Abstract
Formin homology proteins (formins) are actin nucleation factors which remain bound to the growing barbed end and processively elongate actin filament (F-actin). Recently, we have demonstrated that a mammalian formin mDia1 rotates along the long-pitch helix of F-actin during processive elongation (helical rotation) by single-molecule fluorescence polarization. We have also shown processive depolymerization of mDia1-bound F-actin during which helical rotation was visualized. In the cell where F-actins are highly cross-linked, formins should rotate during filament elongation. Therefore, when formins are tightly anchored to cellular structures, formins may not elongate F-actin. Adversely, helical rotation of formins might affect the twist of F-actin. Formins could thus control actin elongation and regulate stability of cellular actin filaments through helical rotation. On the other hand, ADP-actin elongation at the mDia1-bound barbed end turned out to become decelerated by profilin, in marked contrast to its remarkably positive effect on mDia1-mediated ATP-actin elongation. This deceleration is caused by enhancement of the off-rate of ADP-actin. While mDia1 and profilin enhance the ADP-actin off-rate, they do not apparently increase the ADP-actin on-rate at the barbed end. These results imply that G-actin-bound ATP and its hydrolysis may be part of the acceleration mechanism of formin-mediated actin elongation.
Keywords: formin homology protein, single-molecule, fluorescence polarization, ATP hydrolysis, filament twist and stability
In response to external stimuli, cells rapidly change the shape by actin polymerization and depolymerization. Formin homology proteins (formins) are responsible for the formation of the actin based structures such as actin stress fibers, actin cables, and the contractile ring during cytokinesis1–5. Formins accelerate actin filament (F-actin) polymerization by enhancing filament nucleation. Formins remain bound to the growing barbed end and processively assemble F-actin6,7. Formins have two conserved domains, formin homology domain 1 and 2 (FH1 and FH2) in the C-terminal half3. Formin FH1 is composed of poly-proline repeats and binds to an actin monomer binding protein profilin. The interaction between FH1 and profilin-actin complex accelerates formin-mediated actin polymerization8. FH2 forms a ring-like dimer and binds around the barbed end of actin9 which forms a long-pitch double-helical filament10.
Our recent study has revealed using the single-molecule fluorescence polarization method that mDia1 rotates along the long-pitch helix of F-actin during processive elongation (referred to here as helical rotation)11. Although it had long been speculated that formins might rotate during processive elongation, helical rotation of formins had not been observed before our study. Our data indicate tight coupling of helical rotation of mDia1 with actin elongation. In addition, we for the first time visualized processive depolymerization of ADP-F-actin bound to mDia1. We further found that pro-filin accelerates depolymerization of ADP-F-actin bound to mDia1, which led to our discovery of the opposite effects of ATP- and ADP-actin on the rate of filament elongation catalyzed by formins and profilin.
In this review, we first describe the visualization method of helical rotation using single-molecule fluorescence polarization. Next we explain the observed coupling between helical rotation of mDia1 and the processive actin elongation. We discuss implications of our finding of the requirement of ATP in the acceleration of formin-mediated elongation by profilin. We also describe questions raised by our findings in relation to the barbed end structure. Finally, we discuss possible roles of helical rotation of formins in the regulation of actin turnover in cells.
Visualization of helical rotation of formins using single-molecule fluorescence polarization
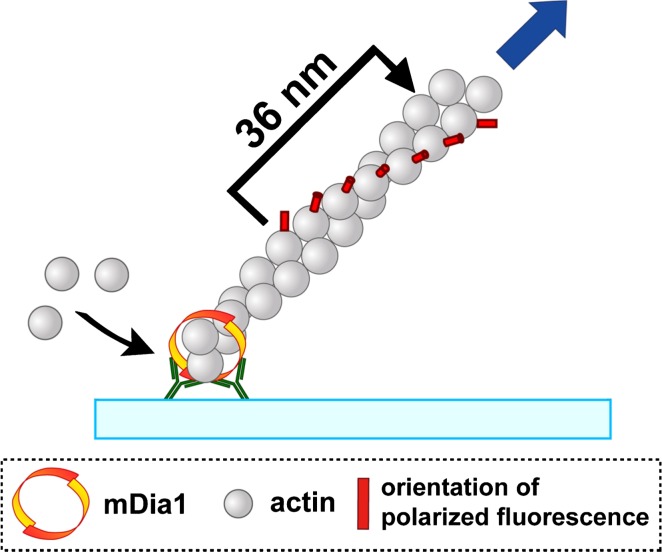
We visualized helical rotation of mDia1 using single-molecule fluorescence polarization11. Actin was labeled with tetramethylrhodamine at Cys-374 (TMR-actin). For the observation of single-molecules, we took images of F-actin containing TMR-actin at a low density. TMR-actin in the filament emits polarized fluorescence at an angle of 45 degree to the filament axis12 (Fig. 1). GST-mDia1 FH1-FH2 was anchored to the glass in protein aggregates composed of anti-GST antibodies and secondary antibodies. During processive elongation, incorporated actin subunits to the barbed end move directionally away from mDia1. We excited TMR-actin by non-polarized epi-fluorescence illumination, and the emitted light was separated through a polarizing beam splitter to the vertically polarized fluorescence (FLV) and the horizontally polarized fluorescence (FLH). When F-actin lying at an angle of 45 degrees in the view field rotates around the filament axis, FLV and FLH of TMR-actin alternatively become bright. We simultaneously measured the elongated filament length by displacement of single-molecule TMR-actin.
Figure 1.
Overview of visualization of rotational movement of an actin filament elongating from immobilized mDia1. GST-mDia1 FH1–FH2 was fixed in the protein aggregate composed of anti-GST antibodies and fluorescent secondary antibodies. The protein aggregate was adsorbed on the glass surface, and processive filament elongation was initiated by the addition of G-actin composed of TMR-actin (≈0.3%) and excess unlabeled actin. The vertically polarized fluorescence (FLV) and the horizontally polarized fluorescence (FLH) from TMR-actin in the filament were recorded after separated through a polarizing beam splitter.
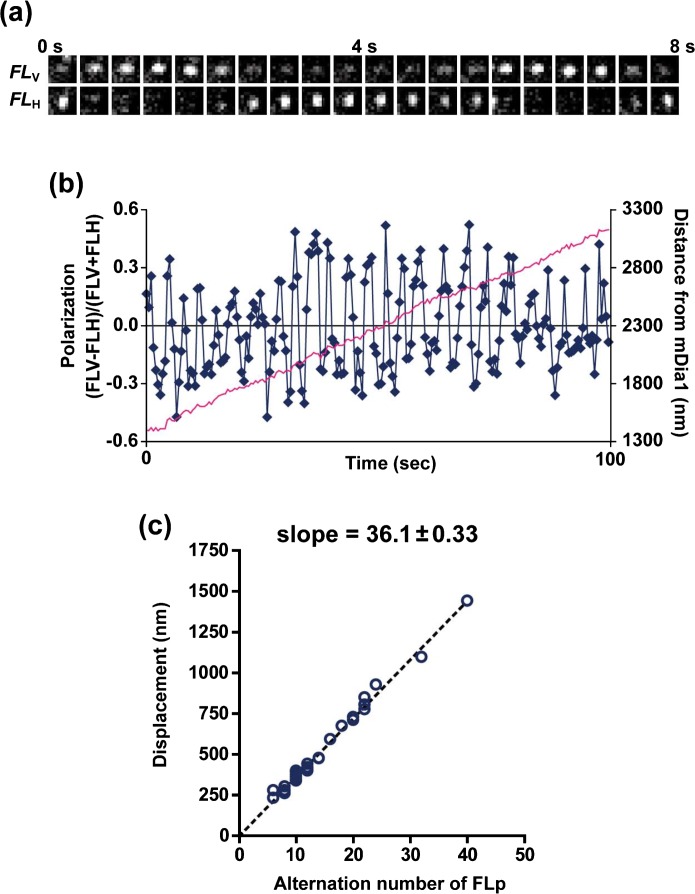
Figure 2 shows an example of fluorescence polarization measurement of an ATP-actin filament elongating from immobilized mDia1. The intensity of FLV and FLH of a TMR-actin alternated periodically (Fig. 2a). Figure 2b shows the time resolved measurement of fluorescence polarization (FLP) calculated by the intensity ratio of FLV and FLH (blue line) and displacement of a TMR-actin (pink line). The average distance per an alternation of FLP was 36.1 nm (Fig. 2c) which corresponds with the half-pitch length of F-actin10. These results indicate that mDia1 rotates along the long-pitch helical structure of F-actin during processive elongation. Helical rotation of mDia1 was also observed in the presence of profilin and during ADP-actin elongation.
Figure 2.
Alternation of fluorescence polarization (FLP) of TMR-actin in the filament processively elongating from mDia1. (a) Time-lapse images of a single-molecule TMR-actin. Upper and lower panels show images of the vertically polarized fluorescence (FLV) and the horizontally polarized fluorescence (FLH), respectively. (b) Time-course of FLP (blue) and displacement (pink) of a TMR-actin. FLP was calculated according to the equation, FLP= (FLV−FLH)/(FLV+FLH). (c) The average distance per an alternation of FLP was 36.1 nm with ATP-G-actin. Modified from ref. 11 with permission.
Cellular F-actin is highly cross-linked with each other as evidenced by the observations that relative positions of fluorescence labeled-actin subunits in the actin network remain nearly constant along the retrograde actin flow13. Therefore, formins should rotate when they execute processive actin elongation in the cell.
Filament elongation is blocked by interference of helical rotation of mDia1
In contrast to our conclusion, Kovar, et al. had concluded that formins slip around the barbed end like a bearing during processive actin elongation14. They challenged this issue by fixing the pointed end side of F-actin processively elongating from a yeast formin, Bni1p. Bni1p was adsorbed nonspecifically to the glass surface. They assumed that if glass-adsorbed Bni1p rotates along the long-pitch helix of a pointed end-fixed filament, torsional strain would accumulate in the filament and the filament should form a supercoil similar to a DNA supercoil. In their experiments, however, F-actin elongating from immobilized Bni1p continued to elongate for several microns after trapped by the glass surface and eventually buckled, forming a bent loop without supercoiling. These observations may result from slippage either between Bni1p and the filament barbed end or between Bni1p and the glass surface. The previous study did not discern which of these two possibilities facilitated the buckled filament elongation14.
We reinvestigated where in the F-actin the torsional stress generated by helical rotation of FH2 could be relaxed11. Also in the case of mDia1, continuous elongation of buckled F-actin from mDia1 was often observed upon capture of the pointed end side. We therefore compared the different methods for immobilization of FH2 on the glass surface. One used the mDia1 nonspecifically adsorbed to the glass and the other used the mDia1 anchored in protein aggregates composed of anti-GST and secondary antibodies bound to the glass (Ab-trapped mDia1). We measured the ratio of buckled elongating filaments and the total pointed end-trapped filaments among those processively elongated by mDia1. The buckling frequency with Ab-trapped mDia1 was substantially smaller than that with mDia1 nonspecifically adsorbed to the glass. Thus, continuous elongation of buckled F-actin from FH2 upon stuck of the pointed side on the glass surface is attributable to slippage between FH2 and the glass surface.
Notably, filament elongation from Ab-trapped mDia1 often arrested after the filament bent slightly when the pointed end side was trapped. Incorporation of actin monomer to the barbed end of double helical F-actin can impose two types of forces, pushing force and torsional force on F-actin. Processive elongation by single FH2 can generate force sufficient to buckle an actin filament14. Because elongation stopped after the filament bent only slightly with Ab-trapped, rigidly anchored mDia1, it is unlikely that arrest of F-actin elongation from Ab-trapped mDia1 is caused by impeding pushing force. We postulate that torsional stress imposed on F-actin by helical rotation arrested elongation.
Regardless of ATP- or ADP-actin elongation and presence or absence of profilin, mDia1 appears to faithfully follow the helical F-actin twist during processive elongation. Formins may therefore change their actin elongation speeds when anchored tightly to cellular structures. It is tempting to speculate that changing the degree of F-actin cross-linking might have an impact on actin polymerization activities of formins. The ‘screw capping’ of formins might provide a new type of controlling mechanisms for actin assembly in the cell.
The effects of profilin and nucleotides on formin-mediated actin elongation
In addition to helical rotation, our study revealed a pivotal role of ATP in formin-mediated actin elongation. Form-ins have the astounding property of elongating F-actin faster than the theoretical limit. Elongation of F-actin is a diffusion-limited reaction at the barbed end15. The elongation rate is equal to the frequency of collisions of diffusing G-actin with the end. Profilin-actin can assemble to the free barbed end as fast as G-actin. Formins accelerate actin elongation in the presence of profilin 5–15 fold8, which requires breakage of the diffusion limit of profilin-actin to the barbed end. The acceleration of elongation of formin-bound F-actin has been ascribed to an increase in the local concentration of profilin-actin at the barbed end through the binding of profilin and multiple poly-proline stretches in FH116. This could give rise to an increased frequency of collisions of profilin-actin complex with the barbed end.
Using ATP-actin, we observed marked acceleration of actin elongation with mDia1 and profilin in consistent with the previous studies8,17. However, we noted that profilin decelerates elongation of ADP-actin from mDia111. This finding differed from the previous results showing acceleration of filament elongation with both ATP- and ADP-actin8. We therefore reinvestigated the effect of profilin on ADP-actin elongation from mDia1.
Although previous studies including our own7,8,17 demonstrated processive actin elongation, none had observed the processive depolymerization of formin-bound F-actin under the microscope. Our study11 for the first time visualized processive depolymerization of formin-attached filaments. Using this system, we found that profilin accelerates depolymerization of mDia1-bound ADP-F-actin ≈5 fold. Thus, profilin has an ability to enhance the off-rate at the barbed end of ADP-F-actin bound to mDia1. The acceleration of depolymerization by profilin has also been observed at the free barbed end of ADP-F-actin18. Depolymerization of mDia1-bound ADP-F-actin, however, reaches maximum speed at the much lower profilin concentration (5 μM) than that required for the free barbed end18 (≈40 μM). Thus, depolymerization of mDia1-bound ADP-F-actin is effectively enhanced through the interaction between FH1 and profilin.
We further confirmed using inorganic phosphate (Pi) that profilin enhances the off-rate of ADP-F-actin also at the growing mDia1-bound barbed end. Pi binds the nucleotide binding pocket of actin subunits and is exchangeable in the filament. Pi binding abrogates actin dissociation at the free barbed end of ADP-F-actin19. Also in the case of mDia1-bound ADP-F-actin, a low concentration of Pi (≈1 mM) inhibits the accelerated depolymerization by profilin. During elongation of mDia1-bound F-actin, Pi recovers the elongation rate in the presence of profilin to the level in the absence of profilin. Thus, enhancement of the ADP-actin off-rate gives rise to the observed deceleration of mDia1-mediated ADP-actin elongation by profilin. Nucleotides bound to G-actin switch the function of profilin from an enhancer to a suppressor of formin-mediated actin elongation.
Interestingly, the on-rate calculated from the results with 5 μM ADP-actin and 6 μM profilin does not apparently differ from the on-rate of ADP-actin. For this calculation, we used 2.4 μM−1 s−1 for the on-rate of free ADP-actin (calculated from the rate of mDia1-mediated actin elongation with 0, 3 and 5 μM ADP-actin) and the dose-response curve of the depolymerization rate of mDia1-bound ADP-F-actin with various profilin concentrations11. We adopted various values (1–5 μM) for the dissociation constant of profilin and ADP-actin because this may vary depending on species and measuring methods20,21. With a wide range of the dissociation constants of profilin and ADP-actin from 1 to 5 μM, the on-rates of profilin-ADP-actin were calculated to be between 2.0 and 2.6 μM−1 s−1. Our data thus show that profilin-ADP-actin is incorporated to the mDia1-bould barbed end at a similar rate to ADP-actin.
Although biochemical parameters such as dissociation constants between profilin-ADP-actin and FH1 are unknown, ADP-actin does not seem to utilize the local concentration mechanism16 to drive faster actin elongation. Further analysis over the wide range of profilin-ADP-actin concentrations is required for precise understanding of the ineffectiveness of ADP-actin in this system. If the on-rate accelerates exclusively by ATP-actin, the formin-profilin system might accelerate actin elongation using a mechanism different from the local concentration mechanism.
Emerging questions about barbed end structures
ATP bound to G-actin is rapidly hydrolyzed after incorporation of G-actin to the barbed end while Pi is released slowly at the rate of 0.0022 s−1 after assembly22. It is unknown where ATP is hydrolyzed in F-actin. Several EM studies have investigated the difference between the structures of filaments polymerized from ATP- and ADP-actin. In one study, F-actin polymerized from ADP-actin was found partly thicker than the other portions23. However, the other studies pointed out that the observed disorganization of actin structures could be attributable to the prolonged depletion of ATP during preparation of ADP-G-actin which could denature actin irreversibly. Indeed, using ADP-G-actin quickly prepared, the other studies demonstrated that structures of F-actin polymerized from ATP- and ADP-actin are not markedly different24,25. On the other hand, these EM studies did not observe F-actin shortly after polymerization. A recent EM study reported that individual subunits in F-actin within 2 min after polymerization were substantially tilted compared to those in typical F-actin26,27. Notably, F-actin short after polymerization was slightly more twisted than typical F-actin.
From our published results11, we recollected the data for TMR-actins within 2 min after they assembled on the mDia1-bound barbed end. The distance per half-rotation was 36.2±0.84 nm (n=7 filaments, total 134 alternations). The structural periodicity of mDia1-assembled filaments from ADP-G-actin was 36.1 nm11. Thus, so far our analysis has not detected any change in the filament twist in association with both aging of F-actin and the difference between ATP and ADP.
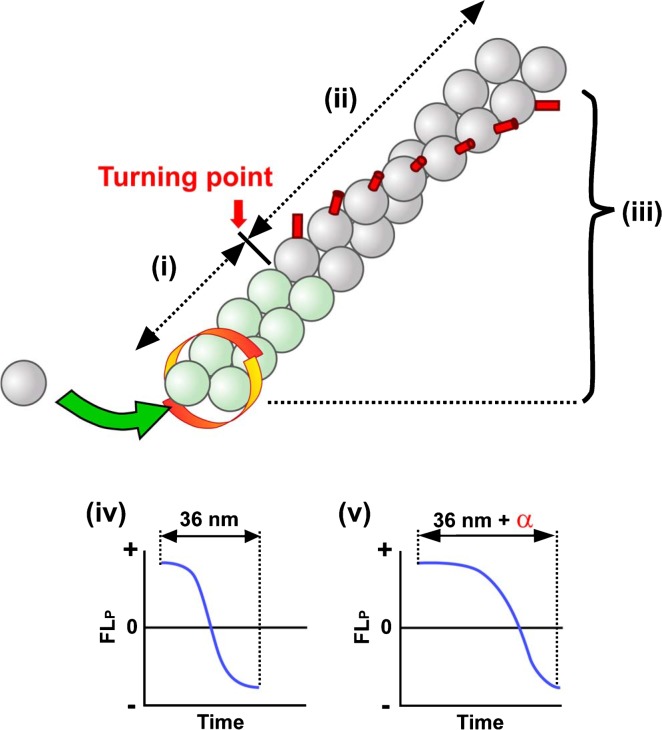
One remaining question is whether the filament twist very close to the barbed end is normal or deviates from typical one. It is of note that our FLP data do not necessarily reflect the local barbed end structure. For instance, if we assume that the filament forms a relaxed double-helical structure near the barbed end and this locally relaxed helix shifts to a typical helix at a certain turning point (Fig. 3), TMR-actins would move without periodical rotation (i) until they reach to the turning point (ii). However, after TMR-actins pass over the turning point (iii), they would rotate along the typical long-pitch helix. Thus, observing alternation of FLP away from the turning point does not provide any information regarding the local filament twist near the barbed end. To solve this issue (iv or v), one needs to detect FLP in a very short period after TMR-actins assembled to the barbed end. The time to detect the first alternation of FLP after assembly would be delayed if filament twist is relaxed locally. In practice, FLP detection with high time resolution and averaging of a large number of observations would be required for resolving the local barbed end structure. Our previous study11 analyzed FLP with time resolution of 0.5 sec intervals. The standard deviation of the position of immobile TMR-actin was as large as ±24.7 nm and ±23.7 nm in horizontal and vertical directions, respectively. To detect local FLP near the barbed end, much improved spatiotemporal resolution is required.
Figure 3.
FLP measurement near the barbed end bound is required for revealing local twist of the processively elongation filament. The diagram shows an assumptive structure of a formin-bound filament which forms a relaxed helical structure locally near the barbed end (green actins). In this scheme, it is assumed that the filament structure shifts from the relaxed helix to the typical actin long-pitch helix at the turning point. It is also assumed that the distance between the barbed end and the turning point is constant. When TMR-actin exists in the region of green actins (i), alternation of FLP would not be observed. When TMR-actin is distal to the turning point (ii), FLP from TMR would alternate periodically according to the local twist of distal potion of the filament (iii). To detect the barbed end filament twist, one needs to precisely measure the timing of FLP alternations after TMR-actin is incorporated to the barbed end. If the local filament twist near the barbed end is similar to the typical F-actin twist (i.e. zero length for the relaxed green actin portion), FLP would start alternations without any delay after assembly (iv). The time to detect the first alternation of FLP would be extended if the barbed end filament is locally relaxed for certain distance (v). In practice, one cannot know the exact initial direction of TMR-actin at assembly, and might therefore need to measure a large number of FLP delay times to see whether filament twist near the barbed end deviates from the typical long-pitch actin helix.
Electron microscopy (EM) and X-ray diffraction analysis are useful for detecting the conformational change of F-actin in detail. Indeed, two recent structural studies using cryo-EM28 and X-ray fiber diffraction29 have revealed flatting between two major domains of actin subunits upon transition from G- to F-actin. The structure of barbed ends bound to capping protein has also been solved by cryo-EM30 which showed the typical long-pitch helix. Combination of our FLP observation, which enables observing a live actin filament, with high resolution EM observation might elucidate the conformational change of the newly assembled barbed end filament associated with ATP hydrolysis and other regulatory processes.
On the other hand, several studies reported that binding of formins may induce the long-range allosteric effect on the F-actin. These studies observed the mobility of the probes attached to Cys-374 on actin using FRET31, fluorescence anisotropy decay32 and electron paramagnetic resonance33. The binding of formins to the barbed end was shown to enhance the flexibility of F-actin. However, these studies compared native F-actin with that assembled with high concentrations of mDia1. The latter condition could lead to the massive formation of short actin fragments. In addition, these studies included high concentrations of Cys-374-modified actin (>90%). For example, Cys-374 modification by TMR prevents actin from polymerization at the abundance ratio of >50%34. mDia1-assembled and the control actin might have contained different amounts of F-actin. Therefore, it is arguable that differences in the length or the amount of filaments might give rise to the distinct mobility of the actin attached probes.
In the co-crystal composed of FH2 and TMR-actin, actin is aligned without a helical twist, which differs from a left-handed short-pitch helix of the Holmes model9. The sheet-like actin ribbon was also observed in the co-crystal composed of profilin and β-actin35, although its geometrical feature was further deviated from F-actin. In the profilin-actin ribbon, a subdomain 2 of an actin monomer contacts with a subdomain 1 of the symmetrically-located neighbor actin monomer, but subdomains 3 and 4 lose their intra-strand contacts. It is tempting to speculate that the transient structure near the polymerizing formin-bound barbed end could be different from normal F-actin twist. In addition, mDia1 rapidly incorporates ATP-G-actin to the barbed end in the presence of profilin, at 5 times faster than spontaneous actin elongation8 and at 720 subunits/s in the cell7. Thus, formins can be used as a trick to obtain F-actin comprising more ATP-bound subunits near the barbed end if formins do not accelerate ATP hydrolysis. It is interesting to see whether the rapidly elongating formin-bound barbed end might exhibit unique morphologies.
Screw capping by formins in the cellular context
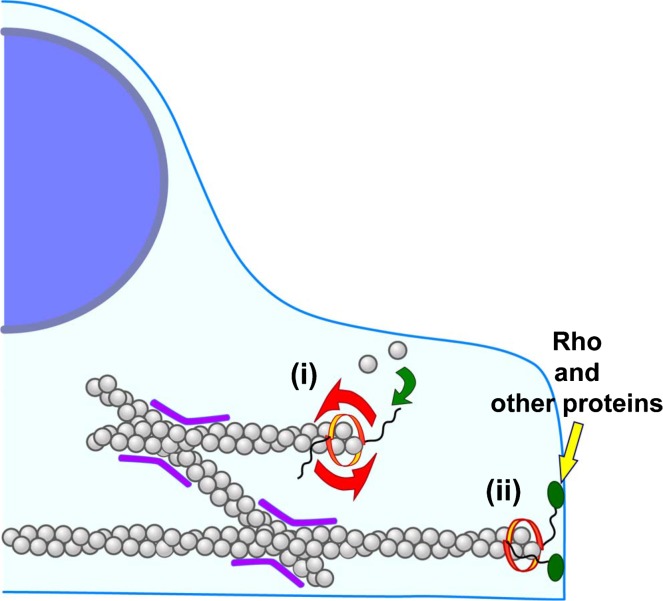
F-actin rotates when processively assembled by formins trapped in protein aggregates on the glass surface. Processive actin elongation is stopped when helical rotation is artificially arrested. Thus helical rotation of formins appears to tightly couple with processive actin elongation. Our results imply several potential functions of ‘screw capping’ of actin filaments by formins in the cell (Fig. 4).
Figure 4.
How screw capping of formins might function in the cell. F-actin cannot move freely in the cytosol by cross-linking (purple) and bundling. When freely moving formins elongate F-actin processively, formins rotate along the long-pitch helix of F-actin (i). When formins are anchored to the cellular structure through the binding of Rho or other proteins (ii), both processive actin elongation and stability of F-actin might be influenced. For example, in yeast, Bni1p and for3p are localized at the cell tip and thought to processively assemble actin cables2,40. Because actin cables continuously flow from the cell tip to the cell interior, Bni1p and for3p must be anchored loosely at the cell tip. In the case of animal cells, actin also undergoes continuous retrograde flow at the cell periphery. Although both mDia1 and mDia2 accumulate at the filopodium tip when expressed as activated truncated forms, full length proteins scarcely localized to the filopodium tip7,36,44,45. It remains unknown whether mammalian formins processively assemble F-actin while anchored to cellular structures. In vitro F-actin elongation from Ab-trapped mDia1 arrests without buckling when the pointed end side of F-actin is trapped11. We suggest that processive actin elongation may be regulated by the rigidity of anchorage of formins in the cell. On the other hand, incorporation of G-actin to the formin-bound barbed end may generate the torsional force to untwist the long-pitch helix of F-actin. The untwisting torsional strain may inhibit the binding of cofilin to F-actin because cofilin twists the long-pitch helix of F-actin42. Helical rotation of formins may thus have the potential to enhance the F-actin stability.
Among wild-type mDia1 visualized as single-molecules in XTC cells, a half is stationary (47.5%), and processively moving mDia1 constitutes a minor population36 (4.3%). It is also noteworthy that most of fluorescence signals of EGFP-wild-type mDia1 distribute diffusely in the cytoplasm. We currently do not know to which cellular structures stationary mDia1 is anchored and whether stationary mDia1 processively polymerizes F-actin or not. mDia1 has a Rho binding domain (RBD) in the N-terminus. Rho has the CAAX-motif and is anchored to the plasma membrane. mDia also binds anillin37 and Liprin α38 in the N-terminus downstream of RBD. Therefore, wild-type mDia1 may be anchored to the plasma-membrane through the binding to these regulatory proteins. Bni1p and for3p, which are responsible for actin cable assembly in budding yeast and fission yeast, form protein complexes with proteins such as tea1p and bud6p39 at the cell tip40,41. Therefore, if helical rotation of formins is inhibited by interactions within such protein complexes, these formins may stop or retard processive actin elongation.
Adversely, helical rotation may impose torsional force on F-actin. When a formin and the pointed end of F-actin are trapped, incorporation of G-actin to the barbed end should generate the torsional force to untwist the long-pitch helix of F-actin. Because the major actin depolymerizing factor cofilin binds the side of F-actin and twists the helical structure of filament42, the untwisting torsional strain imposed by helical rotation of formins might inhibit the binding of cofilin, which may in turn enhance the stability of F-actin. Recently, the binding of cofilin to F-actin was shown to be reduced upon stretch of F-actin, which results in the inhibition of severing activity of cofilin43. It is currently unknown whether F-actin stability is modulated by screw capping by formins both in vitro and in vivo.
Concluding remarks
We have revealed helical rotation of formins associated with processive actin polymerization and depolymerization. Formins might exert previously unrecognized functions by ‘screw capping’. The screw capping may affect not only the speed of processive actin elongation but also actin filament turnover in cells. On the other hand, our FLP detection method is a useful tool for revealing the barbed end structure of live actin filaments. The structural change in the proximity of the barbed end may provide important clues for understanding the role of ATP in actin treadmilling as well as the acceleration mechanism of formin/profilin-catalyzed fast actin elongation.
Acknowledgments
This work was supported by the Cabinet Office, Government of Japan through Funding Program for Next Generation World-Leading Researchers (LS013) and by grants from the Human Frontier Science Program and the Takeda Science Foundation.
References
- 1.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 2.Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- 3.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2838. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe N. Inside view of cell locomotion through single-molecule: fast F-/G-actin cycle and G-actin regulation of polymer restoration. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:62–83. doi: 10.2183/pjab.86.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- 7.Higashida C, Miyoshi T, Fujita A, Ocequera-Yanez F, Monypenny J, Andou Y, Narumiya S, Watanabe N. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- 8.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 9.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 10.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 11.Mizuno H, Higashida C, Yuan Y, Ishizaki T, Narumiya S, Watanabe N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 12.Sase I, Miyata H, Ishiwata S, Kinosita K., Jr Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proc Natl Acad Sci USA. 1997;94:5646–5650. doi: 10.1073/pnas.94.11.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 14.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drenckhahn D, Pollard TD. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J Biol Chem. 1986;261:12754–12758. [PubMed] [Google Scholar]
- 16.Vavylonis D, Kovar DR, O’Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero S, Didry D, Larguet E, Boisset N, Pantaloni D, Carlier MF. How ATP hydrolysis controls filament assembly from profilin-actin: implication for formin processivity. J Biol Chem. 2007;282:8435–8445. doi: 10.1074/jbc.M609886200. [DOI] [PubMed] [Google Scholar]
- 18.Bubb MR, Yarmola EG, Gibson BG, Southwick FS. Depolymerization of actin filaments by profilin. Effects of profilin on capping protein function. J Biol Chem. 2003;278:24629–24635. doi: 10.1074/jbc.M302796200. [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara I, Vavylonis D, Pollard TD. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc Natl Acad Sci USA. 2007;104:8827–8832. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perelroizen I, Carlier MF, Pantaloni D. Binding of divalent cation and nucleotide to G-actin in the presence of profilin. J Biol Chem. 1995;270:1501–1508. doi: 10.1074/jbc.270.4.1501. [DOI] [PubMed] [Google Scholar]
- 21.Vinson VK, De La Cruz EM, Higgs HN, Pollard TD. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 1998;37:10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- 22.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Ann Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 23.Janmey PA, Hvidt S, Oster GF, Lamb J, Stossel TP, Hartwig JH. Effect of ATP on actin filament stiffness. Nature. 1990;347:95–99. doi: 10.1038/347095a0. [DOI] [PubMed] [Google Scholar]
- 24.Pollard TD, Goldberg I, Schwarz WH. Nucleotide exchange, structure, and mechanical properties of filaments assembled from ATP-actin and ADP-actin. J Biol Chem. 1992;267:20339–20345. [PubMed] [Google Scholar]
- 25.Newman J, Zaner KS, Schick KL, Gershman LC, Selden LA, Kinosian HJ, Travis JL, Estes JE. Nucleotide exchange and rheometric studies with F-actin prepared from ATP- or ADP-monomeric actin. Biophys J. 1993;64:1559–1566. doi: 10.1016/S0006-3495(93)81525-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlova A, Shvetsov A, Galkin VE, Kudryashov DS, Rubenstein PA, Egelman EH, Reisler E. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc Natl Acad Sci USA. 2004;101:17664–17668. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kueh HY, Mitchison TJ. Structural plasticity in actin and tubulin polymer dynamics. Science. 2009;325:960–963. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 29.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 30.Narita A, Takeda S, Yamashita A, Maeda Y. Structural basis of actin filament capping at the barbed-end: a cryoelectron microscopy study. EMBO J. 2006;25:5626–5633. doi: 10.1038/sj.emboj.7601395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bugyi B, Papp G, Hild G, Lõrinczy D, Nevalainen EM, Lappalainen P, Somogy B, Nyitrai M. Formins regulate actin filament flexibility through long range allosteric interactions. J Biol Chem. 2006;281:10727–10736. doi: 10.1074/jbc.M510252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papp G, Bugyi B, Ujfalusi Z, Barkó S, Hild G, Somogy B, Nyitrai M. Conformational changes in actin filaments induced by formin binding to the barbed end. Biophys J. 2006;91:2564–2572. doi: 10.1529/biophysj.106.087775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kupi T, Gróf P, Nyitrai M, Belágyi J. The uncoupling of the effects of formins on the local and global dynamics of actin filaments. Biophys J. 2009;96:2901–2911. doi: 10.1016/j.bpj.2008.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudryashov DS, Phillips M, Reisler E. Formation and destabilization of actin filaments with tetramethylrhodamine-modified actin. Biophys J. 2004;87:1136–1145. doi: 10.1529/biophysj.104.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schutt CE, Myslik JC, Roxycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 36.Higashida C, Suetsugu S, Tsuji T, Monypenny J, Narumiya S, Watanabe N. G-actin regulates rapid induction of actin nucleation by mDia1 to restore cellular actin polymers. J Cell Sci. 2008;121:3403–3412. doi: 10.1242/jcs.030940. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, Arakawa T, Kinoshita M, Ishizaki T, Narumiya S. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell. 2010;21:3193–3204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakamoto S, Ishizaki T, Okawa K, Watanabe S, Arakawa T, Watanabe N, Narumiya S. Liprin-α controls stress fiber formation by binding to mDia and regulating its membrane localization. J Cell Sci. 2012;25:108–120. doi: 10.1242/jcs.087411. [DOI] [PubMed] [Google Scholar]
- 39.Feierbach B, Verde F, Chang F. Regulation of a formin complex by the microtubule plus end protein tea1p. J Cell Biol. 2004;165:697–707. doi: 10.1083/jcb.200403090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 41.Buttery SM, Yoshida S, Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell. 2007;18:1826–1838. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;95:721–727. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol. 2003;13:534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- 45.Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, Lappalainen P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]