Abstract
Extensins (EXTs) are hydroxyproline-rich glycoproteins (HRGPs) that are structural components of the plant primary cell wall. They are basic proteins and are highly glycosylated with carbohydrate accounting for >50% of their dry weight. Carbohydrate occurs as monogalactosyl serine and arabinosyl hydroxyproline, with arabinosides ranging in size from ~1 to 4 or 5 residues. Proposed functions of EXT arabinosylation include stabilizing the polyproline II helix structure and facilitating EXT cross-linking. Here, the involvement of arabinosylation in EXT cross-linking was investigated by assaying the initial cross-linking rate and degree of cross-linking of partially or fully de-arabinosylated EXTs using an in vitro cross-linking assay followed by gel permeation chromatography. Our results indicate that EXT arabinosylation is required for EXT cross-linking in vitro and the fourth arabinosyl residue in the tetraarabinoside chain, which is uniquely α-linked, may determine the initial cross-linking rate. Our results also confirm the conserved structure of the oligoarabinosides across species, indicating an evolutionary significance for EXT arabinosylation.
Keywords: HRGP, extensin, arabinosylation, in vitro cross-linking, RSH, tomato P1, peroxidase
Introduction
Plant cells are surrounded by a thin primary cell wall comprised of interpenetrating networks of polysaccharide (cellulose, hemicellulose, and pectin) and structural glycoproteins (~10% dicot cell wall dry weight).1 Structural proteins are often rich in hydroxyproline (Hyp) and are hence named hydroxyproline-rich glycoprotein (HRGP).2 HRGPs are categorized into three major types: the extensins (EXTs), the arabinogalactan proteins (AGPs), and the proline-rich proteins (PRPs) based on their repetitive peptide motifs and the type and extent of glycosylation, which occurs mainly on the Hyp residues. Despite being a relatively minor component in the primary cell wall compared with the matrix polysaccharides, HRGPs play an important role in wall architecture,3,4 plant development,5–9 embryogenesis,10,11 stress responses,12–14 and defense against pathogen attacks.15–18
EXTs are a major class of HRGP. Hyp accounts for >30 mol% of an EXT’s amino acids19,20 and occurs in short peptide repeats that alternate with Hyp-poor repeats containing hydrophobic and basic residues. Hydrophobic motifs often contain Tyr residues that participate in intra and intermolecular cross-linking. Ser-(Hyp)4 is the “signature” repeat motif in EXTs with Ser being monogalactosylated21 and all the Hyp residues O-arabinosylated (Hyp–O-Ara1–4/5).22–24 EXTs are highly basic25 due to abundant His and Lys content and thus carry a positive net charge at cell wall physiological pH (~5.0).26 This positive charge enables ionic interactions between EXTs and acidic wall polysaccharides such as pectin,27,28 although covalent cross-links between EXTs and pectins occur.29–31 Cross-linking motifs contain Tyr residues, commonly in the sequence of Tyr–X–Tyr (X usually = Lys, Tyr, Leu, or Val)32 and possibly Val–Tyr–Lys.33 The Tyr–X–Tyr motif gives rise to intramolecular isodityrosine (Idt),34,35 which can itself undergo further cross-linking to produce intermolecular di-isodityrosine (Di-Idt)36,37 or pulcherosine38 cross-links. The cross-linking of EXTs (with other EXTs or possibly other wall structural proteins) is likely catalyzed by wall-associated peroxidases.33,39 Cross-linking leads to the formation of a covalently linked protein network that is somehow independent of wall polysaccharide networks, since this protein network remains intact after treatment of the wall with anhydrous hydrogen fluoride (HF),40–42 which cleaves glycosidic bonds but not peptide bonds.40
Previously, Schnabelrauch et al33 reported that deglycosylated EXT monomers were not cross-linked in vitro by EXT peroxidase. This observation suggested a role for arabinosylation in EXT self-assembly and cross-linking, one of the most fundamental functions of EXTs. In this study, we investigated the involvement of arabinosylation in EXT cross-linking and provide evidence suggesting that arabinosylation controls the initial rate and the extent of EXT cross-linking in vitro. We also found that Arabidopsis oligoarabinosides share a common structure with arabinosides from other species, indicating that these glycosides play a significant role in the evolutionary progression of EXT molecular function.
Material and Methods
Chemicals and reagents
All general chemical reagents were purchased from Sigma-Aldrich, and all general lab equipment were from VWR unless otherwise indicated.
Isolation of EXT precursors from suspension culture cells
Monomeric EXTs from tomato (TOMP1, ~400 aa protein, MW ~110 kDa) or Arabidopsis (RSH, 404 aa protein, MW ~ 100 kDa) were isolated from suspension culture cells using the intact cell elution method as described earlier by Smith et al.19 Briefly, cells at 25% packed cell volume (Vcell/Vculture) were harvested by filtration on a course-sintered funnel and eluted with 80 mM AlCl3 for 15 minutes. The AlCl3 eluate was collected by filtration, lyophilized, and re-dissolved in 90 mL of water, and 10 mL cold 100% (w/v) trichloroethanoic acid (TCA) was added to the eluate. The mixture was incubated overnight at 4°C and then centrifuged at 30,000 × g. The supernatant was collected, dialyzed using 3500-Da cut-off dialysis tubing (Spectrum Laboratories) against distilled water for 48 hours, and then lyophilized. The resulting TCA-free eluate powder was dissolved at 5 mg/mL in 30 mM sodium phosphate buffer (pH 7.6), and EXTs were purified using a SP Sepharose Fast Flow (GE Healthcare) strong cation exchange chromatography followed by reverse-phase HPLC chromatography using a PRP-1 column (Hamilton) as described by Terneus.20
Amino acid composition of purified EXTs
The amino acid compositions of protein samples were determined by phenylisothiocyanate (PITC) derivatization of acid hydrolyzed proteins as described by Janssen et al.43
Partial dearabinosylation by α-l-Arabinofuranosidase
RSH and TOMP1 were dissolved in 100 mM NaOAc buffer (pH 5.5) at a concentration of 5 mg/mL. α-l-arabinofuranosidase (lot 10401, 40 U/mg, Megazyme) was added at a 1:100 weight ratio of enzyme: EXT. The solutions were incubated at 50°C for 20 hours with 2 mM NaN3 to prevent microbial growth. Dearabinosylated EXTs were isolated by reverse-phase HPLC fractionation as described above. The resulting dearabinosylated EXTs were named RSHAra and P1Ara, respectively.
Partial dearabinosylation by treatment with pH 2.0 HCl21,44
RSH and TOMP1 were dissolved (5 mg/mL) in 0.01 M HCl (pH 2.0) and incubated at 95°C for 1 hour. The reactions were then cooled on ice and neutralized with 0.01 M NaOH. Partially dearabinosylated RSH and TOMP1 were isolated by reverse-phase HPLC fractionation as described earlier, and the products were designated as RSHpH2 and P1pH2, respectively.
Complete dearabinosylation by treatment with pH 1.0 HCl
The arabinosyl residues of RSH and TOMP1 were removed using pH 1.0 HCl. EXT samples were dissolved (5 mg/mL) in 0.1 M HCl (pH 1.0) and incubated at 100°C for one hour as described by Schnabelrauch et al.33 The reactions were cooled on ice and neutralized with 0.1 M NaOH. Dearabinosylated RSH and TOMP1 were isolated by reverse-phase HPLC fractionation as described earlier, and the products were designated as RSHpH1 and P1pH1, respectively.
Complete deglycosylation using anhydrous HF
One hour HF deglycosylations were performed as described by Mort and Lamport.40 After quenching, the HF was removed by blowing down the aqueous mixtures under a steam of N2 gas twice and then lyophilized. The deglycosylated EXTs were isolated by reverse-phase HPLC fractionation followed by lyophilization. The products were designated as RSHHF and P1HF, respectively.
Colorimetric arabinose/galactose assay
The remaining arabinose and galactose contents on EXTs after deglycosylation were measured colorimetrically as described by Fry.45
Hyp-arabinoside profiling
Hyp-arabinoside profiling was carried out using methods described by Lamport and Miller.23 Native and partially dearabinosylated (α-l-arabinofuranosidase or pH 2.0 HCl treated, 2 mg) EXTs were dissolved (5 mg/mL) in saturated (~0.2 M) Ba(OH)2 in sealed tubes and hydrolyzed at 105°C for 18 hours. The reactions were then cooled on ice, and the Ba(OH)2 was neutralized with ice-cold 1 M H2SO4 to a pH of 7.0 (monitored by pH paper). BaSO4 generated by neutralization was removed from the reactions by centrifugation, and the neutralized reaction mixtures were then loaded onto a strong cation exchange Chromobeads C-2 column (Technicon) equilibrated with water. The column was then washed with a linear gradient of HCl generated by mixing 100 mL of water with 100 mL of 0.5 M HCl at 0.5 mL/minute. Fractions eluted from the C-2 column were collected at 4 minutes/tube, and 0.5 mL of each fraction was assayed for Hyp using the colorimetric Hyp assay as described previously.23
Purification of EXT peroxidase from tomato cell suspension culture
A pI 4.6 peroxidase was isolated from tomato suspension cell culture using a combination of diethylamino-ethyl anion-exchange chromatography and isoelectric focusing as described by Dong et al.46 The resulting protein fraction was analyzed by SDS–PAGE. Peroxidase activity was determined by reaction with 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) as described by Everdeen et al.47
In vitro cross-linking of deglycosylated EXTs
Partially or fully dearabinosylated and fully deglycosylated RSH and TOMP1 samples were cross-linked in vitro with a crude preparation of the pI 4.6-extensin peroxidase from tomato suspension culture medium. All protein samples were first made into 10 mg/mL stock solutions. Stock solutions (60 μL) were added to 40 μL of 0.1 M citrate buffer (pH 6.0) to form working solutions. The cross-linking reactions contained 10 μL of extensin working solution (60 μg of EXT), 5 μL diluted crude peroxidase containing 0.05 ng of peroxidase based on the ABTS assay, and 5 μL 0.24 mM H2O2 (the final concentration of H2O2 in reaction system was 60 μM) in a total reaction volume of 20 μL. Native RSH and TOMP1 were cross-linked in the same way as controls. Timing of the reactions was started upon the addition of H2O2 and terminated by the addition of the stop reagents: 10 μL of 50 mM β-mercaptoethanol. The initial cross-linking rates were determined using the first-order rate equation ν0 = kC0 presented in terms of microgram EXT cross-linked in 1 mL reaction solution per minute as described by Everdeen et al,47 where ν0 is the initial rate, k is the first-order reaction rate constant, and C0 is the initial EXT concentration at zero minute of cross-linking. The rate constant k was determined by equation k = ln(A0/A)/t, where A0 is the peak area of monomeric EXTs cross-linked for zero minute, and A is the peak area of monomeric EXTs after certain reaction time. The monomeric EXT peak areas before and after the cross-linking reactions were assayed by gel filtration chromatography using a Superose 6 10/300 GL column (GE Healthcare), monitored at 220 nm absorbance. Three parallel reactions were run for each sample to determine the initial cross-linking rate (cross-linking for five minutes) and the data averaged. For zero-minute cross-linking reactions, the stop reagents were added before the addition of H2O2. Each EXT sample was also cross-linked for 30 minutes for complete reaction. Completely dearabinosylated (treated with pH 1.0 HCl or HF) samples had very slow cross-linking rates, and thus only the 30-minute cross-linking experiment was performed. Additionally, these reactions were succinylated before injecting onto the column by adding 1 mg succinic anhydride into the reaction mixture and incubated for 60 minutes. Succinylation reverses the positive charges on the proteins, which interact with the Superose column matrix and retard protein migration.
Hyp-arabinosyl linkage analysis of RSH
Hyp-arabinosyl linkage analysis was performed on RSH at the Center for Plant and Microbial Complex Carbohydrates at the Complex Carbohydrate Research Center (CCRC), University of Georgia. RSH was permethylated, depolymerized, reduced, and acetylated. The resulting partially methylated alditol acetates (PMAAs) were analyzed by gas chromatography-mass spectrometry (GC-MS) as described by York et al.48
Isolation of Hyp-oligoarabinosides from RSH
RSH was hydrolyzed with 0.2 M Ba(OH)2 at 105°C for 18 hours, and the reaction was neutralized with H2SO4 before the Hyp-arabinosides were isolated by cation exchange Chromobeads C-2 column chromatography as described earlier. However, gradient elution was performed by mixing 100 mL of water (eluent A) with 110 mL of 0.7 M trifluoroacetic acid (TFA, eluent B) for >300 minutes. The C-2 column was washed after TFA gradient with 0.7 M TFA to elute Hyp-diarabinoside (Hyp-Ara2) for 30 minutes, and TFA was replaced with 0.5 M HCl to elute Hyp-monoarabinoside and non-glycosylated Hyp. The column eluates were collected in four-minute fractions, and 0.1 mL of each fraction was assayed for Hyp using the colorimetric Hyp assay. The fractions containing Hyp-tetraarabinosides (Hyp-Ara4) and Hyp-triarabinosides (Hyp-Ara3) were lyophilized for three times with water to remove the excess TFA before sending for NMR structural analysis.
Structure elucidation of Hyp-oligoarabinosides by NMR spectroscopy
Hyp-Ara4 (1.7 mg) and Hyp-Ara3 (2 mg) were each dissolved in 180 mL of D2O. Each solution was transferred into a 3 mm NMR tube. NMR data were collected at 25°C on a VNMRS 800 instrument equipped with a 3 mm cold probe at the CCRC Facilities. The 1D1H NMR and 2D1H homonuclear COSY, TOCSY (DIPSI2 mixing time 60 millisecond), and NOESY (NOE mixing time 100 millisecond), 2D heteronuclear1H–13C HSQC, and HMBC experiments were performed as described earlier.49
Results
Monomeric EXT isolation
SP Sepharose fractionation of tomato eluates yielded three peaks (Supplementary Fig. 1A). The void peak and peak 2 (Supplementary Fig. 1A) were relatively poor in Hyp judging by colorimetric Hyp assay (Hypvoid = 2.4 μg Hyp/mg protein and Hyppeak 2 = 1.6 μg Hyp/mg protein, respectively). Peak 1 contained 38.2 μg Hyp/mg protein and was further fractionated by reverse-phase HPLC, yielding a single peak (Supplementary Fig. 1B). The amino acid composition identified the protein in this peak as EXT precursor 1 (TOMP1) as described earlier by Smith et al19 (Supplementary Table 1).
Similarly, SP Sepharose fractionation of Arabidopsis eluates (Supplementary Fig. 1C) resulted in one major peak, peak 1, containing 33.7 μg Hyp/mg protein, in addition to the void peak (8.3 μg Hyp/mg protein). Peak 1 was further fractionated by reverse-phase HPLC to produce one major peak (Supplementary Fig. 1D). Through the analysis of amino acid composition (Supplementary Table 1), the protein in this peak was identified as RSH, as described earlier.20 All minor Sepharose peaks (the small peak group and peak 2, Supplementary Fig. 1C) in the fractionated eluate of Arabidopsis suspension culture were relatively rich in Hyp (Hyps-mall peak group = 23.7 μg Hyp/mg protein and Hyppeak 2 = 11.7 μg Hyp/mg protein, respectively). The protein components in those Sepharose peaks formed precipitate with β-Yariv reagent, indicating the presence of AGP-type structural proteins that may potentially contribute to the high Hyp content; thus, they were not fractionated or analyzed further.
Characterization of deglycosylated EXTs
The removal of Ara residues by each deglycosylation was first assayed using colorimetric arabinose assays. Treatment with pH 1.0 HCl and HF deglycosylation removed almost all arabinosyl residues on the EXTs (Table 1), which was judged by comparison with native RSH and TOMP1. Arabinofuranosidases removed about 30% of Ara residues from RSH and TOMP1, while the HCl treatment at pH 2.0 removed about half the Ara residues from both EXTs. These data are also supported by the decrease in the weight percentage of Hyp-oligoarabinosyl residues in Hyp-Arabinoside (Hyp-Ara) profiles (Table 2).
Table 1.
Remaining arabinose and galactose on deglycosylated RSH and TOMP1.
| SAMPLE (µg) | ARABINOSE (µg) | ARABINOSE CONTENT (W/W) | |
|---|---|---|---|
| RSH | 30 | 13 | 43.3% |
| RSHAra | 30 | 8.7 | 29.0% |
| RSHpH2 | 30 | 6.6 | 21.7% |
| RSHpH1 | 30 | 0.4 | 1.3% |
| RSHHF | 30 | 0.2 | 0.6% |
| TOMP1 | 30 | 13.8 | 46.0% |
| P1Ara | 30 | 9.9 | 33.0% |
| P1pH2 | 30 | 7.2 | 24.0% |
| P1pH1 | 30 | 0.4 | 1.3% |
| P1HF | 30 | 0.2 | 0.6% |
| SAMPLE (µg) | GALACTOSE (µg) | GALACTOSE CONTENT (W/W) | |
| RSHpH1 | 100 | 5.1 | 5.1% |
| RSHHF | 100 | 0.2 | 0.2% |
| P1pH1 | 100 | 3.5 | 3.5% |
| P1HF | 100 | 0.3 | 0.3% |
Table 2.
Hyp-Ara profiles of partially deglycosylated EXTs.
| Hyp-Aran | RSH | RSHAra | RSHpH2 | TOMP1 | P1Ara | P1pH2 |
|---|---|---|---|---|---|---|
| n = 4 (%) | 21.4 | 0.0 | 5.8 | 25.4 | 0.0 | 9.7 |
| n = 3 (%) | 20.7 | 16.7 | 4.9 | 10.2 | 25.2 | 5.7 |
| n = 2 (%) | 13.2 | 13.3 | 8.0 | 10.7 | 15.5 | 8.2 |
| n = 1 (%) | 17.7 | 23.2 | 23.0 | 28.1 | 25.7 | 30.2 |
| n = 0 (%) | 27.0 | 46.8 | 58.3 | 25.6 | 33.6 | 46.2 |
Notes: Hyp-Aran: the number of arabinoses attached to Hyp, n = 0 means non-glycosylated Hyp. Ratio of different arabino-oligosaccharides was expressed as percentage of Hyp bearing those oligosaccharides to total Hyp.
EXT galactosyl residues are much less susceptible to acid hydrolysis than arabinosyl residues. A previous study21 has showed that Ser-Gal remained intact after hydrolysis for one hour at 100°C by pH 1.0 HCl. The partial dearabinosylation approaches used here were milder and thus unlikely to remove EXT Gal residues. Thus only the residual Gal content of the pH 1.0 HCl and HF-treated EXTs were determined (Table 1). HF removed almost all the Gal on RSH and TOMP1, while galactose was still present in pH 1.0 HCl-treated EXTs, as expected.
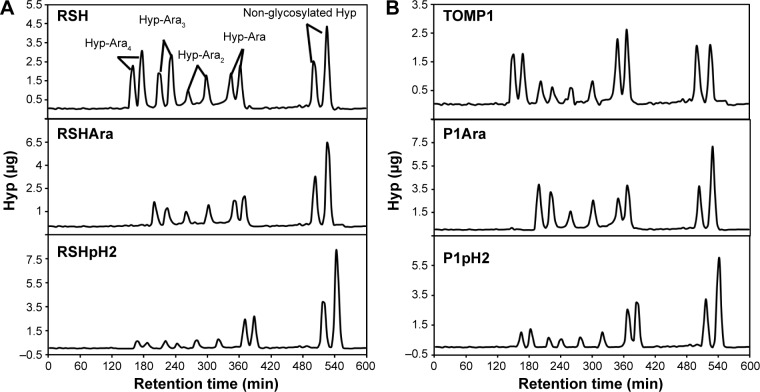
Hyp-Ara profiles of EXTs treated with arabinofuranosidase and pH 2.0 HCl (native RSH and TOMP1 as controls, Fig. 1 and Table 2) were determined, and the results revealed that no Hyp-tetraarabinosides were observed after treatment with arabinofuranosidases, while some Hyp-Ara4 remained after pH 2.0 treatments (Fig. 1).
Figure 1.
Hyp-Ara profiling of partially deglycosylated EXTs. (A) Top to bottom: RSH, RSHAra, and RSHpH2; (B) top to bottom: TOMP1, P1Ara, and P1pH2. Each Hyp-arabinosides were present in two peaks due to two configurations of Hyp (cis or trans) generated during base hydrolysis.50 The Hyp-Ara4 peaks disappeared after α-arabinofuranosidase treatment, while some Hyp-Ara4 glycans were still present after pH 2.0 HCl deglycosylation. See Table 2 for Hyp-oligoarabinan content quantification in each sample.
In vitro cross-linking of deglycosylated EXTs
The initial cross-linking reaction rates of partially dearabinosylated RSH and TOMP1 were determined by gel filtration chromatography using the first-order rate equation employed earlier by Everdeen et al47: ν0 = kX0 with native RSH and TOMP1 as controls. The results are summarized in Table 3.
Table 3.
Initial cross-linking (XL) reaction rates of deglycosylated EXTs.
| EXTS | INITIAL CROSS-LINKING RATE (µg/mL⋅min) | PEAK AREA LOSS (5~30 min XL, %) |
|---|---|---|
| Native RSH | 268.8 ± 9.4 | 80.3 |
| RSHAra | 85.0 ± 3.3 | 40.1 |
| RSHpH2 | 223.2 ± 12.4 | 74.2 |
| RSHpH1 | 0 | 0 |
| RSHHF | 0 | 0 |
| Native TOMP1 | 213.3 ± 6.3 | 73.9 |
| P1Ara | 28.5 ± 2.5 | 24.9 |
| P1pH2 | 95.3 ± 1.7 | 54.2 |
| P1pH1 | 0 | 0 |
| P1HF | 0 | 0 |
Notes: Three technical replicas averaged to obtain the rate of native extensins and extensins deglycosylated with arabinofuranosidases and pH 2.0 HCl. Area loss is presented as (monomer area five minutes–monomer area 30 minutes)/monomer area five minutes. Area losses were calculated once from profiles in Figure 2 (RSH) and Figure 3 (TOMP1).
Samples that essentially lacked Ara did not cross-link at all (pH1 or HF treated), even after 30 minutes of incubation with the pI 4.6 extensin peroxidase (Supplementary Fig. 2). Thus, the initial cross-linking rates of these samples were documented as zero (Table 3). Initial cross-linking rates of the partially dearabinosylated samples were lower than that of the native EXTs. For partially dearabinosylated RSH samples, initial rate decreases of 17% and 68% were observed for RSHpH2 and RSHAra, respectively, compared with fully arabinosylated RSH. TOMP1 showed a similar trend, as initial cross-linking rates decreased after partial dearabinosylation by 50% and 86% for P1pH2 and P1Ara samples, respectively (Table 3). Between partially dearabinosylated EXTs, those with no Hyp-tetraarabinosides (arabinofuranosidase treated) showed much lower initial rates than the samples subjected to pH 2.0 HCl treatment, which retained considerably more Hyp-tetraarabinosides (Table 2, Fig. 1).
The loss of EXT monomer peak area from five minutes to 30 minutes of cross-linking (shown as the loss of the percentage of the five minutes monomer peak area) also indicates the extent of EXT cross-linking after deglycosylation. With a higher initial cross-linking rate, more monomers were converted to higher molecular weight oligomers (Figs. 2 and 3) after 30 minutes. The results are also summarized in Table 3.
Figure 2.
Superose 6 size exclusion chromatography profiles of the cross-linking of deglycosylated RSH. Left to right: native RSH (control), RSH treated with arabinofuranosidase (RSHAra) and pH 2.0 HCl (RSHpH2), respectively. A conversion of monomers to oligomers with the increase of cross-linking time is observed. The two peaks after 100 minutes were contributed by buffer and 2-mercaptoethanol.
Figure 3.
Superose 6 size exclusion chromatography profiles of the cross-linking of deglycosylated TOMP1. Left to right: native TOMP1 (control), TOMP1 treated with arabinofuranosidase (P1Ara) and pH 2.0 HCl (P1pH2), respectively. A conversion of monomers to oligomers with the increase of cross-linking time is observed. The two peaks after 100 minutes were contributed by buffer and 2-mercaptoethanol.
RSH Hyp-oligoarabinosides structure determination
In order to determine the structures of RSH Hyp-Ara4 and Hyp-Ara3, we first analyzed the glycosyl linkages of the RSH glycans (Table 4). RSH showed a simple glycan profile that contained mainly terminally linked Gal residues (presumably attached to the serine residues) and twice as much 2-linked as 3-linked arabinofuranosyl residues (2-Araf and 3-Araf). There was also a trace of 2, 4-linked Ara (0.3%) and a small amount of ribose; undoubtedly, a contamination as ribose is not a component of extracellular molecules. Further, an earlier neutral sugar composition of RSH indicated it contained mainly Ara and Gal (Table 4), but clearly no ribose.
Table 4.
Monosaccharide composition and glycosyl-linkage analysis of RSH.
| MONOSUGAR | COMPOSITION (mole%) | GLYCOSYL LINKAGE | % PEAK AREA |
|---|---|---|---|
| Arabinose (Ara) | 79 | Terminally linked arabinofuranosyl residue (t-Araf) | 8 |
| 3-linked arabinofuranosyl residue (3-Araf) | 21 | ||
| 2-linked arabinofuranosyl residue (2-Araf) | 54 | ||
| Galactose (Gal) | 15 | Terminally linked galactopyranosyl residue (t-Gal) | 15 |
| Rhamnose (Rha) | 2 | ||
| Xylose (Xyl) | 2 | ||
| Glucose (Glc) | 2 | 2-linked ribofuranosyl residue (2-Ribf) | 2 |
Notes: Monosugar analysis results were obtained from Terneus.20 Trace amount (0.3%) of 2,4-linked Ara was also observed.
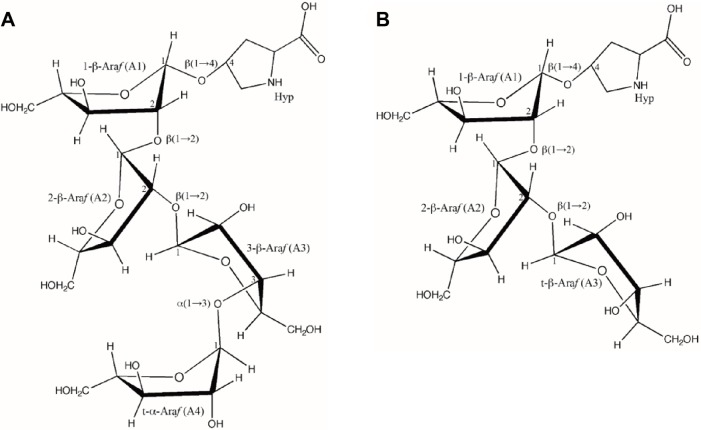
TFA elution of RSH Hyp-oligoarabinosides yielded a profile similar to that of HCl elution (Fig. 1A, first panel). Peaks containing Hyp-Ara4 and Hyp-Ara3 were collected (cis and trans peaks pooled for NMR analysis), and through NMR spectra (COSY, TCOSY, HSQC, and HMBC) the glycosyl residues, ring systems, and ordered linkages of the components of RSH Hyp-Ara4 (Fig. 4) and Hyp-Ara3 (Fig. 5) were identified. The results corroborate the chemical shift obtained earlier by Akiyama et al50,51 for Hyparabinosides isolated from tobacco suspension culture cell walls (Table 5) and identified the structures of RSH Hyp-Ara4 and Hyp-Ara3 as (Fig. 6).
Hyp-Ara4: α-l-Araf-(1→3)-β-l-Araf-(1→2)-β-l-Araf-(1→2)-β-l-Araf-(1→4)-Hyp.
Hyp-Ara3: β-l-Araf-(1→2)-β-l-Araf-(1→2)-β-l-Araf-(1→4)-Hyp.
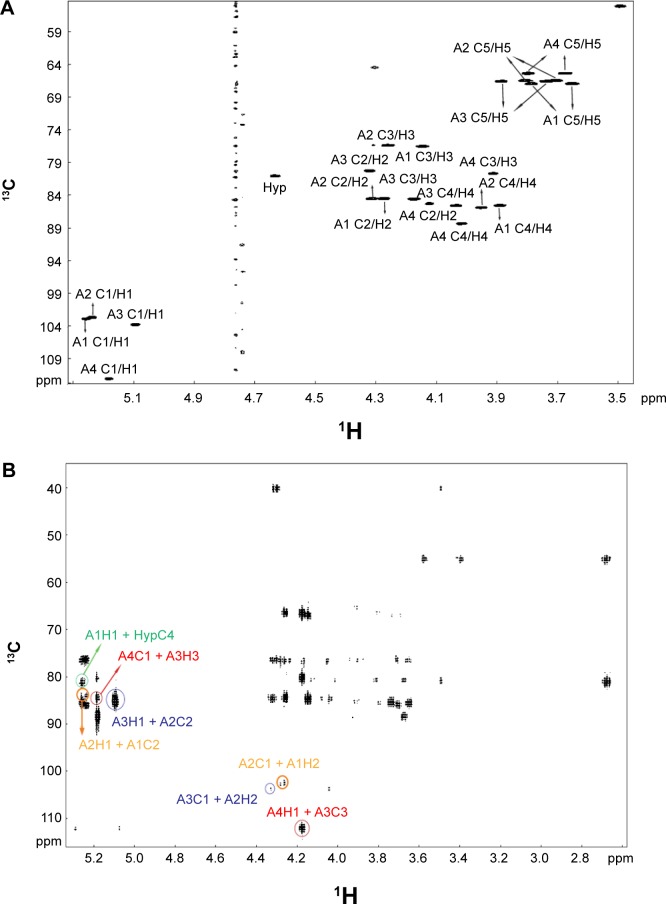
Figure 4.
Structure elucidation of RSH Hyp-Ara4. (A) HSQC spectrum: cross-peaks identified the chemical shifts of each carbon atom and its corresponding hydrogen atom(s) in each arabinose ring system. A4 is the fourth Ara at the non-reducing end of the Hyp-Ara4 chain, while A1 occupies the reducing end and is attached to Hyp (first Ara in the chain). A2 and A3 are the second and third Ara of the chain. The A1 C1/H1 label indicates the cross-peak arising from the chemical shifts of the anomeric carbon (C1) and its corresponding hydrogen (H1) on the A1 residue. The cross-peaks for the other carbon atoms and their corresponding hydrogens of A1 and the cross-peaks for A2 to A4 are similarly labeled. Two cross-peaks are observed for the fifth carbon atoms on each ring system due to their possession of two corresponding hydrogen atoms. (B) HMBC spectrum: the cross-peaks arising from A4H1(5.2 ppm) + A3C3(84.6 ppm) and A4C1(112.1 ppm) + A3H3(4.2 ppm), highlighted by red circles, established the α-Araf-(1→3)-β-Araf linkage between A4 and A3; cross-peaks arising from A3H1(5.1 ppm) + A2C2(84.5 ppm) and A3C1(103.8 ppm) + A2H2(4.3 ppm), highlighted by blue circles, established the β-Araf-(1→2)-β-Araf linkage between A3 and A2; cross-peaks arising from A2H1(5.2 ppm) + A1C2(84.5 ppm) and A2C1(102.7 ppm) + A1H2(4.3 ppm), highlighted by orange circles, established the β-Araf-(1→2)-β-Araf linkage between A2 and A1, and cross-peak arising from A1H1(5.3 ppm) + HypC4 (81.1 ppm), highlighted by the green circle, established the β-Araf-(1→4)-Hyp linkage between A1 and Hyp. The chemical shifts of the cross-peaks are summarized in Table 5.
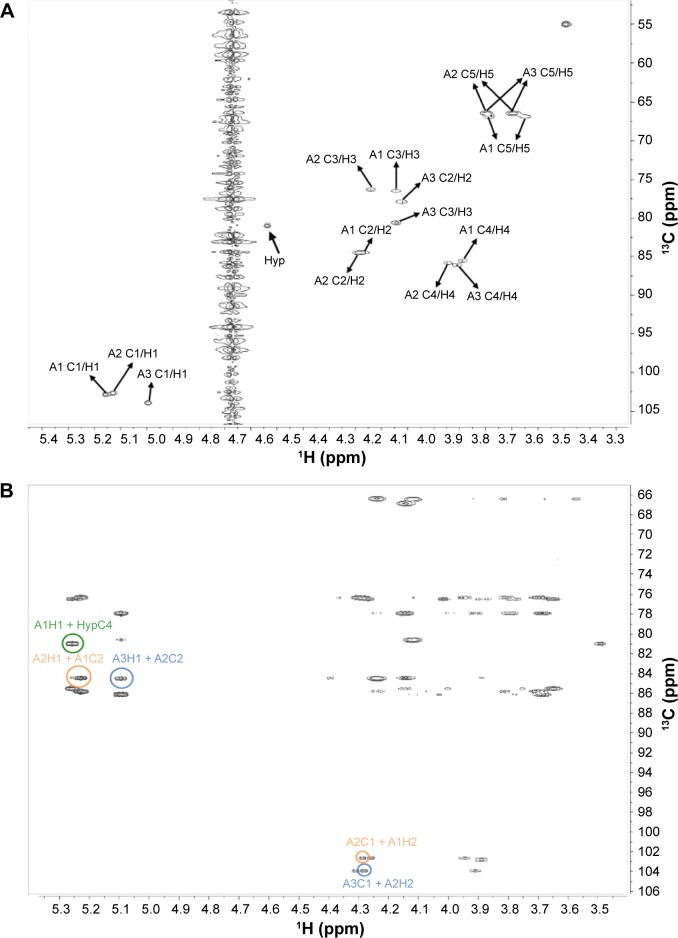
Figure 5.
Structure elucidation of RSH Hyp-Ara3. (A) HSQC spectrum: cross-peaks identified the chemical shifts of each carbon atom and its corresponding hydrogen atom(s) in each arabinose ring system. A3 is the third Ara at the nonreducing end of the Hyp-Ara3 chain, while A1 occupies the reducing end and is attached to Hyp (first Ara in the chain). A2 is the second Ara of the chain. The A1 C1/H1 label indicates the cross-peak arising from the chemical shifts of the anomeric carbon (C1) and its corresponding hydrogen (H1) on the A1 residue. The cross-peaks for the other carbon atoms and their corresponding hydrogens of A1 and the cross-peaks for A2 to A3 are similarly labeled. Two cross-peaks are observed for the fifth carbon atoms on each ring system due to their possession of two corresponding hydrogen atoms. There are overlapping signals between the cross-peaks from A1 C2/H2 and A2 C2/H2 and those from A2 C5/H5 and A3 C5/H5 due to their identical chemical shifts (Table 5). (B) HMBC spectrum: cross-peaks arising from A3H1(5.1 ppm) + A2C2(84.5 ppm) and A3C1(103.9 ppm) + A2H2(4.3 ppm), highlighted by blue circles, established the β-Araf-(1→2)-β-Araf linkage between A3 and A2; cross-peaks arising from A2H1(5.2 ppm) + A1C2(84.5 ppm) and A2C1(102.7 ppm) + A1H2(4.3 ppm), highlighted by orange circles, established the β-Araf-(1→2)-β-Araf linkage between A2 and A1, and cross-peak arising from A1H1(5.3 ppm) + HypC4(81.0 ppm), highlighted by the green circle, established the β-Araf-(1→4)-Hyp linkage between A1 and Hyp. The chemical shifts are summarized in Table 5.
Table 5.
Chemical shifts (ppm) of RSH arabinose C/H atoms in Hyp-Ara4 and Hyp-Ara3.
| RESIDUES | C1/H1 | C2/H2 | C3/H3 | C4/H4 | C5/H5 |
|---|---|---|---|---|---|
| Hyp-Ara4 | ppm | ||||
| α-Araf (A4) | 112.1/5.2 | 85.3/4.1 | 80.7/3.9 | 88.3/4.0 | 65.4/3.8, 3.7 |
| β-Araf (A3) | 103.8/5.1 | 80.2/4.3 | 84.6/4.2 | 85.6/4.0 | 66.6/3.9, 3.7 |
| β-Araf (A2) | 102.7/5.2 | 84.5/4.3 | 76.3/4.3 | 85.9/3.9 | 66.5/3.8, 3.7 |
| β-Araf (A1) | 102.9/5.3 | 84.5/4.3 | 76.5/4.1 | 85.6/3.9 | 66.9/3.8, 3.6 |
| Hyp | 175.4 | 61.1 | 38.7 | 81.1 | 54.2 |
| Hyp-Ara3 | ppm | ||||
| β-Araf (A3) | 103.9/5.1 | 77.9/4.1 | 80.6/4.1 | 86.1/3.9 | 66.4/3.8, 3.7 |
| β-Araf (A2) | 102.7/5.2 | 84.5/4.3 | 76.3/4.2 | 85.8/3.9 | 66.4/3.8, 3.7 |
| β-Araf (A1) | 102.8/5.3 | 84.5/4.3 | 76.5/4.1 | 85.5/3.9 | 66.9/3.8, 3.7 |
| Hyp | 175.4 | 61.1 | 38.7 | 81.0 | 54.2 |
Notes: A4 in Hyp-Ara4 and A3 in Hyp-Ara3 are the terminal arabinose residues at the non-reducing end. A1 in both Hyp-Ara4 and Hyp-Ara3 is the reducing end arabinose that attached to Hyp. Values corresponding to the cross-peaks in Figures 4 and 5. Anomeric C chemical shift obtained from Akiyama et al51 identified A4 to be α-Ara while the remaining β-Ara in Hyp-Ara4 and all the Ara in Hyp-Ara3 as β-Ara. Two H shift values for the fifth carbons due to their possession of two corresponding hydrogen atoms. The difference in chemical shifts between the fourth C of Hyp in Hyp-oligoarabinan and that of free Hyp (81 ppm vs 71.3 ppm, Akiyama et al51) identified a linkage on the fourth C of Hyp in Hyp-oligoarabinan.
Figure 6.
Characterized primary structure of RSH Hyp-Ara4 and Hyp-Ara3. Each arabinose residue is labeled corresponding to those in Table 5 with their anomeric configurations. The glycosidic linkages between arabinose residues and with Hyp are labeled according to the HMBC data (Figs. 4 and 5) with carbon numbers indicating the C atoms involved in the linkages.
Discussion
In EXTs, glycans comprise ~55% of the glycoprotein on a dry weight basis.52 In the cell, these glycans are attached to EXTs via a series of complex posttranslational modifications prior to the delivery of “mature” EXTs to the cell wall by Golgi vesicles,53 where they carry out their molecular function by self-assembling into positively charged scaffolds that guide the correct deposition of wall polysaccharides (mainly pectin) during growth and mitosis.54 Several glycosyltransferases are responsible for EXT glycosylation after hydroxylation of certain Pro residues to Hyp (eg, in Ser-(Hyp)4 motifs) by prolyl hydroxylase.55,56 One galactosyltransferase has been identified in Chlamydomonas reinhardtii (C. reinhardtii)57 that is responsible for adding the Gal to some EXT Ser residues, while several genes in Arabidopsis genome (RRA1—AT1G75120 and RRA2—AT1G75110;58 RRA3—AT1G19360;56 XEG113—AT2G35610)59 have been identified to potentially involve in EXT arabinosylation. XEG113 was proposed to add the third β-Araf onto the Hyp-Ara4 and Hyp-Ara3 chains,59 while RRA3 adds the second β-Araf to EXT oligoarabinosyl chains.56
Despite their high proportion in the protein, especially the major arabino-oligosaccharides that contribut ~95% (w/w) of the total EXT carbohydrate,60,61 the function of EXT glycans remains unclear. Earlier evidence has indicated that EXT arabinosylation may play a role in the protein–glycan network in the primary cell walls.29,44 Arabinosylation was also proposed to be important for conferring resistance to proteolysis44 and stabilizing the extended EXT polyproline-II (ppII) conformation.60–62 A loss of ppII conformation was observed in deglycosylated EXT monomers by circular dichroism spectroscopy60 and electron microscopy.61
The role of arabinosylation in EXT cross-linking was first suggested by the observation that completely dearabinosylated TOMP1 could not be cross-linked by the extensin peroxidase from tomato cell culture medium.33 Later, by gene knockout experiments, it was found that mutants-lacked EXT arabino-syltransferase genes showed a decreased EXT content in their cell walls.56,59 To test the effect of arabinosylation on EXT in vitro cross-linking, we partially dearabinosylated RSH and TOMP1 with α-l-arabinofuranosidase (ideally removes only the fourth Ara on Hyp-Ara4 due to its unique α configuration) and pH 2.0 HCl (less selective and removes Ara on all arabino-oligosaccharide chains). RSH and TOMP1 were also completely dearabinosylated by pH 1.0 HCl (preserved the Gal residues) and completely deglycosylated with anhydrous HF (removed all the sugar residues). EXT samples after each treatment were cross-linked in vitro using tomato extensin peroxidase, and their initial cross-linking rates were determined.
Treatment of RSH with α-l-arabinofuranosidase removed all the fourth Ara on Hyp-Ara4 as expected (Table 2). However, this treatment did not increase the percentage of the Hyp-Ara3, but instead a twofold increase in non-glycosylated Hyp was observed according to the Hyp-Arabinoside profile (Table 2). This observation indicated that either the Hyp-Ara4 oligosaccharides in RSH had more α-linked arabinofuronsides than only the fourth Ara in Hyp-Ara4 as that of tobacco50,51 or the enzyme used was contaminated with β-l-arabinofuranosidase activity. The fact that Arabidopsis and tobacco are only distantly related raises the possibility that the structure of the arabinosides may not be monolithic. Thus Hyp-Ara4 and Hyp-Ara3 were isolated from RSH and their structures determined by NMR. The structures of RSH Hyp-Ara4 and Hyp-Ara3 were identical to those from tobacco (Figs. 4–6, Table 5). Thus, it is likely that the old α-l-arabinofuranosidase preparation might contain β-l-arabinofuranosidase activity (also confirmed by a communication from Megazyme Company).
Besides the structural conservation of Hyp-Ara4 and Hyp-Ara3 between Arabidopsis and tobacco, a recent study of the Hyp-Ara4 and Hyp-Ara3 on EXTs of Boswellia serrata and B. carteri also showed similar structures.63 These findings indicate a conserved glycosylation pattern among these dicot species. Interestingly, Waffenschmidt’s group isolated Hyp-oligosaccharides from green algae C. reinhardtii and determined the glycosidic linkages and configurations in those glycans. Their proposed C. reinhardtii glycan structure were α-D-Galf-(1→2)-β-l-Araf-(1→2)-β-l-Araf-(1→4)-Hyp and 3-methyl-β-l-Araf-(1→5)-α-D-Galf-(1→2)-β-l-Araf-(1→2)-β-l-Araf-(1→4)-Hyp.64 The algae Hyp-oligosaccharides showed similarity in the inner-core glycan structure (β-l-Araf-(1→2)-β-l-Araf-(1→4)-Hyp) with those from dicots,51,63 which are undoubtedly due to some common steps in their biosynthetic evolution.64
Such conserved arabino-oligosaccharide structures across the evolutionary progression implies an important functional role for EXT oligoarabinosides. Data from this study support the notion that this structural conservation could play a role in EXT cross-linking.
First of all, the completely dearabinosylated RSH and TOMP1 (pH 1.0 HCl and HF treatment) could not be cross-linked by the tomato extensin peroxidase (initial rate = 0, Table 3). This result agreed with that from a previous study33 and indicates that arabinosylation is indeed required for EXT cross-linking.
Second, EXTs partially dearabinosylated by α-arabinofuranosidase (RSHAra and P1Ara) and pH 2.0 HCl (RSHpH2, and P1pH2) all have lower Ara content (Table 1) and slower initial cross-linking rates than the native EXTs (Table 3). This observation indicates a positive correlation between the cross-linking rate and the Ara content.
Finally, RSHpH2 and P1pH2 showed a much faster initial cross-linking rate (Table 3), and at the end of a 30-minute cross-linking, a much greater loss in the monomeric EXT content (Figs. 2 and 3, Table 3) compared with RSHAra and P1Ara was observed. The cross-linking profiles also revealed that monomeric RSHpH2 were converted to a larger molecular weight oligomer (40-minute retention time peak) after 30-minute cross-linking while a portion of the monomeric RSHAra was converted to a smaller molecular weight oligomer (60-minute retention time peak, bottom panel, Fig. 2), indicating that it was more difficult for RSHAra to reach complete cross-linking than RSHpH2. Similarly, after 30-minute cross-linking, the major product of P1pH2 was the 60-minute oligomer peak, with a less portion of P1pH2 converted to the 40-minute oligomer. Only a 60-minute oligomer peak was observed after cross-linking of P1Ara (bottom panel, Fig. 3), suggesting that P1Ara was more difficult to cross-link than P1pH2. Residual sugar analysis showed that pH 2.0 dearabinosylated EXTs retained less Ara than arabinofuranosidase dearabinosylated EXTs (Table 1). Furthermore, the former cross-linked faster and easier than the latter. This seems contradictory to the positive correlation between the cross-linking rate and the Ara content. However, the Hyp-arabinoside profiles showed that all the fourth Ara on the Hyp-Ara4 chains were removed in RSHAra and P1Ara while a certain amount of the intact Hyp-Ara4 chains were still preserved after the pH 2.0 HCl treatment (Table 2). This observation suggests that the fourth α-Ara on the Hyp-Ara4 chains may be crucial for the cross-linking of EXTs.
Taken together, we propose that the arabinosylation may play an important role in EXT cross-linking and the arabino-oligosaccharide structures, especially the α-configured fourth Ara on the Hyp-Ara4 chains, are conserved, in part, to facilitate rapid cross-linking of EXTs.
It is also worth noting that P1pH2 contains relatively more residual Hyp-arabinosides (even more Hyp-Ara4) than RSHpH2 (Table 2). However, the initial cross-linking rate of P1pH2 is significantly lower than that of RSHpH2 (Table 3). After 30-minute cross-linking, P1pH2 was mainly converted to a smaller molecular weight oligomer (60-minute retention time peak, Fig. 3, bottom right panel), while RSHpH2 was more efficiently converted to a larger molecular weight oligomer (40-minute retention time peak, Fig. 2, bottom right panel). This suggests that in addition to the extent of arabinosylation, the primary protein sequence (RSH has protein sequence resemble to P3-type EXT,35 while TOMP1 has a P1-type EXT sequence65), the alignment of polypeptides, and especially the type and number of cross-linking sites are also important for in vitro cross-linking of different EXTs. RSH has 16 Idt motifs in its protein sequence that can cross-link to form pulcherosine by combining with peptidyl Tyr residues (major in vitro cross-linking product) or Idt residues can cross-link to form Di-Idt.54 Notably, TOMP1 only has three potential Idt motifs at the protein terminus that can cross-link with its abundant Val-Tyr-Lys motifs to potentially form pulcherosine (deduced from TOMP1 protein sequence in the tomato genomic database—Sol Genomic Network, accession SGN-U315189). Thus, greater abundance in cross-linking sites may account for the better cross-linking of RSHpH2.
Conclusion
The cross-linking of partially or fully deglycosylated EXTs led to the first conclusion that EXT arabinosylation, especially the α-configured fourth Ara on the Hyp-Ara4 chains, is crucial for the initial rate and extent of EXT cross-linking in vitro by peroxidase. Determination of glycan structures of RSH Hyp-Ara4 and Hyp-Ara3 showed structural conservancy of those glycans between different dicot species and even with green algae, indicating the importance of the Hyp-arabino-oligosaccharides throughout the evolutionary progression of different plant species.
Supplementary Materials
Supplementary figure 1. EXTs isolation by cation exchange and reveres-phase chromatography. Dialyzed tomato culture eluate was fractionated by SP Sepharose (A) chromatography and Peak 1 (collected between markers) was purified by reveres-phase (B) that yielded TOMP1. Similarly, dialyzed Arabidopsis culture eluate was fractionated by SP Sepharose (C) chromatography and Peak 1 was purified by reveres-phase (D) that yielded RSH.
Supplementary figure 2. Cross-linking of deglycosylated extensins. Deglycosylated TOMP1 (A) and RSH (B) cross-linked for 30 minutes by peroxidase. No change in monomer areas were observed in all cases. pH 1.0 HCl deglycosylated sample runs were recorded for a shorter time (120 minutes); thus, the succinic acid peaks were not included in those profiles. Monomer peak shoulders could be due to the uneven succinylation of the monomers.
Supplementary table 1. Amino acid composition (mol%) of extensin precursors TOMP1 and RSH.
Footnotes
ACADEMIC EDITOR: Gabor Mocz, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,199 words, excluding any confidential comments to the academic editor.
FUNDING: The authors thank NFS (Grant to MJK) for the funding of this study, andthis study was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-014575-01 from the National Center for Research Resources, National Institutes of Health. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Designed the research: YC, MJK. Performed the experiments: YC, WD, LT. Analyzed the data: YC, MAH, MJK. Wrote the paper: YC, LT, MAH, MJK. All authors read and approved the final manuscript.
REFERENCES
- 1.Lamport DTA, Kieliszewski MJ, Chen Y, Cannon MC. Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 2011;156:11–19. doi: 10.1104/pp.110.169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamport DTA, Northcote DH. Hydroxyproline in primary cell walls of higher plants. Nature. 1960;188:665–666. [Google Scholar]
- 3.Goodenough UW, Heuser JE. The Chlamydomonas cell wall and its constituent glycoprotein analyzed by the quick-freeze, deep-etch technique. J Cell Biol. 1985;101:1550–1568. doi: 10.1083/jcb.101.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodenough UW, Gebhart B, Mecham RP, Heuser JE. Crystals of the Chlamydomonas reinhardtii cell wall: polymerization, depolymerization and purification of glycoprotein monomers. J Cell Biol. 1986;103:405–417. doi: 10.1083/jcb.103.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadava D, Chrispeels MJ. Hydroxyproline-rich cell wall protein (extensin): role in the cessation of elongation in excised pea epicotyls. Dev Biol. 1973;30:49–55. doi: 10.1016/0012-1606(73)90047-x. [DOI] [PubMed] [Google Scholar]
- 6.Hong JC, Nagao RT, Key JL. Developmentally regulated expression of soybean proline-rich cell wall protein genes. Plant Cell. 1989;1:937–943. doi: 10.1105/tpc.1.9.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knox JP, Linstead PJ, Peart J, Cooper C, Roberts K. Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1991;1:317–326. doi: 10.1046/j.1365-313X.1991.t01-9-00999.x. [DOI] [PubMed] [Google Scholar]
- 8.Pennell RI, Janniche L, Kjellbom P, Scofield GN, Peart JM, Roberts K. Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell. 1991;3:1317–1326. doi: 10.1105/tpc.3.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman MH, Pezzotti M, Seurinck J, Mariani C. Developmental expression of tobacco pistil-specific genes encoding novel extensin-like proteins. Plant Cell. 1992;4:1041–1051. doi: 10.1105/tpc.4.9.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreuger M, van Holst GJ. Arabinogalactant proteins are essential in somatic embryogenesis of Daucus carota L. Planta. 1993;189:243–248. [Google Scholar]
- 11.Hall Q, Cannon MC. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell. 2002;14:1161–1172. doi: 10.1105/tpc.010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Varner JE. Isolation and characterization of cDNA clones for carrot extensin and A proline-rich 33-kDa protein (wound response) Proc Natl Acad Sci U S A. 1985;82:4399–4403. doi: 10.1073/pnas.82.13.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Showalter AM, Zhou J, Rumeau D, Worst SG, Varner JE. Tomato extensin and extensin-like cDNAs: structure and expression in response to wounding. Plant Mol Biol. 1991;16:547–565. doi: 10.1007/BF00023421. [DOI] [PubMed] [Google Scholar]
- 14.Bradley DJ, Kjellbom P, Lamb CJ. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- 15.Hammerschmidt R, Lamport DTA, Muldoon EP. Cell wall hydroxyproline enhancement and lignin deposition as an early event in the resistance of cucumber to Cladosporium cucumerinum. Physiol Plant Pathol. 1984;24:43–47. [Google Scholar]
- 16.Showalter AM, Bell JN, Cramer CL, Bailey JA, Varner JE, Lamb CJ. Accumulation of hydroxyproline-rich glycoprotein mRNAs in response to fungal elicitor and infection. Proc Natl Acad Sci U S A. 1985;82:6551–6555. doi: 10.1073/pnas.82.19.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazau D, Esqueré-Tugayé MT. Hydroxyproline-rich glycoprotein accumulation in the cell walls of plants infected by various pathogens. Physiol Mol Plant Pathol. 1986;29:147–157. [Google Scholar]
- 18.Lawton MA, Lamb CJ. Transcriptional activation of plant defense genes by fungal elicitor, wounding, and infection. Mol Cell Biol. 1987;7:335–341. doi: 10.1128/mcb.7.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JJ, Muldoon EP, Lamport DTA. Isolation of extensin precursors by direct elution of intact tomato cell suspension cultures. Phytochemistry. 1984;23:1233–1239. [Google Scholar]
- 20.Terneus KA. The Isolation and Characterization of a Tobacco Extensin Precursor and Two Arabidopsis Hydroxyproline-Rich Glycoproteins [Ph.D dissertation] Athens, OH: Ohio University; 2006. [Google Scholar]
- 21.Lamport DTA, Katona L, Roerig S. Galactosylserine in extensin. Biochem J. 1973;133:125–131. doi: 10.1042/bj1330125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamport DTA. Hydroxproline-O-glycosidic linkage of the plant cell wall glycoprotein extensin. Nature. 1967;216:1322–1324. [Google Scholar]
- 23.Lamport DTA, Miller DH. Hydroxyproline arabinosides in the plant kingdom. Plant Physiol. 1971;48:454–456. doi: 10.1104/pp.48.4.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campargue C, Lafitte C, Esquerré-Tugayé MT, Mazau D. Analysis of hydroxyproline and hydroxyproline-arabinosides of plant origin by high-performance anion-exchange chromatography ± pulsed amperometric detection. Anal Biochem. 1998;257:20–25. doi: 10.1006/abio.1997.2526. [DOI] [PubMed] [Google Scholar]
- 25.Cassab GI. Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- 26.Cosgrove DC. Growth of the Plant Cell Wall. Nature. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- 27.Cooper JB, Varner JE. Cross-linking of soluble extensin in isolated cell walls. Plant Physiol. 1984;76:414–417. doi: 10.1104/pp.76.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper JB, Chen J, van Holst GJ, Varner JE. Hydroxyproline-rich glycoproteins of plant cell walls. Trends Biochem Sci. 1987;12:24–27. [Google Scholar]
- 29.Keegstra K, Talmadge KW, Bauer WD, Albersheim P. The structure of plant cell walls. III. a model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 1973;51:188–196. doi: 10.1104/pp.51.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi X, Behrens BX, West PR, Mort AJ. Solubilization and partial characterization of extensin fragments from cell walls of cotton suspension cultures. Evidence for a covalent cross-link between extensin and pectin. Plant Physiol. 1995;108:1691–1701. doi: 10.1104/pp.108.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuñez A, Fishman ML, Fortis LL, Cooke PH, Hotchkiss AT., Jr Identification of extensin protein associated with sugar beet pectin. J Agric Food Chem. 2009;57:10951–10958. doi: 10.1021/jf902162t. [DOI] [PubMed] [Google Scholar]
- 32.Kieliszewski MJ, Lamport DTA. Extensin: repetitive motifs, functional sites, posttranslational codes, and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- 33.Schnabelrauch LS, Kieliszewski M, Upham BL, Alizedeh H, Lamport DT. Isolation of pI 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 1996;9:477–489. doi: 10.1046/j.1365-313x.1996.09040477.x. [DOI] [PubMed] [Google Scholar]
- 34.Fry SC. Isodityrosine, a new cross-linking amino acid from plant cell wall glycoprotein. Biochem J. 1982;204:449–455. doi: 10.1042/bj2040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epstein L, Lamport DTA. An intramolecular linkage involving isodityrosine in extensin. Phytochemistry. 1984;23:1241–1246. [Google Scholar]
- 36.Brady JD, Sadler IH, Fry SC. Di-isodityrosine, a novel tetrameric derivative of tyrosine in plant cell wall proteins: a new potential cross-link. Biochem J. 1996;315:323–327. doi: 10.1042/bj3150323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Held MA, Tan L, Kamyab A, Hare M, Shpak E, Kieliszewski MJ. Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J Biol Chem. 2004;279:55474–55482. doi: 10.1074/jbc.M408396200. [DOI] [PubMed] [Google Scholar]
- 38.Brady JD, Sadler IH, Fry SC. Pulcherosine, an oxidatively coupled trimer of tyrosine in plant cell wall: its role in cross-link formation. Phytochemistry. 1998;47:349–353. doi: 10.1016/s0031-9422(97)00592-x. [DOI] [PubMed] [Google Scholar]
- 39.Heckman JWJ, Terhune BT, Lamport DTA. Characterization of native and modified extensin monomers and oligomers by electron microscopy and gel filtration. Plant Physiol. 1988;86:848–856. doi: 10.1104/pp.86.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mort AJ, Lamport DTA. Anhydrous hydrogen fluoride deglycosylates glycoproteins. Anal Biochem. 1977;82:289–309. doi: 10.1016/0003-2697(77)90165-8. [DOI] [PubMed] [Google Scholar]
- 41.Frueauf JB, Dolata M, Leykam JF, et al. Peptides isolated from cell walls of Medicago truncatula nodules and uninfected root. Phytochemistry. 2000;55:429–438. doi: 10.1016/s0031-9422(00)00336-8. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Ye D, Held MA, et al. Identification of the abundant hydroxyproline-rich glycoproteins in the root walls of wild-type arabidopsis, an ext3 mutant line, and its phenotypic revertant. Plants. 2015;4:85–111. doi: 10.3390/plants4010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janssen PSL, van Nispen JW, Melgers PATA, van den Bogaart HWM, Hamelinck RLAE, Goverde BC. HPLC analysis of phenylthiocarbamyl (PTC) amino acids. I. Evaluation and optimization of the procedure. Chromatographia. 1986;22:7–12. [Google Scholar]
- 44.Lamport DTA. The isolation and partial characterization of hydroxyproline-rich glycopeptides obtained by enzymatic degradation of primary cell wall. Biochemistry. 1969;8:1115–1163. doi: 10.1021/bi00831a049. [DOI] [PubMed] [Google Scholar]
- 45.Fry SC. Chapter 3, wall polymers: extraction and fractionation. In: Wilkens M, editor. The Growing Plant Cell Wall: Chemical and Metabolic Analysis. Essex UK: Longman Scientific & Technical; John Wiley and Sons Inc; 1988. p. 97. [Google Scholar]
- 46.Dong W, Kieliszewski MJ, Held MA. Identification of the pI 4.6 extensin peroxidase from Lycopersicon esculentum using proteomics and reverse-genomics. Phytochemistry. 2015;112:151–159. doi: 10.1016/j.phytochem.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Everdeen DS, Kiefer S, Willard JJ, et al. Enzymic cross-linkage of monomeric extensin precursors in vitro. Plant Physiol. 1988;87:616–621. doi: 10.1104/pp.87.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.York WS, Darvill AG, McNeil M, et al. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1985;118:3–40. [Google Scholar]
- 49.Tan L, Qiu F, Lamport DT, Kieliszewski MJ. Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive ala-hyp expressed in transgenic Nicotiana tabacum. J Biol Chem. 2004;279:13156–13165. doi: 10.1074/jbc.M311864200. [DOI] [PubMed] [Google Scholar]
- 50.Akiyama Y, Katõ K. Hydroxyproline arabinosides from suspension-cultured tobacco cell wall. Agric Biol Chem. 1976;40:2343–2348. [Google Scholar]
- 51.Akiyama Y, Mori M, Katõ K. 13C-NMR analysis of hydroxyproline arabinosides from Nicotiana tabacum. Agric Biol Chem. 1980;44:2487–2489. [Google Scholar]
- 52.Albersheim P, Darvill A, Roberts K, et al. Biochemistry of the cell wall molecules. In: Sigrid Masson., editor. Plant Cell Walls from Chemistry to Biology. New York, NY: Garland Science, Taylor & Francis Group, LLC; 2010. pp. 67–118. [Google Scholar]
- 53.Carpita N, McCann MC. The cell wall. In: Bob B Buchanan., editor. Biochemistry & Molecular Biology of Plants. Rockville, MD: Wiley; 2000. pp. 52–108. [Google Scholar]
- 54.Cannon MC, Terneus K, Hall Q, et al. Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci U S A. 2008;105:2226–2231. doi: 10.1073/pnas.0711980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadava D, Chrispeels MJ. Hydroxyproline biosynthesis in plant cells peptidyl proline hydroxylase from carrot disks. Biochim Biophys Acta. 1971;227:278–287. doi: 10.1016/0005-2744(71)90060-x. [DOI] [PubMed] [Google Scholar]
- 56.Velasquez SM, Ricardi MM, Dorosz JG, et al. O-glycosylated cell wall Proteins are essential in root hair growth. Science. 2011;332:1401–1403. doi: 10.1126/science.1206657. [DOI] [PubMed] [Google Scholar]
- 57.Saito F, Suyama A, Oka T, et al. Identification of novel peptidyl serine α-galactosyltransferase gene family in plants. J Biol Chem. 2014;289:20405–20420. doi: 10.1074/jbc.M114.553933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Egelund J, Obel N, Ulvskov P, et al. Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol. 2007;64:439–451. doi: 10.1007/s11103-007-9162-y. [DOI] [PubMed] [Google Scholar]
- 59.Gille S, Hänsel U, Ziemann M, Pauly M. Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci U S A. 2009;106:14699–14704. doi: 10.1073/pnas.0905434106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Holst GJ, Varner JE. Reinforced polyproline II conformation in a hydroxyproline-rich cell wall glycoprotein from carrot root. Plant Physiol. 1984;74:247–251. doi: 10.1104/pp.74.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stafstrom JP, Staehelin LA. The role of carbohydrate in maintaining extensin in an extended conformation. Plant Physiol. 1986;81:242–246. doi: 10.1104/pp.81.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferris PJ, Woessner JP, Waffenschmidt S, Kilz S, Drees J, Goodenough UW. Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry. 2001;40:2978–2987. doi: 10.1021/bi0023605. [DOI] [PubMed] [Google Scholar]
- 63.Herrmann A, König S, Lechtenberg M, et al. Proteoglycans from Boswellia ser-rata Roxb and B. carteri Birdw and Identification of a proteolytic plant basic Secretory protein. Glycobiology. 2012;22:1424–1439. doi: 10.1093/glycob/cws107. [DOI] [PubMed] [Google Scholar]
- 64.Bollig K, Lamshöft M, Schweimer K, Marner FJ, Budzikiewicz H, Waffenschmidt S. Structural analysis of linear hydroxyproline-bound O-glycans of Chlamydomonas reinhardtii— conservation of the inner core in chlamydomonas and land plants. Carbohydr Res. 2007;342:2557–2566. doi: 10.1016/j.carres.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 65.Smith JJ, Muldoon EP, Willard JJ, Lamport DTA. Tomato extensin precursors P1 and P2 are highly periodic structures. Phytochemistry. 1986;5:1021–1030. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. EXTs isolation by cation exchange and reveres-phase chromatography. Dialyzed tomato culture eluate was fractionated by SP Sepharose (A) chromatography and Peak 1 (collected between markers) was purified by reveres-phase (B) that yielded TOMP1. Similarly, dialyzed Arabidopsis culture eluate was fractionated by SP Sepharose (C) chromatography and Peak 1 was purified by reveres-phase (D) that yielded RSH.
Supplementary figure 2. Cross-linking of deglycosylated extensins. Deglycosylated TOMP1 (A) and RSH (B) cross-linked for 30 minutes by peroxidase. No change in monomer areas were observed in all cases. pH 1.0 HCl deglycosylated sample runs were recorded for a shorter time (120 minutes); thus, the succinic acid peaks were not included in those profiles. Monomer peak shoulders could be due to the uneven succinylation of the monomers.
Supplementary table 1. Amino acid composition (mol%) of extensin precursors TOMP1 and RSH.