Abstract
Human chlamydiosis is a zoonotic disease of avian origin caused by Chlamydia psittaci. The highest infection rates have been detected in parrots (Psittacidae) and pigeons (Columbiformes), the latter most frequently carry the genotypes B and E. These genotypes have been shown to also infect humans. Because pigeons (Columba livia) cohabit with humans in urban areas, C. psittaci present in the dust from dry feces of infected pigeons may be transmitted by inhalation and represent a significant public health problem. Between 2012 and 2013 a total of 120 fecal samples were collected from pigeons at four public places (Plaza de la Cultura, Parque Morazán, Parque Central de Guadalupe, Plaza de las Garantías Sociales) in San José, Costa Rica. A nested polymerase chain reaction (PCR) was used to amplify a region of the outer membrane protein A gene of C. psittaci. Only one sample was positive in PCR and the positive sample was further subjected to sequencing and genotyping. Sequencing identified this sample as C. psittaci genotype B. This study is the first report to show the presence of this organism in pigeons of Costa Rica, and shows that the infected pigeons may represent a significant risk for humans who visit public places that are inhabited by pigeons.
Keywords: Chlamydia psittaci, Costa Rica, Genotype B, Pigeons, Zoonosis
Introduction
Avian chlamydiosis is caused by Chlamydia psittaci (C. psittaci), a gram-negative and obligate intracellular bacterium, with nine (A to F, E/B, M56, and WC) known genotypes (Van Lent et al., 2012). C. psittaci has been identified in 465 different bird species (Kaleta and Taday, 2003), but the highest rate of infection was found in parrots (Psittacidae) and pigeons (Columbiformes) (de Freitas et al., 2002; Dovc et al., 2005; Dovc et al., 2007).
C. psittaci is shed regularly or intermittently in feces, lacrimal fluids, nasal discharges, oropharyngeal mucus and crop milk of the infected birds. Extreme environmental changes can trigger the onset of clinical disease in the infected birds; however prolonged subacute clinical forms of the disease are most common (Gerlach, 1986). Avian chlamydiosis presents in the form of nonspecific clinical signs to severe systemic disease, especially in young animals (Andersen and Vanrompay, 2003).
All genotypes of C. psittaci can be transmitted to humans and cause psittacosis or parrot fever (Andersen and Vanrompay, 2003); this transmission can occur either through inhalation, ingestion or via direct contact with the infected birds (Longbottom and Coulter, 2003). In humans, the disease can vary from nonspecific flu-like symptoms to severe pneumonia. Also, cases of endocarditis and encephalitis have been reported (Crosse, 1990).
C. psittaci is the most common pathogen found in domestic pigeons and mainly belong to the genotypes B and E (Vanrompay et al., 1993; Andersen and Vanrompay, 2000; Magnino et al., 2009). Because pigeons spread chlamydial infection and usually fly distances of about 5 km, these birds represent a major risk to public health as they cohabit with humans in urban and rural areas, in public places, parks and even gardens. It is believed that pigeons have been underestimated for a long time as an important source of human chlamydial infections (Heddema et al., 2006a; Vázquez et al., 2010).
A study carried out in Costa Rica in 2001 found antibodies against C. psittaci in 12.4% of 129 captive macaws (Ara macao and Ara ambigua) (Herrera et al., 2001). Recently C. psittaci was detected in four (3.4%) out of 117 captive psittacine birds, which cohabited in human households and in two of those cases the organism was found to be of genotype A (Sheleby-Elias, 2010).
The objective of the present study was to determine the presence of C. psittaci in feces of pigeons of urban public places and parks that are frequently visited by children, elderly and tourists.
Materials and Methods
Size, type of sample and sampling method
Between October 2012 and May 2013 a total of 120 pigeons were tested in four different public parks (Plaza de la Cultura, Plaza de las Garantías Sociales, Parque Morazán and Parque Central Guadalupe) in San José, by cookie crumbs.
Feces were collected from the floor with sterile swabs from defecating birds, and transferred into tubes containing 0.5 ml Minimal Essential Medium. The swabs were stored at 4°C for 24 hours until further processing.
Polymerase chain reaction (PCR)
For DNA extraction DNeasy® Blood & Tissue Kit of QIAGEN (Venlo, Netherlands) was used and the extraction was performed according to the manufacturer’s instructions. For the detection of C. psittaci, a nested PCR described by Sachse and Hotzel (2003) was used, which partially amplifies the gene outer membrane protein A (ompA) to identify the genus Chlamydia. The primers used were: 191CHOMP (5’-GCIYTITGGGARTGYGGITGYGCI AC-3’) and CHOMP371 (5’-TTAGAAICKGAATTGIGCRTTIAYGTGIGCIGC-3’). All amplification products were subjected to a second PCR to identify C. psittaci, using the primers CHOMP 336s (5’-CCRCAAGMTTTTCTRGAYTTCAWYTTGTTRAT-3’) and 218PSITT (5’-GTAATTTCIAGCCCAGCACAATTYGTG-3’). Samples with bands of 389-404 bp were considered positive for C. psittaci. DNA of C. psittaci was donated by the Clinic of Birds, Reptiles, Amphibians and Fish, Justus Liebig University, Giessen, Germany and was used as a positive control; nuclease free water (molecular biology grade, Thermo Scientific, Waltham, USA) was used as a negative control.
Sequencing, genotyping and construction of the phylogenetic tree
One positive sample identified during nested PCR was genotyped through analysis of ompA sequences (Heddema et al., 2006b).
The primers used were CPsittGenoFor (5’-GCTACGGGTTCCGCTCT-3’) and CPsittGenoRev (5’-TTTGTTGATYTGAATCGAAGC-3’), which amplify conserved regions of the ompA gene covering four variable domains. The size of the amplified fragment was 1041bp. PCR product was purified using the QIAquick® kit (QIAGEN, Venlo, Netherlands), according to the manufacturer’s instructions.
The DNA sample was sent to Macrogen (Seoul, Korea) for sequencing. Partial sequence was aligned with BioEdit Sequence Aligment Editor® (Hall, 1999) and compared using the BLASTn algorithm with the database of National Center for Biotechnology Information. Then the sequences were imported into MEGA 5 (Tamura et al., 2011), using the Jukes & Cantor algorithm (Jukes and Cantor, 1969) and the UPGMA method (Sneath and Sokal, 1973), for the design of the phylogenetic tree. A total of 10000 replicates were calculated (Felsenstein, 1985). The analysis included the reference sequences of the nine C. psittaci genotypes available in the database of GenBank, A (accession number AY762608), B (AF269265), C (L25436), D (AF269266), E (X12647), F (AF269259), E/B (AY762613), M56 (AF269268) and WC (AF269269) (Sachse et al., 2008), the phylogenetic tree was rooted with the C. caviae strain (GPIC, GenBank AF269282) (Zhang et al., 1989).
Results
Only one out of 120 samples yielded positive results during the first PCR (576-597pb), and the specific band (389-404pb) was subsequently amplified using the nested PCR (Fig. 1).
Fig. 1.
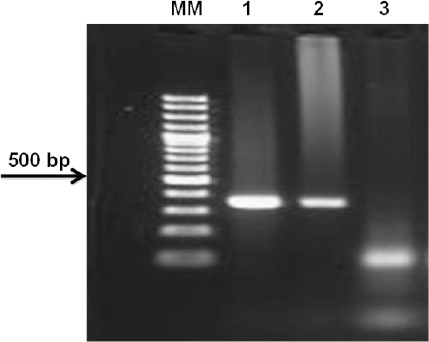
Gel electrophoresis of the amplified PCR products of ompA gene of C. psittaci (MM: molecular marker; 1: positive control; 2: pigeon from Costa Rica; 3: negative control).
The sample that tested positive for C. psittaci was collected from a pigeon from the public park Plaza de la Cultura. Sequencing confirmed the results, and identified this sample as C. psittaci genotype B (Fig. 2).
Fig. 2.
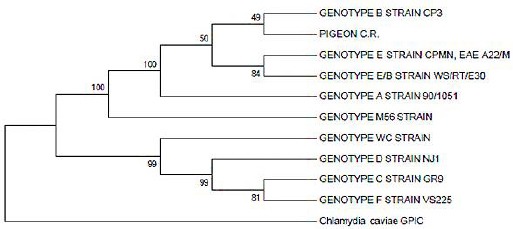
Phylogenetic tree of gene ompA sequence of one pigeon sample (PIGEON C.R.) of Costa Rica that tested positive for C. psittaci.
The sequence analysis of the product of PCR using BLASTn, found 99.9% (1029/1030) nucleotide identity with sequences of C. psittaci genotype B deposited in GenBank (CP003797.1, AY762609.1, AF269265.1). Comparison of the partial sequence of the fecal pigeon sample of Costa Rica with a C. psittaci sequence isolated in 1958 from air sacs of pigeons in California, USA by Bush and Everett (2001) is shown in Figure 3.
Fig. 3.
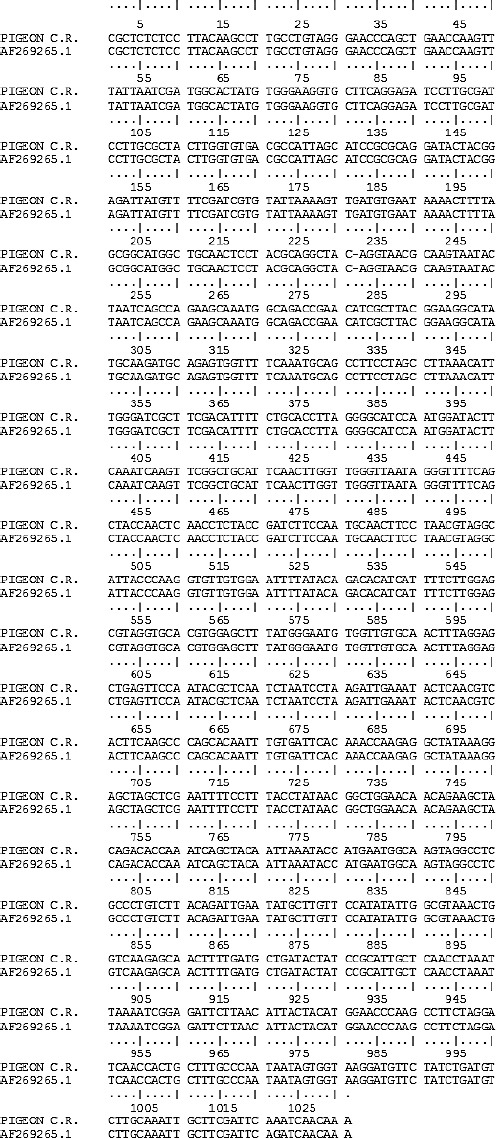
Alignment of the nucleotide sequences of the ompA gene sequence of C. psittaci obtained from a pigeon in Costa Rica (PIGEON C.R.) with that of C. psittaci genotype B (AF269265.1) isolated in 1958 from air sacs of pigeons in California, USA. The nucleotide difference between the sequences is underlined at the position.
Discussion
This is the first study in Costa Rica that reported the presence of C. psittaci genotype B in a pigeon of a public park in Costa Rica, and is consistent with the international literature which mentions that this genotype is found more frequently in domestic and feral pigeons (Vanrompay et al., 1993; Andersen and Vanrompay, 2003; Magnino et al., 2009).
Only one bird was found to be positive for the pathogen (3.3%), in contrast to studies carried out in Madrid (61/116, 52.6%), Zagreb (30/232, 12.9%) and Amsterdam (26/331, 7.8%) (Pruckner-Radovcic et al., 2005; Heddema et al., 2006a; Vázquez et al., 2010), but our results are in accordance with studies carried out in Ghent (1/61, 1.6%) (Dickx et al., 2010). The reasons could be due to several factors such as type of the study, sample size, or the sampling method.
In addition, false negative results could not be ruled out, due to polymerase inhibitors in the samples or because of intermittent shedding of C. psittaci from cloacal samples that could have resulted in an underestimation of the agent (Pruckner-Radovcic et al., 2005).
Nevertheless, it may also indicate a low infection rate of C. psittaci in pigeons of Costa Rica, which would be consistent with studies carried out by Sheleby-Elias (2010), who detected a low infection rate (3.4%) of C. psittaci in captive psittacine birds inhabiting the households. The genotype present in captive parrots was determined as genotype A.
The finding of this zoonotic agent in the feces of a pigeon shows that C. psittaci is found in places that are regularly visited by human population, especially on weekends by families with young children, who feed the pigeons. This poses a significant risk of transmission of the agent by inhalation or direct contact with the infected birds (Longbottom and Coulter, 2003).
The low infection rate determined in the present study is beneficial to the public health (Pruckner-Radovcic et al., 2005), nevertheless it is recommended that more comprehensive and representative studies are required to determine the prevalence of C. psittaci in urban pigeons, establish the genotypes circulating in these birds, because certain genotypes (A and D) can cause serious infections in people, while others, such as genotype B, usually cause mild respiratory symptoms (Dickx et al., 2010).
Finally, we recommend implementation of control programs for decreasing the prevalence of this disease in pigeons, coupled with environmental educational programs, to prevent a possible spread of C. psittaci to people visiting public places of Costa Rica.
Acknowledgements
This research was conducted within the framework of the project titled “Detection of infectious agents in wild birds of Costa Rica”; project number NFEG05 of the Universidad Nacional.
Special thanks to Prof. Dr. Michael Lierz, Clinic of Birds, Reptiles, Amphibians and Fish, Justus Liebig University, Giessen, Germany, who kindly donated DNA of C. psittaci that was used as a positive control in the PCR in our investigation. Ethical guidelines were followed during this investigation, as animals were used in our study.
References
- Andersen A.A, Vanrompay D. Avian chlamydiosis. Rev. Sci. Tech. Off. Int. Epiz. 2000;19:396–404. [PubMed] [Google Scholar]
- Andersen A.A, Vanrompay D. Avian chlamydiosis (psittacosis, ornithosis) In: Saif Y.M, Barnes H.J, Fadly A.M, Glisson J.R, McDougald L.R, Swayne D.E, editors. Diseases of Poultry. 11th edition. Ames, Iowa: State University Press; 2003. pp. 863–887. [Google Scholar]
- Bush R.M, Everett K.D. Molecular evolution of the Chlamydiaceae. Int. J. Syst. Evol. Microbiol. 2001;51:203–220. doi: 10.1099/00207713-51-1-203. [DOI] [PubMed] [Google Scholar]
- Crosse B. Psittacosis: a clinical review. J. Infect. 1990;21:251–259. doi: 10.1016/0163-4453(90)93909-c. [DOI] [PubMed] [Google Scholar]
- de Freitas R.T, Júnior A.B, Pinto A.A. Evidence of Chlamydophila psittaci infection in captive Amazon parrots in Brazil. J. Zoo Wildl. Med. 2002;33:118–121. doi: 10.1638/1042-7260(2002)033[0118:EOCPII]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Dickx V, Beeckman D.S, Dossche L, Tavernier P, Vanrompay D. Chlamydophila psittaci in homing and feral pigeons and zoonotic transmission. J. Med. Microbiol. 2010;59:1348–1353. doi: 10.1099/jmm.0.023499-0. [DOI] [PubMed] [Google Scholar]
- Dovc A, Dovc P, Kese D, Vlahovic K, Pavlak M, Zorman-Rojs O. Long-term study of Chlamydophilosis in Slovenia. Vet. Res. Commun. 2005;29:23–36. doi: 10.1007/s11259-005-0834-2. [DOI] [PubMed] [Google Scholar]
- Dovc A, Slavec D, Lindtner-Knific R, Zorman-Rojs O, Racnik J, Golja J, Vlahovic K. the Proceedings of the 5th Annual Workshop of COST Action 855 Animal Chlamydioses and Zoonotic Implications; 2007. The study of a Chlamydophila psittaci outbreak in budgerigars; p. 24. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gerlach H. Chlamydia. In: Harrison G.J, Harrison L.R, editors. Clinical Avian Medicine and Surgery. Philadelphia, London, Toronto: WB Saunders Co; 1986. pp. 457–463. [Google Scholar]
- Hall T.A. BioEdit: to user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Heddema E.R, Ter Sluis S, Buys J.A, Vandenbroucke-Grauls C.M, van Wijnen J.H, Visser C.E. Prevalence of Chlamydophila psittaci in fecal droppings from feral pigeons in Amsterdam, The Netherlands. Appl. Environ. Microbiol. 2006a;72:4423–4425. doi: 10.1128/AEM.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddema E.R, van Hannen E.J, Duim B, Vandenbroucke-Grauls C.M, Pannekoek Y. Genotyping of Chlamydophila psittaci in human samples. Emerg. Infect. Dis. 2006b;12:1989–1990. doi: 10.3201/eid1212.051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera I, Khan S.R, Kaleta E.F, Müller H, Dolz G, Neumann U. Serological status for Chlamydophila psittaci, Newcastle disease virus, avian polyoma virus, and Pacheco disease virus in scarlet macaws (Ara macao) kept in captivity in Costa Rica. J. Vet. Med. B. Infec. Dis. Vet. Public Health. 2001;48:721–726. doi: 10.1046/j.1439-0450.2001.00485.x. [DOI] [PubMed] [Google Scholar]
- Jukes T.H, Cantor C.R. Evolution of protein molecules. In: Munro H.N, editor. Mammalian Protein Metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Kaleta E.F, Taday E.M. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 2003;32:435–461. doi: 10.1080/03079450310001593613. [DOI] [PubMed] [Google Scholar]
- Longbottom D, Coulter L.J. Animal chlamydioses and zoonotic implications. J. Comp. Pathol. 2003;128:217–244. doi: 10.1053/jcpa.2002.0629. [DOI] [PubMed] [Google Scholar]
- Magnino S, Haag-Wackernagel D, Geigenfeind I, Helmecke S, Dovc A, Prukner-Radovcic E, Residbegovic E, Ilieski V, Laroucau K, Donati M. Chlamydial infections in feral pigeons in Europe: Review of data and focus on public health implications. Vet. Microbiol. 2009;135:54–67. doi: 10.1016/j.vetmic.2008.09.045. [DOI] [PubMed] [Google Scholar]
- Pruckner-Radovcic E, Horvatek D, Gottstein Y, Grozdanic I.C, Mazija H. Epidemiological investigation of Chlamydophila psittaci in pigeons and free living birds in Croatia. Vet. Res. Commun. 2005;29:17–21. doi: 10.1007/s11259-005-0083-4. [DOI] [PubMed] [Google Scholar]
- Sachse K, Hotzel H. Detection and differentiation of Chlamydiae by nested PCR. In PCR Detection of Microbial Pathogens: Methods and protocols. Meth. Mol. Biol. 2003;216:123–136. doi: 10.1385/1-59259-344-5:123. [DOI] [PubMed] [Google Scholar]
- Sachse K, Laroucau K, Hotzel H, Schubert E, Ehricht R, Slickers P. Genotyping of Chlamydophila psittaci using a new DNA microarray assay based on sequence analysis of ompA genes. BMC Microbiol. 2008 doi: 10.1186/1471-2180-8-63. doi: 10.1186/1471-2180-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheleby-Elias J. M. S. thesis. Heredia, Costa Rica: Universidad Nacional; 2010. Detección molecular de Circovirus, Poliomavirus y Chlamydophila psittaci en psitácidas en cautiverio de Costa Rica. [Google Scholar]
- Sneath P.H.A, Sokal R.R. Numerical taxonomy. The principles and practice of numerical classification. Freeman. 1973 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. E. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lent S, Piet J.R, Beeckman D, Van Der Ende A, Van Nieuwerburgh F, Bavoil P, Myers G, Vanrompay D, Pannekoek Y. Full genome sequences of all nine Chlamydia psittaci genotype reference strains. J. Bacteriol. 2012;194:6930–6931. doi: 10.1128/JB.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrompay D, Andersen A.A, Ducatelle R, Haesebrouck F. Serotyping of European isolates of Chlamydia psittaci from poultry and other birds. J. Clin. Microbiol. 1993;31:134–137. doi: 10.1128/jcm.31.1.134-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez B, Esperón F, Neves E, López J, Ballesteros C, Muñoz M.J. Screening for several potential pathogens in feral pigeons (Columba livia) in Madrid. Acta Vet. Scand. 2010;52:45. doi: 10.1186/1751-0147-52-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.X, Morrison S.G, Caldwell H.D, Baehr W. Cloning and sequence analysis of the major outer membrane protein genes of two Chlamydia psittaci strains. Infect. Immun. 1989;57:1621–1625. doi: 10.1128/iai.57.5.1621-1625.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]


