the simulated correlation matrix. Following the proposed framework, we constructed the sparse correlation networks and constructed a Betti-plot. Then performed the Jackknife resampling and constructed 20 Betti-plots in each group. The rank sum test was applied and obtained the p-value less than 0.001. This significance corresponds to the horizontal gap between the Betti-plots after the filtration value 0.7 (Fig. 3 right).
Fig. 3.
Simulation study 2. Left: the simulated correlation matrix for Group 2, where the first 5 nodes are connected (white square). Group 1 has no connection. Middle: The resulting β0-plot showing group differences. Right: Leave-one-out Jackknife resampled β0-plots of Group 1(solid line) and Group 2 (dotted line). The rank-sum test is performed on the area differences under β0-curves between the groups (p-value < 0.001). The statistically significant result corresponds to the horizontal gap in the Betti numbers after filtration value 0.7.
III. Application
A. Imaging Data Set and Preprocessing
The study consists of 23 children who experienced documented maltreatment early in their lives, and 31 age-matched normal control (NC) subjects. Additional details on subjects can be found in [35], [60]. All the children were recruited and screened at the University of Wisconsin. The maltreated children were raised in institutional settings, where the quality of care has been documented as falling below the standard necessary for healthy human development. For the controls, we selected children without a history of maltreatment from families with similar current socioeconomic statuses. The exclusion criteria include, among many others, abnormal IQ (< 78), congenital abnormalities (e.g., Down syndrome or cerebral palsy) and fetal alcohol syndrome (FAS). The average age for maltreated children was 11.26 ± 1.71 years while that of controls was 11.58 ± 1.61 years. This particular age range is selected since this development period is characterized by major regressive and progressive brain changes [35], [49]. There are 10 boys and 13 girls in the maltreated group and 18 boys and 13 girls in the control group. Groups did not differ on age, pubertal stage, sex, or socio-economic status [35]. The average amount of time spent in institutional care by children was 2.5 years ± 1.4 years, with a range from 3 months to 5.4 years. Children were on average 3.2 years ± 1.9 months when they adopted, with a range of 3 months to 7.7 years. Table I summarizes the participant characteristics.
TABLE I.
Study Participant Characteristics
| Maltreated | Normal controls | |
|---|---|---|
| Sample size | 23 | 31 |
| Sex (males) | 10 | 18 |
| Age (years) | 11.26 ± 1.71 | 11.58 ± 1.61 |
| Duration (years) | 2.5 ± 1.4 (0.25 to 5.4) | |
| Time of adoption (years) | 3.2 ± 1.9 (0.25 to 7.7) |
T1-weighted MRI were collected using a 3T General Electric SIGNA scanner (Waukesha, WI), with a quadrature birdcage head coil. DTI were also collected in the same scanner using a cardiac-gated, diffusion-weighted, spin-echo, single-shot, EPI pulse sequence. The details on image acquisition parameters are given in [35]. Diffusion tensor encoding was achieved using twelve optimum non-collinear encoding directions with a diffusion weighting of 1114 s/mm2 and a non-DW T2-weighted reference image. Other imaging parameters were TE = 78.2 ms, 3 averages (NEX: magnitude averaging), and an image acquisition matrix of 120 × 120 over a field of view of 240 × 240 mm2. The acquired voxel size of 2 × 2 × 3 mm was interpolated to 0.9375 mm isotropic dimensions (256 × 256 in plane image matrix). To minimize field inhomogeneity and image artifacts, high order shimming and fieldmap images were collected using a pair of non-EPI gradient echo images at two echo times: TE1 = 8 ms and TE2 = 11 ms.
For MRI, a study specific template was constructed using the diffeomorphic shape and intensity averaging technique through Advanced Normalization Tools (ANTS) [5]. Image normalization of each individual image to the template was done using symmetric normalization with cross-correlation as the similarity metric. The 1 mm resolution inverse deformation fields are then smoothed out with Gaussian kernel with bandwidth σ = 4 mm, which is equivalent to the full width at half maximum (FWHM) of 4 mm. Then the Jacobian determinants of the inverse deformations from the template to individual subjects were computed at each voxel. The Jacobian determinants measure the amount of voxel-wise change from the template to the individual subjects. White matter was also segmented into tissue probability maps using template-based priors, and registered to the template [9]. For DTI, images were corrected for eddy current related distortion and head motion via FSL software (http://www.fmrib.ox.ac.uk/fsl) and distortions from field inhomogeneities were corrected using custom software based on the method given in [43] before performing a non-linear tensor estimation using CAMINO [21]. Subsequently, we have used iterative tensor image registration strategy for spatial normalization [44], [78]. Then fractional anisotropy (FA) were calculated for diffusion tensor volumes diffeomorphically registered to the study specific template.
B. Results: Proposed Sparse Correlation
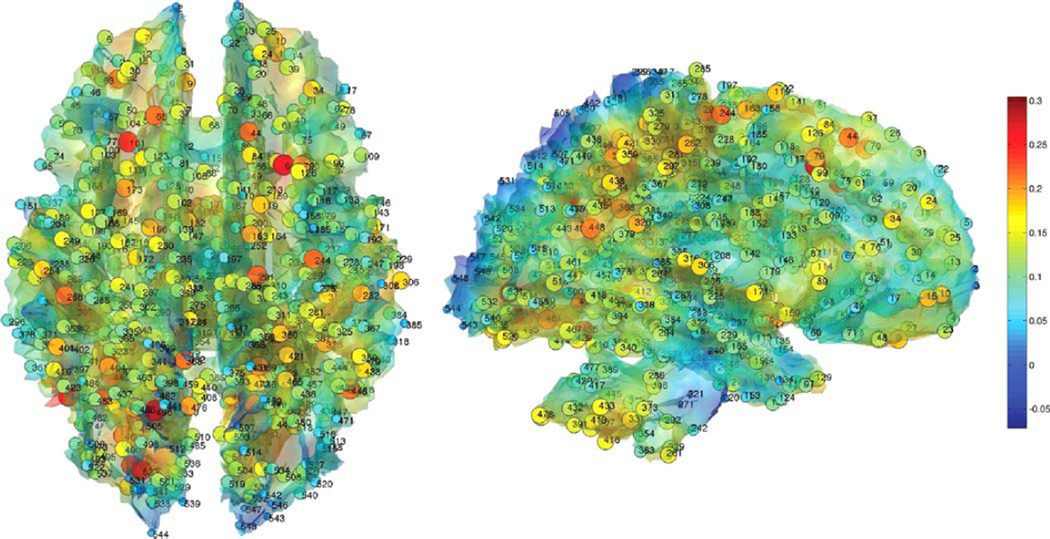
We thresholded the white matter density at 0.7 and obtained the isosurface. The resulting isosurface is not the gray and white matter tissue boundary and it is located inside the white matter. We are interested in the white matter changes along the tissue boundary. The surface mesh has 189536 mesh vertices and the average inter-nodal distance of 0.98 mm. Since Jacobian determinant and FA values at neighboring voxels are highly correlated, 0.3% of the total mesh vertices are uniformly sampled to produce p = 548. This gives average inter-nodal distance of 15.7 mm, which is large enough to avoid spurious high correlation between two adjacent nodes (Fig. 4). The isosurface of the white matter template was extracted using the marching cube algorithm [53]. The number of nodes are still larger than most region of interest (ROI) approaches in MRI and DTI, which usually have around 100 regions [77]. This resulted in 548 × 548 sample covariances and correlation matrices, which are not full rank. We constructed the proposed sparse correlation based network filtrations from the soft-thresholding method (Fig. 5). Subsequently, Betti-plots are computed (Fig. 6). Since each group produces one Betti-plot, the leave-one-out Jackknife resampling technique was performed to produce 23 and 31 resampled Bettiplots respectively for the two groups. We then computed the areas under the Betti-plots. Using the rank-sum test, we detected the statistical significance of the area differences between the groups (p-value < 0.001). The Betti-plots for normal controls show much higher Betti numbers at any given threshold.
Fig. 4.
548 uniformly sampled nodes along the white matter surface where the sparse correlations and covariances are computed. The nodes are sparsely sampled on the template surface to guarantee there is no spurious high correlation due to proximity between nodes. Color scales are the Jacobian determinant of a subject. The same nodes are taken in both MRI and DTI for comparison between the two modalities.
Fig. 5.
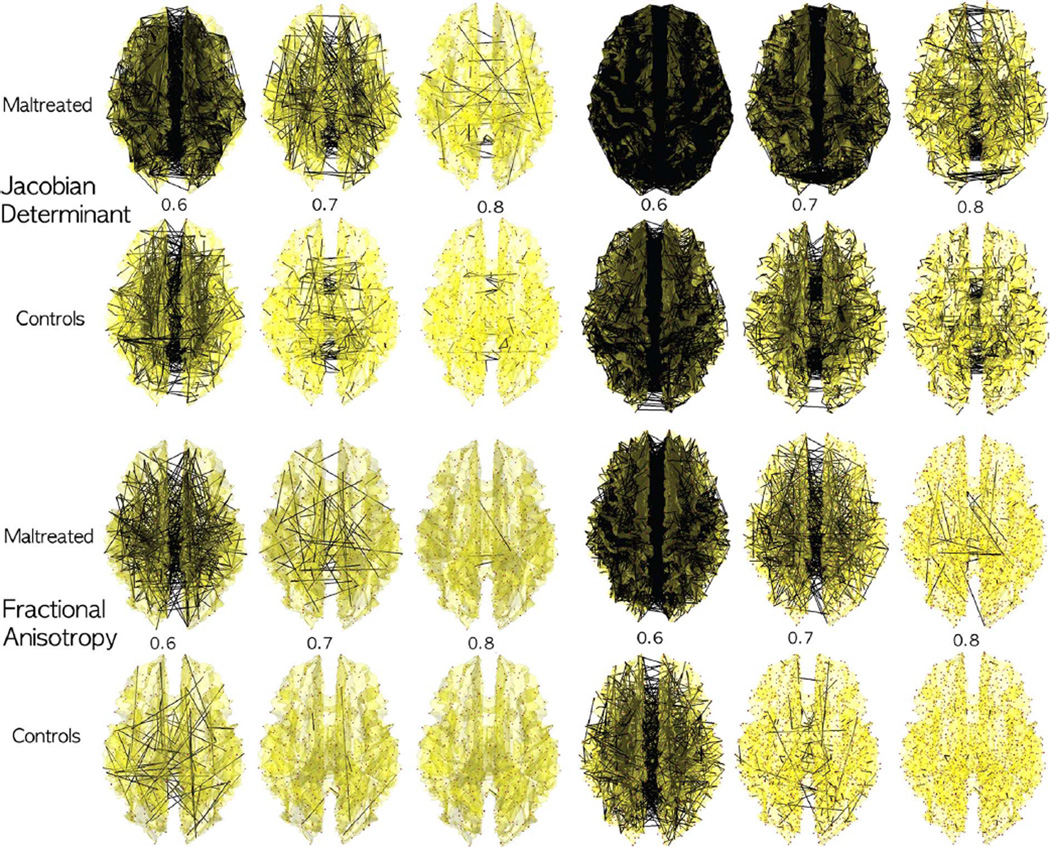
Networks 𝒢(λ) obtained by thresholding sparse correlations for the Jacobian determinant from MRI and fractional anisotropy (FA) from DTI at different λ values (λ = 0.6, 0.7, 0.8) for 548 nodes (left three columns) 1856 nodes (right three columns). The collection of the thresholded graphs forms a filtration. The children exposed to early life stress and maltreatment show more dense network at the given λ value. Since the maltreated children are more homogenous in the white matter region, there are more dense high correlations between nodes. The over all pattern of dense connections in the maltreated children is similar between the networks of different node sizes and across the different imaging modalities.
Fig. 6.
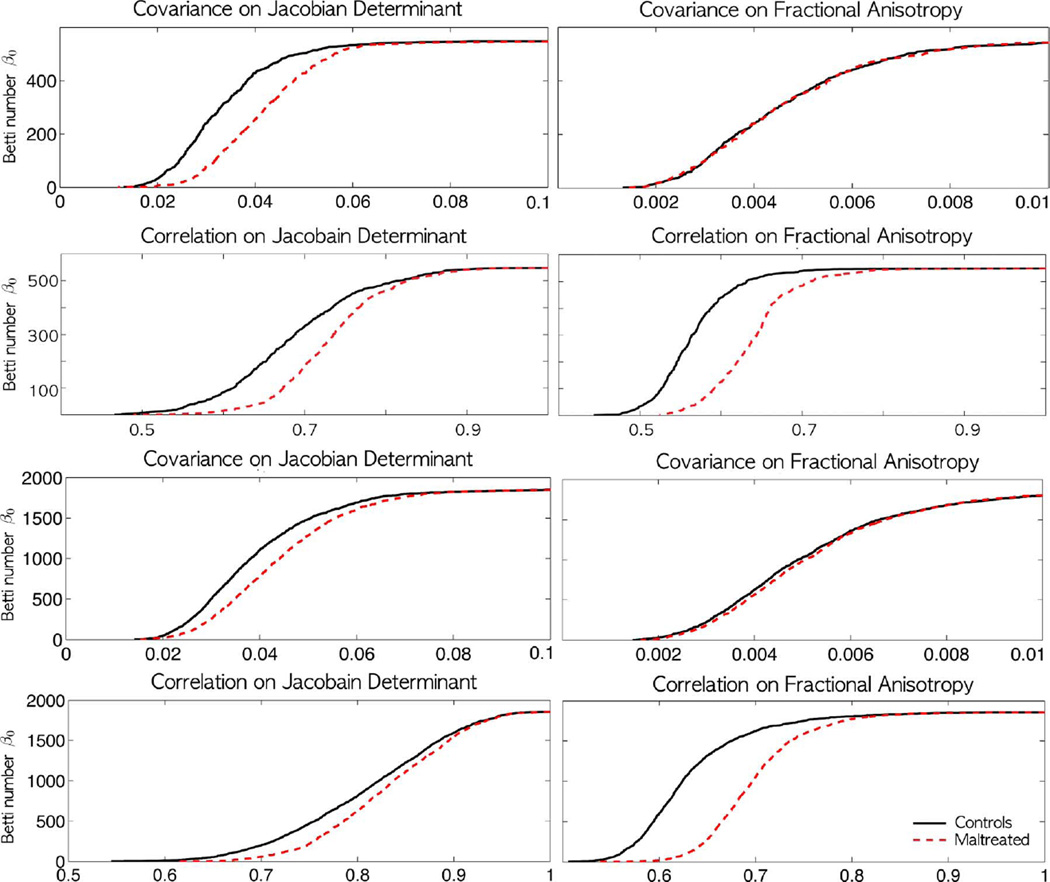
The Betti-plots on the sparse covariance and the proposed sparse correlation for Jacobian determinant (left column) and FA (right column) on 548 (top two rows) and 1856 (bottom two rows) node studies. Unlike the sparse covariance, the sparse correlation seems to shows huge group separation between normal and stress-exposed children visually. However, in all 7 cases except top right (548 nodes covariance for FA), we detected statistically significant differences using the rank-sum test on the areas under the Betti-plots (p-value < 0.001). The shapes of Betti-plots are consistent between the studies with different node sizes indicating the robustness of the proposed method over changing number of nodes.
1) Biological Interpretation
In the Betti-plots (Fig. 6), we obtain more disconnected components for controls than for children in the early stress group for any specific λ value. It can only happen if Jacobian determinants have higher correlations in the maltreated children across the white matter voxels compared to the controls. So when thresholded at a specific correlation value, more edges are preserved in the maltreated children resulting in more connected components. Thus, the children exposed to early life stress and maltreatment show more dense network at a given λ value. This is clearly demonstrated visually in Fig. 5. If the variations of Jacobian determents are similar across voxels, we would obtain higher correlations. This suggests more anatomical homogeneity across whole brain white matter regions in the maltreated children. Our finding is consistent with the previous study on neglected children that shows disrupted white matter organization, which results in more diffuse connections between brain regions [35]. Lower white matter directional organization in white matter may correspond to the increased homogeneity of Jacobian determinants and FA-values across the brain regions. Similar experiences may cause some areas to be connected to other regions of the brain at a higher degree inducing higher homogeneity in the regions. This type of relational interpretation cannot be obtained from the traditional univariate TBM.
C. Comparison Against Sparse Covariance
We compared the performance of the proposed sparse correlation technique to the widely used penalized log-likelihood method [6], [7], [30], [41], [55], where the log-likelihood is regularized with a sparse penalty:
| (18) |
Σ = (σij) is the covariance matrix and S is the sample covariance matrix. ‖·‖1 is the sum of the absolute values of the elements. The penalized log-likelihood is maximized over the space of all possible symmetric positive definite matrices. Equation (18) is a convex problem and it is numerically optimized using the graphical-LASSO (GLASSO) algorithm [6], [7], [30], [41]. The tuning parameter λ > 0 controls the sparsity of the off-diagonal elements of the covariance matrix. By increasing λ > 0, the estimated covariance matrix becomes more sparse.
We also performed the graph filtration technique to the estimated sparse covariance matrix Σ̂ = (σ̂ij). Let A = (aij) be the adjacency matrix defined from the estimated sparse covariance:
| (19) |
The adjacency matrix A induces graph 𝒢(λ) consisting of κ(λ) number of partitioned subgraphs:
| (20) |
where Vl and El are vertex and edge sets of the subgraph Gl respectively. Unlike the sparse correlation case, we do not have full persistency on the induced graph 𝒢. The partitioned graphs can be proven to be partially nested in a sense that only the partitioned node sets are persistent [17], [41], [55], i.e.,
| (21) |
for λ1 < λ2 < λ3 < ⋯ and all l. Subsequently the collection of partitioned vertex set is also persistent. On the other hand, edge sets El may not be persistent.
From (21), it is unclear if there exists a unique maximal filtration on the vertex set. The maximal filtration can be obtained as follows. Let B(λ) = (bij) be another adjacency matrix given by
| (22) |
where ŝij is the sample covariance matrix. It can be shown that the adjacency matrix B similarly induces graph ℋ [17], [55]:
| (23) |
for some edge set Fl(λ). Further, the subgraphs Gl and Hl have identical vertex set but different edge sets. Then from Theorem 2, we have maximal filtration on the graph ℋ, where the edge weights are given by the sample covariances. Theorem 2 requires the edge weights to be all unique, which is satisfied for the study data set. Then similar to Theorem 3, the Betti-plots are determined by ordering the entries of the sample covariance matrices. The resulting barcode is displayed in Fig. 6. The sparse covariance was also able to discriminate the groups statistically (p-value < 0.001). The changes in the first Betti number are occurring in a really narrow window but was still able to detect the group differences using the areas under the Betti number plots (Fig. 6). However, the sparse correlations exhibit slower changes in the Betti number over the wide window, making it easier to discriminate the groups.
D. Comparison Against Fractional Anisotropy in DTI
For children who suffered early stress, white matter microstructures have been reported as more diffusely organized [35]. Therefore, we predicted less white matter variability in both the Jacobian determinants and FA-values. The DTI acquisitions were done in the same 3T GE SIGNA scanner; acquisition parameters can be found in [35]. We applied the proposed persistent homological method in obtaining the filtrations for sparse correlations and covariances in the same 548 nodes on FA values (Fig. 4). The resulting filtration patterns show similar patterns of a rapid increase in disconnected components for sparse correlations (Figs. 5 and 6). The Jackknife resampling followed by the rank-sum test on the area differences shows a significant group difference for sparse correlations (p-value < 0.001). These results are due to a consistent abnormality among the stress-exposed children that is observed in both MRI and DTI modalities. The stress-exposed children exhibited stronger white matter homogeneity and less spatial variability compared to normal controls in both MRI and DTI measurements. However, the covariance results fail to discriminate the groups at 0.01 level (p-value = 0.043) indicative of a poor performance compared to the sparse correlation method.
E. Robustness on Node Size Changes
Depending on the number of nodes, the parameters of graph vary considerably up to 95% and the resulting statistical results will change substantially [29], [34], [77]. On the other hand, the proposed method is very robust under the change of node size. For the node sizes between 548 and 1856 (0.3% and 1% of original 189536 mesh vertices), the choice of node size did not affect the pattern of graph filtrations, the shape of Betti-plots, or the subsequent statistical results significantly. For example, the graph filtration on 1856 nodes shows a similar pattern of dense connections for the maltreated children (Fig. 5). The resulting Betti-plots also show similar pattern of the group separation (Fig. 6). The statistical results are also somewhat consistent. For both the Jacobian determinant and FA values, the group differences in Betti-plots obtained from sparse correlations and covariances are all statistically significant (p-value < 0.001) in both 548 and 1856 nodes except one case. For the case of the 548 nodes covariance on FA values, we did not detect any group differences at 0.01 level (p-value = 0.043). On the other hand, we detected the group difference for the 1856 nodes case at 0.001 level. The proposed framework is very sensitive, so it can detect really narrow but very consistent Betti-plot differences.
F. Effect of Image Registration
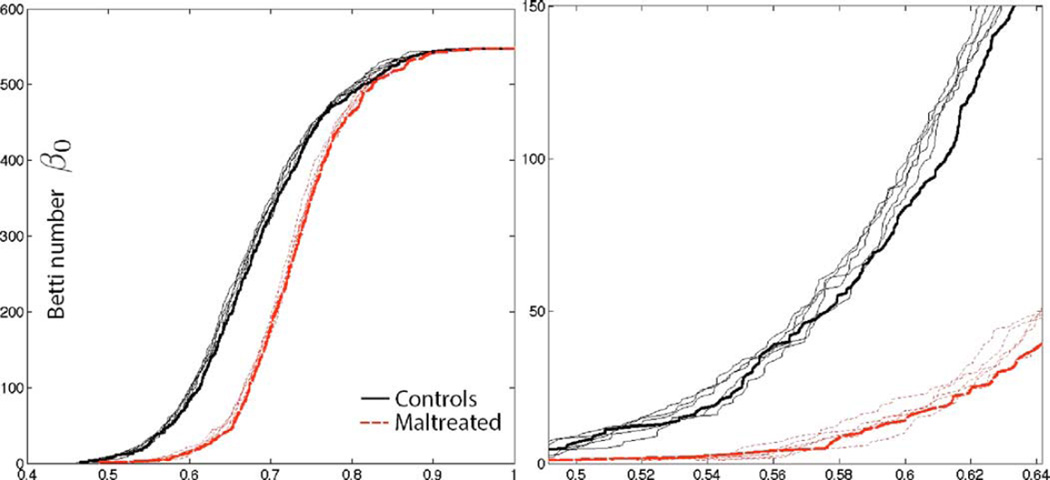
We checked how much impact image registration has on the robustness of the proposed method. Anatomical measurements across neighboring voxels are highly correlated within white matter so we do not expect image misalignment will have much effect on the final results. To determine the variability associated with the image registration, the displacement vector fields from the template to individual brains were randomly perturbed by adding Gaussian noise N(0, 1) to each component. This is sufficiently large noise and causes up to 4 mm misalignment for some nodes. Then following the proposed pipeline, the Jacobian determinants are correlated across 548 nodes and Betti-plots are computed. Fig. 7 shows five perturbation results. The thick line is without any perturbation. The perturbed Betti-plots are very stable and close to the Betti-plots without any perturbation (thick lines). Th height differences in the perturbed Betti-plots are less than 4.4% in average, which is negligible in the subsequent statistical analysis. In fact, the resulting p-values are similar to each other and all the perturbed results detected the group difference (p-value < 0.001). Thus, we conclude that the proposed topological framework is robust under image misalignment.
Fig. 7.
The displacement vector field from the template to individual brain is randomly perturbed. Then the Jacobian determinants are correlated across 548 nodes and Betti-plots are subsequently produced. The process is repeated five times to produce five perturbed Betti-plots. The thick line is without any perturbation. The perturbed Betti-plots are very stable and close to the Betti-plots without any perturbation (thick lines). The proposed topological framework is very robust under sufficiently large image misalignment. Right figure is the enlargement of the left figure.
IV. Conclusions and Discussions
By identifying persistent homological structures in sparse correlations, we were able to exploit them for drastically speeding up computations. A procedure that takes 56 hours was completed in few seconds without utilizing additional computational resources. Although we have only shown how to identify persistent homology in the sparse Pearson correlation, the underlying principle can be directly applicable to other sparse models and image filtering techniques. These include the least angle regression (LARS) implementation in more general LASSO [13], heat kernel smoothing [19], and diffusion wavelets [45], which all guarantee the nested subset structures over the sparse parameters and kernel bandwidth. We will leave the identification of persistent homology in other models for future studies.
We found that Betti-plots on correlations can visually discriminate better than Betti-plots on covariances. In Fig. 6, almost all topological changes associated with the covariance occur in really small range between 0 and 0.1. However, unlike covariances, correlations are normalized by the variances so the topological changes are more spread out between 0 and 1. This has the effect of making the Betti-plots shape differences spread out more uniformly and wide in the unit interval. This is most clearly demonstrated in the covariance vs. correlation on FA (second column). The Betti-plots of covariances are difficult to discriminate visually because the Betti-plots are squeezed into small range between 0 and 0.1 but the Betti-plots of correlations are more discriminative since the Betti-plots are more spread out. The visual discriminative power comes from the normalization associated with the Pearson correlation. The change in the metric affects the filtration process itself since it is based on the sorted edge weights. Subsequently, the shape of Betti-plots and the statistical inference results also change.
While massive univariate approaches can detect signal locally at each voxel, the proposed network approach can detect signal globally over the whole brain region. Even though the information obtained by the two methods are complementary, they are somewhat exclusive. The proposed approach tabulates the changes of the number of connected components in the thresholded networks via Betti-plots, which cannot be done at individual node level. There is no easy straightforward way of combining or comparing the results from the two methods. The Betti-plots is a global index that is defined over a whole graph so it is not directly applicable to node-level analysis. However, just like any global graph theoretic indices such as small-worldness and modularity [10], [63], it can be applied to subgraphs around a given node. This is the beyond the scope of the paper and we left it as future research.
Acknowledgment
The authors like to thank H. Lee of Seoul National University and M. Arnold of University of Bristol for the valuable discussions on persistent homology and sparse regressions, and S.-G. Kim of Max Planck Institute and N. Adluru of University of Wisconsin-Madison for help with image preprocessing.
This work was supported by NIH grants MH61285, MH68858, MH84051, NIH Fellowship DA028087, NIH-NCATS grant UL1TR000427 and the Vilas Associate Award.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Moo K. Chung, Department of Biostatistics and Medical Informatics and Waisman Laboratory for Brain Imaging and Behavior, University of Wisconsin, Madison, WI 53706 USA (mkchung@wisc.edu)
Jamie L. Hanson, Laboratory of Neurogenetics, Duke University, Durham, NC 27710 USA
Jieping Ye, Department of Computational Medicine and Bioinformatics and Department of Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI 48109-2218 USA.
Richard J. Davidson, Waisman Center, University of Wisconsin, Madison, WI 53705 USA
Seth D. Pollak, Waisman Center, University of Wisconsin, Madison, WI 53705 USA
References
- 1.Achard S, Salvador R, Whitcher B, Suckling J, Bullmore ED. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J. Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams H, Tausz A, Vejdemo-Johansson M. Mathematical Software-ICMS 2014. New York: Springer; 2014. javaPlex: A research software package for persistent (co) homology; pp. 129–136. [Google Scholar]
- 3.Ashburner J, Friston K. Voxel-based morphometry–The methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 4.Avants BB, Cook PA, Ungar L, Gee JC, Grossman M. Dementia induces correlated reductions in white matter integrity and cortical thickness: A multivariate neuroimaging study with sparse canonical correlation analysis. NeuroImage. 2010;50:1004–1016. doi: 10.1016/j.neuroimage.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee O, El Ghaoui L, d'Aspremont A. Model selection through sparse maximum likelihood estimation for multivariate Gaussian or binary data. J. Mach. Learn. Res. 2008;9:485–516. [Google Scholar]
- 7.Banerjee O, Ghaoui LE, d'Aspremont A, Natsoulis G. Convex optimization techniques for fitting sparse Gaussian graphical models. Proc. 23rd Int. Conf.Mach. Learn. 2006:96. [Google Scholar]
- 8.Bickel PJ, Levina E. Regularized estimation of large covariance matrices. Ann. Stat. 2008;36:199–227. [Google Scholar]
- 9.Bonner MF, Grossman M. Gray matter density of auditory association cortex relates to knowledge of sound concepts in primary progressive aphasia. J. Neurosci. 2012;32:7986–7991. doi: 10.1523/JNEUROSCI.6241-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullmore E, Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nature Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 11.Cao J, Worsley KJ. The geometry of correlation fields with an application to functional connectivity of the brain. Ann. Appl. Probabil. 1999;9:1021–1057. [Google Scholar]
- 12.Carlsson G, Memoli F. Persistent clustering and a theorem of J. Kleinberg ArXiv. 2008 [Online]. Available: arXiv:0808.2241, to be published. [Google Scholar]
- 13.Carroll MK, Cecchi GA, Irina R, Garg R, Rao AR. Prediction and interpretation of distributed neural activity with sparse models. NeuroImage. 2009;44:112–122. doi: 10.1016/j.neuroimage.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Chung MK. Ph.D. dissertation. Montreal, QC, Canada: McGill Univ.; 2001. Statistical morphometry in neuroanatomy. [Google Scholar]
- 15.Chung MK. Statistical and Computational Methods in Brain Image Analysis. Boca Raton, FL: CRC Press; 2013. [Google Scholar]
- 16.Chung MK, Adluru N, Dalton KM, Alexander AL, Davidson RJ. Scalable brain network construction on white matter fibers. Proc. SPIE. 2011;7962:79624G. doi: 10.1117/12.874245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung MK, et al. MICCAI. Vol. 8149. LNCS; 2013. Persistent homological sparse network approach to detecting white matter abnormality in maltreated children: MRI and DTI multimodal study; pp. 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung MK, Singh V, Kim PT, Dalton KM, Davidson RJ. MICCAI. Vol. 5762. LNCS; 2009. Topological characterization of signal in brain images using min-max diagrams; pp. 158–166. [DOI] [PubMed] [Google Scholar]
- 19.Chung MK, Worsley KJ, Brendon MN, Dalton KM, Davidson RJ. General multivariate linear modeling of surface shapes using SurfStat. NeuroImage. 2010;53:491–505. doi: 10.1016/j.neuroimage.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung MK, et al. A unified statistical approach to deformation-based morphometry. NeuroImage. 2001;14:595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- 21.Cook PA, et al. Camino: Open-source diffusion-MRI reconstruction and processing. Proc. 14th Sci. Meet. Int. Soc. Magn. Reson.Med. 2006 [Google Scholar]
- 22.Davatzikos C, et al. A computerized approach for morphological analysis of the corpus callosum. J. Comput. Assist. Tomogr. 1996;20:88–97. doi: 10.1097/00004728-199601000-00017. [DOI] [PubMed] [Google Scholar]
- 23.de Silva V, Ghrist R. Homological sensor networks. N. Am. Math. Soc. 2007;54:10–17. [Google Scholar]
- 24.Dequeant M-L, et al. Comparison of pattern detection methods in microarray time series of the segmentation clock. PLoS One. 2008;3:e2856. doi: 10.1371/journal.pone.0002856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubb A, Gur R, Avants B, Gee J. Characterization of sexual dimorphism in the human corpus callosum. Neuroimage. 2003;20:512–519. doi: 10.1016/s1053-8119(03)00313-6. [DOI] [PubMed] [Google Scholar]
- 26.Edelsbrunner H, Harer J. Persistent homology–A survey. Contemp. Math. 2008;453:257–282. [Google Scholar]
- 27.Edelsbrunner H, Letscher D, Zomorodian A. Topological persistence and simplification. Discrete Comput. Geomet. 2002;28:511–533. [Google Scholar]
- 28.Efron B. The Jackknife, The Bootstrap and other Resampling Plans. Vol. 38 Philadelphia, PA: SIAM; 1982. [Google Scholar]
- 29.Fornito A, Zalesky A, Bullmore ET. Network scaling effects in graph analytic studies of human resting-state fMRI data. Front. Syst. Neurosci. 2010;4:1–16. doi: 10.3389/fnsys.2010.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical LASSO. Biostatistics. 2008;9:432. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friston KJ, et al. Statistical parametric maps in functional imaging: A general linear approach. Human Brain Map. 1995;2:189–210. [Google Scholar]
- 32.Ghrist R. Barcodes: The persistent topology of data. Bull. Am.Math. Soc. 2008;45:61–75. [Google Scholar]
- 33.Gibbons JD, Chakraborti S. Nonparametric Statistical Inference. Boca Raton, FL: Chapman Hall/CRC Press; 2011. [Google Scholar]
- 34.Gong G, et al. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cerebral Cortex. 2009;19:524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson JL, et al. Early neglect is associated with alterations in white matter integrity and cognitive functioning. Child Develop. 2013;84:1566–1578. doi: 10.1111/cdev.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanson JL, et al. Structural variations in prefrontal cortex mediate the relationship between early childhood stress and spatial working memory. J. Neurosci. 2012;32:7917–7925. doi: 10.1523/JNEUROSCI.0307-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Y, Chen Z, Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J. Neurosci. 2008;28:4756. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cerebral Cortex. 2007;17:2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- 39.Horak D, Maletić S, Rajković M. Persistent homology of complex networks. J. Stat.Mech., Theory Experiment. 2009;2009:03034. [Google Scholar]
- 40.Huang S, et al. Learning brain connectivity of Alzheimer's disease from neuroimaging data. Adv. Neural Inf. Process. Syst. 2009:808–816. [Google Scholar]
- 41.Huang S, et al. Learning brain connectivity of Alzheimer's disease by sparse inverse covariance estimation. NeuroImage. 2010;50:935–949. doi: 10.1016/j.neuroimage.2009.12.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackowski AP, de Araújo CM, de Lacerda ALT, de Jesus M, Kaufman J. Neurostructural imaging findings in children with post-traumatic stress disorder: Brief review. Psychiatry Clin. Neurosci. 2009;63:1–8. doi: 10.1111/j.1440-1819.2008.01906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jezzard P, Clare S. Sources of distortion in functional MRI data. Human Brain Mapp. 1999;8:80–85. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<80::AID-HBM2>3.0.CO;2-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joshi SC, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004;23:151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 45.Kim WH, et al. Wavelet based multi-scale shape features on arbitrary surfaces for cortical thickness discrimination. Adv. Neural Inf. Process. Syst. 2012:1250–1258. [PMC free article] [PubMed] [Google Scholar]
- 46.Lee H, Chung MK, Kang H, Kim B-N, Lee DS. MICCAI. Vol. 6892. LNCS; 2011. Computing the shape of brain networks using graph filtration and Gromov-Haus-dorff metric; pp. 302–309. [DOI] [PubMed] [Google Scholar]
- 47.Lee H, Kang H, Chung MK, Kim B-N, Lee DS. Persistent brain network homology from the perspective of dendrogram. IEEE Trans. Med. Imag. 2012 Dec;31(12):2267–2277. doi: 10.1109/TMI.2012.2219590. [DOI] [PubMed] [Google Scholar]
- 48.Lee H, Lee DS, Kang H, Kim B-N, Chung MK. Sparse brain network recovery under compressed sensing. IEEE Trans. Med. Imag. 2011 May;30(5):1154–1165. doi: 10.1109/TMI.2011.2140380. [DOI] [PubMed] [Google Scholar]
- 49.Lenroot RK, Giedd JN. Brain development in children and adolescents: Insights from anatomical magnetic resonance imaging. Neurosci. Biobehav. Rev. 2006;30:718–729. doi: 10.1016/j.neubiorev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Lerch JP, et al. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. NeuroImage. 2006;31:993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, et al. Brain anatomical network and intelligence. PLoS Computat. Biol. 2009;5(5):e1000395. doi: 10.1371/journal.pcbi.1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loman MM, et al. The effect of early deprivation on executive attention in middle childhood. J. Child Psychiatry Psychol. 2010:224–236. doi: 10.1111/j.1469-7610.2012.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lorensen WE, Cline HE. Marching cubes: A high resolution 3D surface construction algorithm. Proc. 14th Annu. Conf. Comput. Graph. Interactive Tech. 1987:163–169. [Google Scholar]
- 54.Machado AMC, Gee JC. Atlas warping for brain morphometry. SPIE Med. Imag., Image Process. 1998:642–651. [Google Scholar]
- 55.Mazumder R, Hastie T. Exact covariance thresholding into connected components for large-scale graphical LASSO. J. Mach. Learn. Res. 2012;13:781–794. [PMC free article] [PubMed] [Google Scholar]
- 56.Mclntosh AR, Gonzalez-Lima F. Structural equation modeling and its application to network analysis in functional brain imaging. Human Brain Mapp. 1994;2:2–22. [Google Scholar]
- 57.Newman MEJ, Watts DJ. Scaling and percolation in the small-world network model. Phys. Rev. E. 1999;60:7332–7342. doi: 10.1103/physreve.60.7332. [DOI] [PubMed] [Google Scholar]
- 58.Peng J, Wang P, Zhou N, Zhu J. Partial correlation estimation by joint sparse regression models. J. Am. Stat. Assoc. 2009;104:735–746. doi: 10.1198/jasa.2009.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pollak SD. Mechanisms linking early experience and the emergence of emotions: Illustrations from the study of maltreated children. Curr. Direct. Psychol. Sci. 2008;17:17, 370–375. doi: 10.1111/j.1467-8721.2008.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pollak SD, et al. Neurodevelopmental effects of early deprivation in post-institutionalized children. Child Develop. 2010;81:224–236. doi: 10.1111/j.1467-8624.2009.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pothen A, Fan CJ. Computing the block triangular form of a sparse matrix. ACM Trans. Math. Software. 1990;16:324. [Google Scholar]
- 62.Rao A, Aljabar P, Rueckert D. Hierarchical statistical shape analysis and prediction of sub-cortical brain structures. Med. Image Anal. 2008;12:55–68. doi: 10.1016/j.media.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 63.Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 64.Sacan A, Ozturk O, Ferhatosmanoglu H, Wang Y. LFM-PRO: A tool for detecting significant local structural sites in proteins. Bioinformatics. 2007;6:709–716. doi: 10.1093/bioinformatics/btl685. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez MM, Pollak SD. Socio-emotional development following early abuse and neglect: Challenges and insights from translational research. Handbook Develop. Social Neurosci. 2009;17:497–520. [Google Scholar]
- 66.Schäfer J, Strimmer K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genetics Molecular Biol. 2005;4:32. doi: 10.2202/1544-6115.1175. [DOI] [PubMed] [Google Scholar]
- 67.Singh G, et al. Topological analysis of population activity in visual cortex. J. Vis. 2008;8:1–18. doi: 10.1167/8.8.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song C, Havlin S, Makse HA. Self-similarity of complex networks. Nature. 2005;433:392–395. doi: 10.1038/nature03248. [DOI] [PubMed] [Google Scholar]
- 69.Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer's disease. PLoS Computat. Biol. 2008;4(6):e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson PM, et al. Growth patterns in the developing human brain detected using continuum-mechanical tensor mapping. Nature. 2000;404:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- 71.Thompson PM, Toga AW. Anatomically driven strategies for high-dimensional brain image warping and pathology detection. Brain Warp. 1999:311–336. [Google Scholar]
- 72.Tibshirani R. Regression shrinkage and selection via the LASSO. J. R. Stat. Soc.. Ser. B (Methodol.) 1996;58:267–288. [Google Scholar]
- 73.Valdés-Sosa PA, et al. Estimating brain functional connectivity with sparse multivariate autoregression. Phil. Trans. R. Soc. B, Biol. Sci. 2005;360:969–981. doi: 10.1098/rstb.2005.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Witten DM, Friedman JH, Simon N. New insights and faster computations for the graphical LASSO. J. Computat. Graph. Stat. 2011;20:892–900. [Google Scholar]
- 75.Worsley KJ, Charil A, Lerch J, Evans AC. Connectivity of anatomical and functional MRI data. Proc. IEEE Int. Joint Conf. Neural Netw. 2005;3:1534–1541. [Google Scholar]
- 76.Worsley KJ, Chen JI, Lerch J, Evans AC. Comparing functional connectivity via thresholding correlations and singular value decomposition. Phil. Trans. R. Soc. B, Biol. Sci. 2005;360:913. doi: 10.1098/rstb.2005.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zalesky A, et al. Whole-brain anatomical networks: Does the choice of nodes matter? NeuroImage. 2010;50:970–983. doi: 10.1016/j.neuroimage.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 78.Zhang H, van Kaick O, Dyer R. Spectral methods for mesh processing and analysis. Eurographics. 2007:1–22. [Google Scholar]