Abstract
Free-energy landscape is an important quantity to study large-scale motions of a biomolecular system because it maps possible pathways for the motions. When the landscape consists of thermodynamically stable states (low-energy basins), which are connected by narrow conformational pathways (i.e., bottlenecks), the narrowness slows the inter-basin round trips in conformational sampling. This results in inaccuracy of free energies for the basins. This difficulty is not cleared out even when an enhanced conformational sampling is fairly performed along a reaction coordinate. In this study, to enhance the inter-basin round trips we introduced a virtual state that covers the narrow pathways. The probability distribution function for the virtual state was controlled based on detailed balance condition for the inter-state transitions (transitions between the real-state basins and the virtual state). To mimic the free-energy landscape of a real biological system, we introduced a simple model where a wall separates two basins and a narrow hole is pierced in the wall to connect the basins. The sampling was done based on Monte Carlo (MC). We examined several hole-sizes and inter-state transition probabilities. For a small hole-size, a small inter-state transition probability produced a sampling efficiency 100 times higher than a conventional MC does. This result goes against ones intuition, because one considers generally that the sampling efficiency increases with increasing the transition probability. The present method is readily applicable to enhanced conformational sampling such as multi-canonical or adaptive umbrella sampling, and extendable to molecular dynamics.
Keywords: bottleneck, pathway, enhanced sampling, multicanonical, adaptive umbrella
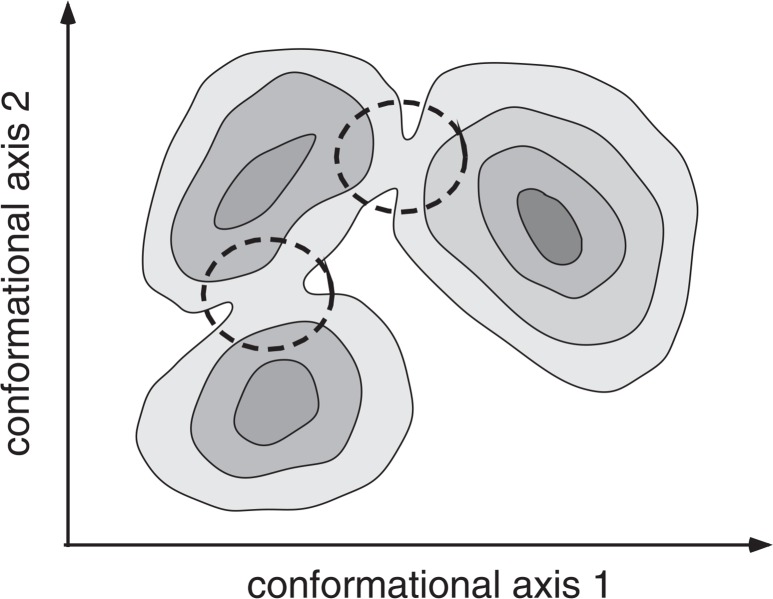
Computer simulation is now widely used to explore the biomolecular conformational space. Free-energy (or energy) landscape is an important quantity obtainable from the simulation. The landscape provides distribution of thermodynamically stable states (low-energy basins) and pathways connecting the stable states. When the sampling is achieved in a wide conformational space, the landscape can be a road map for large-scale motions such as protein folding or protein-ligand binding1–5. Figure 1 shows schematically the landscape, where the conformation passes through regions circled by broken lines, when an inter-basin transition occurs.
Figure 1.
Schematic drawing for free-energy (or energy) landscape. Three low-energy basins are shown with iso-free-energy contour lines, and broken-line circles indicate bottlenecks that connect the low-energy basins.
To accurately estimate the free-energy difference between the low-energy basins, the conformational sampling should substantialize a number of transitions among the low-energy basins. However, when narrow pathways (bottlenecks) connect the low-energy basins, the frequency of transitions decreases, and the conformational sampling takes a long computing time to estimate accurately the probability (free energy) of each basin. Thus, generally, the computing time for the accurate free-energy estimation increases with narrowing the bottlenecks because the bottlenecks prevent inter-basin traveling. This difficulty lies in many sampling problems of biomolecular systems.
A generalized ensemble (GE) method, such as multi-canonical sampling6–10 or adaptive umbrella sampling11,12, generates an even (i.e. flat) probability distribution along a reaction coordinate. The flatness ensures that the conformational space is sampled widely along the reaction coordinate. In other words, the sampling is enhanced along the reaction coordinate. Thus, one may imagine that the GE method can cause the passing through the bottlenecks effectively. However, we have shown that a fairly performed GE method provides even worse sampling efficiency than a conventional method when the conformational space on the reaction coordinate becomes narrow where the entropy suddenly decreases (i.e., the bottleneck appears on the pathway)13. Even the reaction coordinate is well designed so that the conformational changes along the reaction coordinate provide natural passage through the bottleneck, the conformation may be sluggish for long time in a basin before detecting the bottleneck.
In this study, we introduce a virtual state in the conformational space to ease passing the bottlenecks. Although this method is developed for sampling the biological systems, here we apply it to a simple system, which consists of two basins connected by a bottleneck. Because of the simplicity of the system, the free energies of the basins are computable analytically. We perform two Monte Carlo (MC) simulations called a “real-state MC” and a “virtual-state coupled MC”. The real-state MC is a conventional sampling method without the virtual state. In the virtual-state coupled MC, the virtual state covers the bottleneck. We show that the virtual state considerably enhances the inter-basin round trips.
Methods
Figure 1 is a scheme for the free-energy landscape of a biological system: See also Figure 1b of Ref. 3, which is the free-energy landscape of a β-hairpin peptide in explicit water. Thus, to increase the accuracy of the free energies for the low-energy basins, frequency of passing through the bottlenecks should be increased. To develop a useful method, we introduce a simple model explained below. A benefit of the simple model is that one can estimate analytically the free energies of the basins. We impose an important requirement on the model: a transition among the low-energy basins occurs through a narrow pathway. By varying the narrowness of the pathway, we can assess how our method is effective.
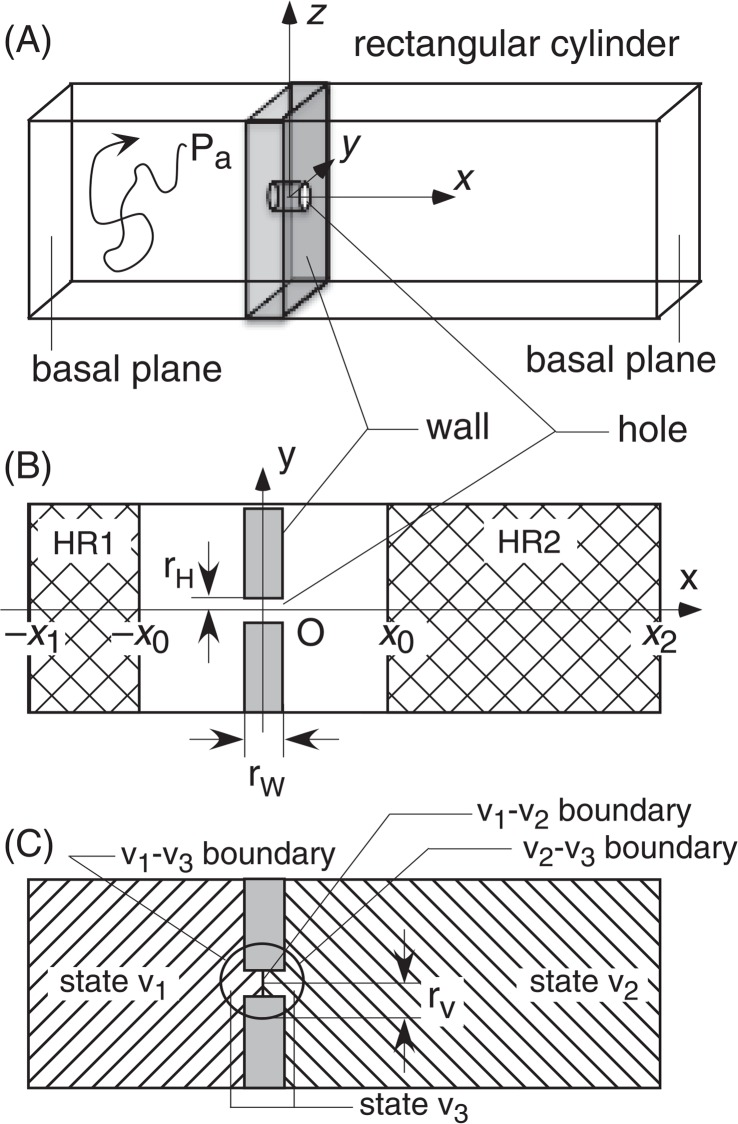
Figure 2 is the simple model, where the inner cavity of a rectangular cylinder is divided into two regions by a wall, and a narrow hole (circular cylinder-shaped hole) is pierced at the center of the wall (see Fig. 2A and B). The two wide regions, denoted as “state v1” and “state v2” in Figure 2C, resemble the low-energy basins, and the narrow hole does the bottleneck. A particle Pa is confined inside a rectangular cylinder and moves during a simulation to estimate free energies of the states v1 and v2. Thickness of the wall and radius of the hole are denoted as rW and rH, respectively (see Fig. 2B). We define a cartesian coordinate system (see Fig. 2) so that the x-axis is the rectangular cylinder axis. The y-axis is parallel to one side of the basal planes of rectangular cylinder and the z-axis to the other side: The origin is set at the body center of the hole (i.e., the body center of the wall). The basal planes for states v1 and v2 are defined by x=−x1 (x1>0) and x =−x2 (x2>0), respectively. The position of Pa is referred as to r = [x, y, z]. For simplicity, we set the potential energy to be constant in the movable region of Pa. Therefore, the free-energy barrier caused by the hole is purely an entropic barrier.
Figure 2.
(A) Overview of system. Winding line illustrates motion of particle Pa. (B) Intersection of the system (panel A) on the x-y plane. (C) Location of the virtual state v3.
First, we performed a conventional MC simulation and estimated numerically the volumes of the states v1 and v2, which are denoted Vv1 and Vv2, respectively. We call this sampling “real-state MC”. Since the potential energy is constant, Pa can move unconditionally in the movable region. To count the round trips of Pa between v1 and v2, we introduced two regions, HR1 and HR2 (see netted areas in Fig. 1B), expressed by inequalities x≤−x0 and x≥x0 (x0>0), respectively. After Pa visits HR1 (or HR2), we wait till Pa returns to HR1, during which Pa visits HR2 at least once. We count this move as a round trip. When Pa has returned to HR1 without visiting HR2 (even with visiting state v2), this move is incomplete as a round trip. Then we wait further till Pa returns after visiting HR2. We denote the number of round trips from a long simulation as NRT.
Analytical values for Vv1 and Vv2 are given as:
| (1) |
where S is the area of the bases planes. The free-energy difference between v1 and v2 is: . We did not involve temperature in this expression by setting as kBT = 1, because the potential energy is always constant in the movable region (i.e., Vv1 and Vv2 are independent of temperature). To check the convergence of sampling, we introduce a quantity, volume-ratio convergence, as:
| (2) |
where is the numerically estimated volume for state vi using a partial simulation trajectory from 0 to t steps. We denote a partial trajectory from t1 to t2 steps as I[t1, t2]. Practically, is replaced by the number of snapshots where Pa exists in vi in I[0, t].
Next, we introduce a spherical state v3 centered at the coordinate origin with radius of rV (Fig. 1C), where the left half (x<0) of v3 overlaps with v1, and the right half (x ≥0) with v2. Therefore, v3 is not a substantial state separated from v1 and v2 but a virtual state. Here, we introduce a virtual-state coupled MC simulation as follows: Suppose that Pa starts from a position in state v1. During an interval I [0, Δt], we confine Pa to stay in v1: When Pa is passing the v1–v2 boundary, this move is rejected, although Pa can pass the v1–v3 boundary freely. If Pa is inside of the v1–v3 boundary at the last step of I[0, Δt], Pa may transition to v3 with a transition probability of p1→3 (the actual values for the transition probability is given later). Note that this inter-state transition alters the attribution of Pa (i.e. the state specifier) from v1 to v3 without changing the position of Pa in the rectangular cylinder. If Pa is outside the v1–v3 boundary at the last snapshot, no transition occurs. The particle Pa is confined again in v1 during the next interval I[Δt, 2Δt], and the state transition is examined at the last step of I[Δt, 2 Δt].
Once the inter-state transition (v1→v3) has been accepted at the end of I[0, Δt], then Pa is confined in v3 during the interval of I[Δt, 2Δt]. Now, Pa can pass the v1–v2 boundary freely. However, moves toward outside of the v1–v3 and v2–v3 boundaries are rejected. At the last step of I[Δt, 2Δt], the state transition is examined as follows: Pa may return back to v1 with a probability of p3→1 when Pa is in the region of x<0. On the other hand, Pa may transition to v2 with a probability of p3→2 when Pa is in the region of x>0. Suppose that Pa has transitioned to v2. Then Pa is confined in v2 during the interval of I[2Δt, 3Δt], where Pa can pass the v2–v3 boundary freely, but moves passing the v1–v2 boundary are rejected. At the last step of I[2Δt, 3Δ], Pa may transition to v3 with a transition probability of p2→3 when Pa is in the region of r≤ rV, where r = [x2+y2+z2]1/2. Otherwise, Pa stays in v2 for the next interval I[3Δt, 4Δt]. By this way, the inter-state transition is examined at steps n Δt (n=1, 2, ...).
Since the motion of Pa from r to r′ within vi is unconditionally accepted, a long simulation yields an equation:
| (3) |
where P(r, vi) is the probability distribution function of Pa at position r in vi. An inter-state transition from vi to vj at a position r is controlled by a transition probability pi→j. Then, the long simulation yields an equation:
| (4) |
Combining Eqs. 3 and 4, the probability distribution function satisfies the following equation:
| (5) |
where r1 and r2 are arbitrary positions in v1 and v2, respectively. Equation 5 indicates that the states v1 and v2 are indirectly linked via the virtual state v3.
We note that Eq. 5 is not an equation to determine the absolute values for the inter-state rate constants. Equation 5 can be rewritten as:
| (6) |
where the inter-state transition probabilities are redefined as:
| (7) |
where the coefficient k is any positive value. With increasing k, the rate constant increases, and the simulation length to reach equilibrium becomes short. In MC scheme, then, the largest rate constant (the quickest convergence) may be result from pi→j = 1. The parameter Δt also changes the rate constants: the larger the Δt, the longer the simulation length to obtained an equilibrated probability distribution function.
Based on the above discussion, the parameter set of [pi→j, Δt]=[1, 1] may be the best to speed up the round trips. Note that this parameter set is similar with the simulation condition of the real-state MC (i.e., the conventional MC). In fact, we show below that the smaller the Δt, the quicker the convergence. However, against our better instincts, the larger the pi→j, the worse the sampling efficiency for a narrow-hole system.
Results and discussion
We set the system parameters (non-dimensional quantities) as: S=2.02=4, rW= 0.2, x1= 1.0, x2= 2.0 and x0= 0.5. Quantities rH, Δt, and pi→j are specific to the virtual state. We examined several values for rH and Δt: rH= {0.005, 0.01, ..., 0.16}={d×20, d×21, ..., d×25} where d=0.005, and Δt= {1, 10, ..., 100000}={100, 101, ..., 105}. For simplicity, the transition probabilities from the virtual state to the real states are set to a single value pt, and those from the real states to the virtual state to 1.0: I.e., p3→1 = p3→2= pt (0< pt≤ 1) and p1→3= p2→3 = 1.0. We examined ten pt values as: pt= {2−9, 2−8, ..., 2−1, 1}. The size of the virtual state rV was set to 0.3. The total MC length (number of trials to move Pa) is 5 × 1011 steps for all simulations.
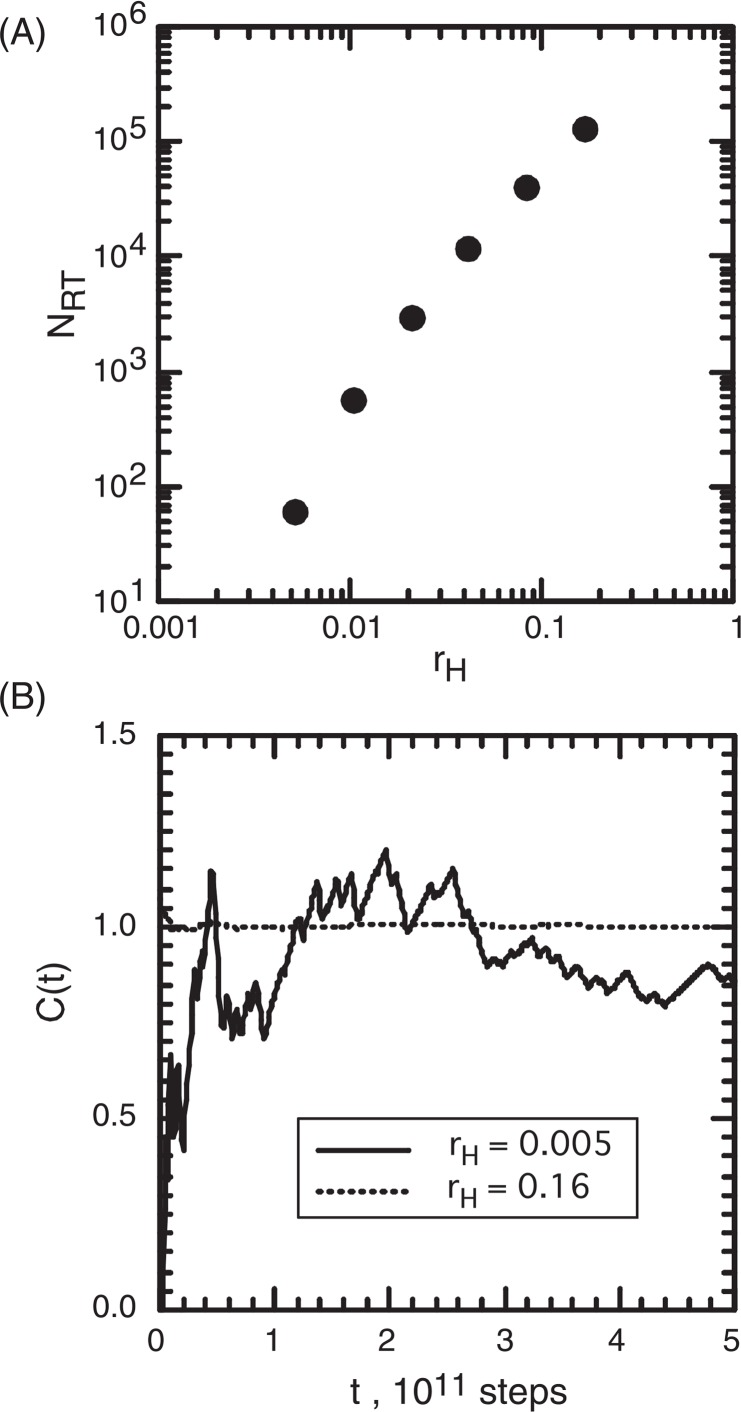
In the real-state MC, NRT rapidly decreased with decreasing rH (Fig. 3A). Figure 3B demonstrates the volume-ratio convergence C(t) for rH= 0.16 and rH= 0.005. The convergence was quick for rH= 0.16 and slow for rH= 0.005.
Figure 3.
Real-state MC. (A) Relation between rH and NRT, and (B) volume-ratio convergence, C(t), for two rH values.
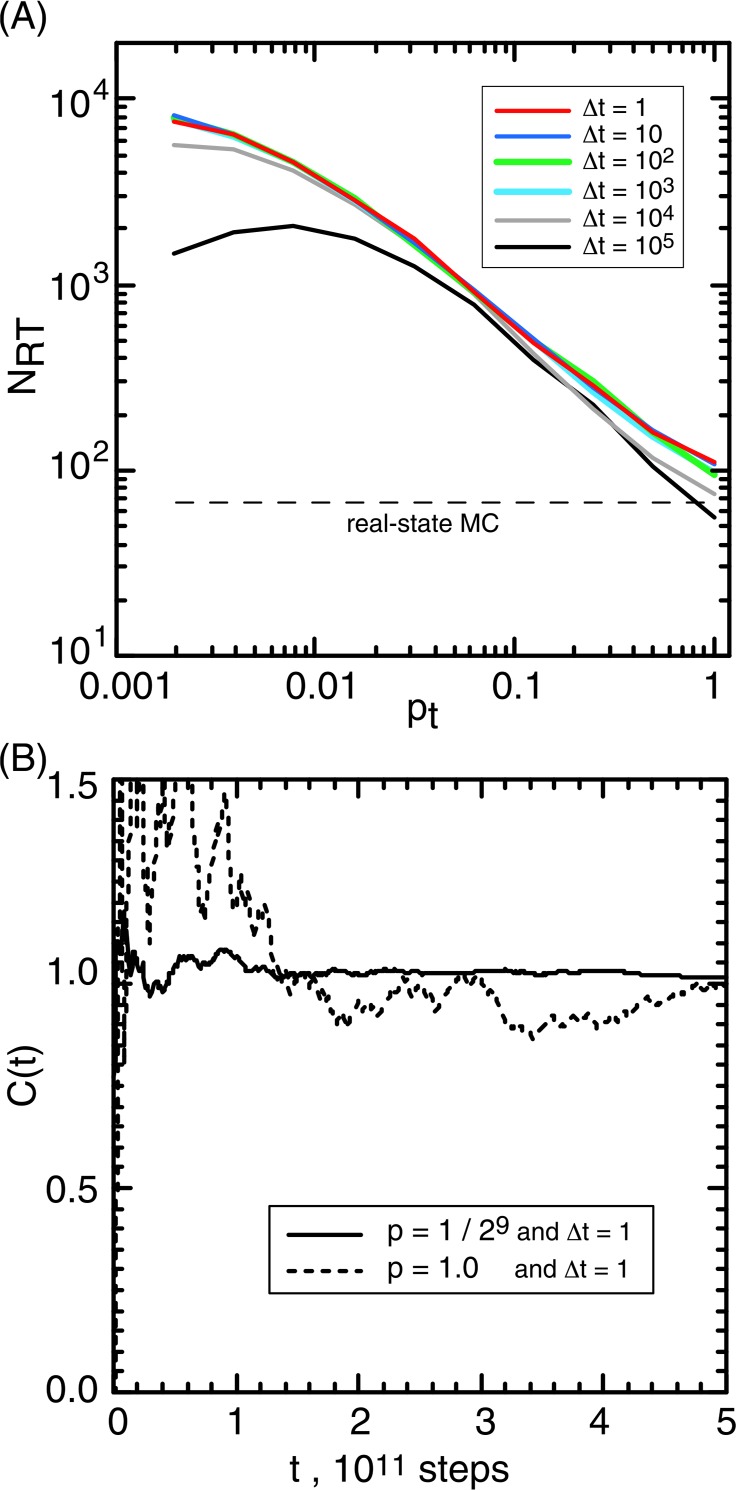
Current study presents a recipe to enhance the sampling by introducing the virtual state for a narrow-hole system. The pt–NRT relation (Fig. 4A) for the narrowest-hole system (rH= 0.005) at various Δt manifests that the virtual state enhances the sampling because NRT is larger than that from the real-state MC (broken line). The only exception was found at [pt, Δt]=[1, 105]. For Δt≤104, NRT increased monotonically with decreasing pt. The highest efficiency was found at pt= 2−9, where NRT was about 100 times larger than that from the real-state MC. Figure 4B plots C(t) from [pt, Δt] = [2−9, 1] and [1, 1]. The convergence was quick for pt= 2−9 and slow for pt= 1. The mechanism for the enhancement is simple: With decreasing pt, the probability P(r, v3) of Pa in the virtual state increases, and accordingly the hole-passing chance increases.
Figure 4.
Virtual-state coupled MC for rH = 0.005. (A) Relation between pt and NRT at the various Δt. Broken line represents NRT from real-state MD. (B) Volume-ratio convergence, C(t), from simulations with [pt, Δt] = [2−9, 1] and [1, 1].
The black solid line (Δt=105) of Figure 4A had different behavior from the other lines (Δt ≤104): NRT had a peak at pt= 2−7. Let us consider a situation that pt falls to zero (pt → 0). In this extremity, NRT should decay to zero (NRT → 0) because Pa cannot escape from v3 once Pa is trapped in v3. Then, we get two inequalities: dNRT/dpt|pt=0 >0 and dNRT/dpt|pt=1<0. These inequalities result in that NRT has a peak. The NRT showed no peak for Δt≤104 because the peak position is below pt= 2−9. Here, one may raise a question: Why does the peak position for Δt =105 was larger than those for the other Δt? This is because increment of Δt decreases the inter-state transitions (the rate constants). In this regime, a large pt (transition probability from v3 to v1 or v2) plays a role of enhancer for inter-state transitions. Accordingly, the peak position shifts positively.
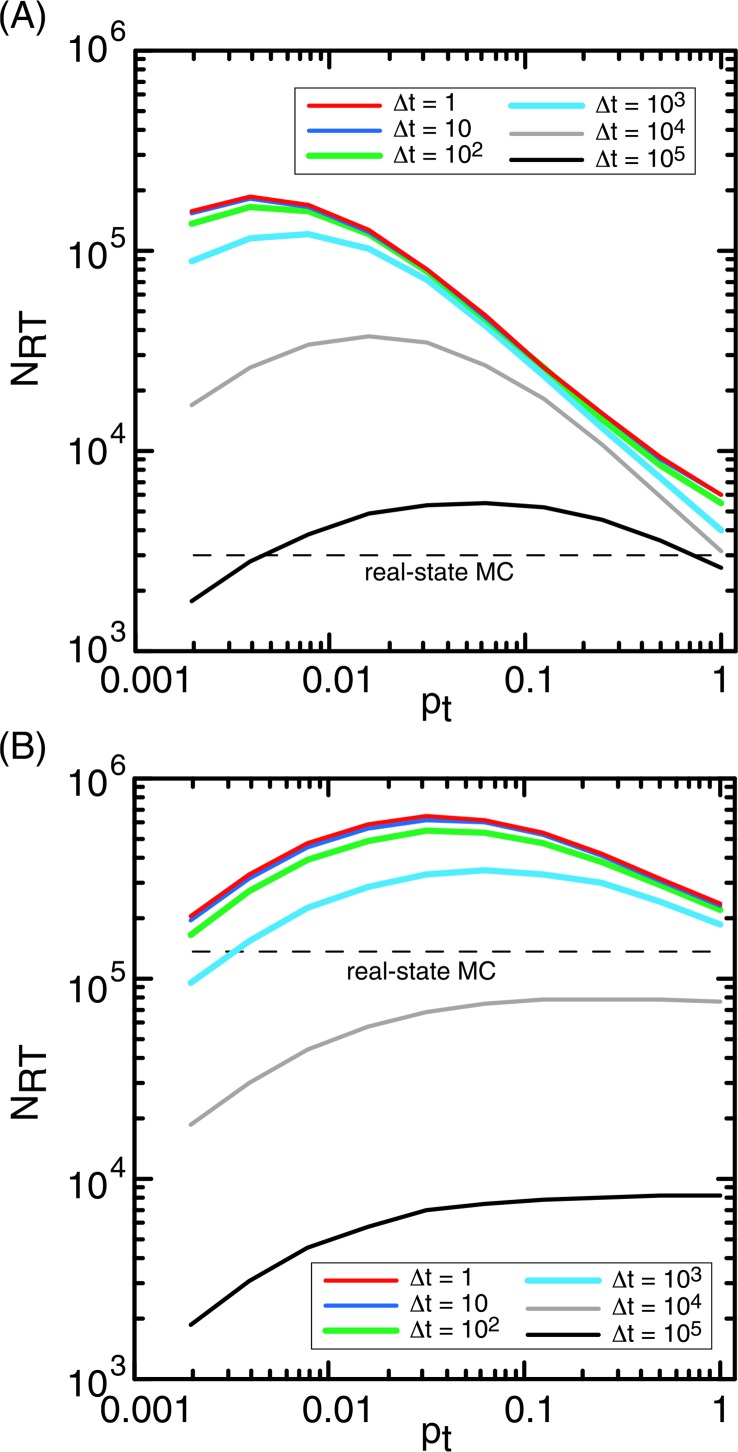
The positive shift of the peak position is clearly shown for a middle hole rH= 0.02 (Fig. 5A): All curves had a peak, and the peak position shifted positively with increasing Δt. Figure 5B demonstrates the pt−NRT relation for the largest hole (rH= 0.16), where the peak position was merge into the edge of pt (pt= 1) for Δt≥104.
Figure 5.
Relation between pt and NRT for rH = 0.02 (A) and rH = 0.16 (B).
The virtual-state coupling is readily applicable to GE methods, such as multicanonical or adaptive umbrella sampling. The GE enhances the sampling along a reaction coordinate x with introducing an effective potential energy Eeff = E+kBT ln[Pc(x, T)], where E is the original potential energy, Pc(x, T) a canonical distribution of x at temperature T, and kB the Boltzmann constant. The reaction coordinate is a function of position (or conformation) r: x=x(r). The positional transition probability from r1 to r2 is simply given by exp[−ΔEeff/kBT], where ΔEeff = Eeff (x(r2)) − Eeff (x(r1)). The phase point fluctuating in the entire conformational space may spend a long time before running into the narrow bottleneck8. One can set the virtual state covering the bottleneck, where the virtual volume is larger than the bottleneck size. Then, the chance that the phase point finds the bottleneck increases. In the virtual state, the phase point can find bottleneck readily because the phase point is confined in the virtual state for a while.
We note that the current method is expandable readily to molecular dynamics (MD) by computing an effective force acting on atom i as: fi = −grad[Eeff (x, T)]9. With introducing the virtual state, a canonical MD simulation at temperature T with becomes virtual-state coupled multi-canonical or adaptive umbrella sampling. Conformational changes in each time interval Δt are done by MD scheme, and the inter-state transitions examined at the end of time intervals (t =nΔt) are achieved by MC scheme.
In the present study, the bottleneck position in the conformational space is known in advance. However, the bottleneck position is generally unknown a priori. Then, before introducing the virtual state, pre-sampling is required, which may be a “real-state” multicanonical or adaptive umbrella sampling. In general, shape and volume of the virtual state are arbitrary depending on the system.
The current model (Fig. 2) was defined in the three-dimensional space. Then, one may doubt if the current method is useful for biological systems, because the biological systems are defined in a high-dimensional space. However, as shown in all-atom model of polypeptides1–5, the bottlenecks are well identified by projecting the high-dimensional distribution in a low-dimensional (2D or 3D) conformational space.
Last, we note that the virtual state acts as a lens to view the probability distribution function at the bottleneck. To increase the events passing through the bottleneck, we set pt to be small, which makes the probability distribution for the virtual state large. Thus, the virtual-state coupled sampling can be used to estimate both free energies of major basins and the bottlenecks (i.e., free-energy barriers).
Acknowledgments
H.N. was supported by Grant-in-Aid for Scientific Research (B) (23370071) from the Japan Society for the Promotion of Science. J. H. was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (21113006) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Japan. J. H. and H. N. were supported by grants from the New Energy and Industrial Technology Development Organization (NEDO) Japan.
References
- 1.Ono S, Nakajima N, Higo J, Nakamura H. The multicanonical weighted histogram analysis method for free energy landscape along structural transition paths. Chem Phys Lett. 1999;312:247–254. [Google Scholar]
- 2.Higo J, Galzitskaya OV, Ono S, Nakamura H. Energy landscape of a β-hairpin peptide in explicit water studied by multicanonical molecular dynamics. Chem Phys Lett. 2001;337:169–175. [Google Scholar]
- 3.Kamiya N, Higo J, Nakamura H. Conformational transition states of a β-hairpin peptide between the ordered and disordered conformations in explicit water. Protein Sci. 2002;11:2297–2307. doi: 10.1110/ps.0213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamiya N, Mitomo D, Shea J-E, Higo J. Folding of the 25 residue Aβ(12–36) peptide in TFE/Water: Temperature dependent transition from a funneled free-energy landscape to a rugged one. J Phys Chem B. 2007;111:5351–5356. doi: 10.1021/jp067075v. [DOI] [PubMed] [Google Scholar]
- 5.Higo J, Nishimura Y, Nakamura H. A free-energy landscape for coupled folding and binding of an intrinsically disordered protein in explicit solvent from detailed all-atom computations. J Am Chem Soc. 2011;133:10448–10458. doi: 10.1021/ja110338e. [DOI] [PubMed] [Google Scholar]
- 6.Berg BA, Neuhaus T. Multicanonical ensemble: A new approach to simulate first-order phase transitions. Phys Rev Let. 1992;68:9–12. doi: 10.1103/PhysRevLett.68.9. [DOI] [PubMed] [Google Scholar]
- 7.Hansmann UHE, Okamoto Y. Prediction of peptide conformation by multicanonical algorithm: New approach to the multiple-minima problem. J Comp Chem. 1993;14:1333–1338. [Google Scholar]
- 8.Kidera A. Enhanced conformational sampling in Monte Carlo simulations of proteins: application to a constrained peptide. Proc Natl Acad Sci USA. 1995;92:9886–9889. doi: 10.1073/pnas.92.21.9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima N, Nakamura H, Kidera A. Multicanonical ensemble generated by molecular dynamics simulation for enhanced conformational sampling of peptides. J Phys Chem B. 1997;101:817–824. [Google Scholar]
- 10.Iba Y, Chikenji G, Kikuchi M. Simulation of Lattice Polymers with multi-self-overlap ensemble. J Phys Soc Jpn. 1998;67:3327–3330. [Google Scholar]
- 11.Paine GH, Scheraga HA. Prediction of the native conformation of a polypeptide by a statistical-mechanical procedure. I. Backbone structure of enkephalin. Biopolymers. 1985;24:1391–1436. doi: 10.1002/bip.360240802. [DOI] [PubMed] [Google Scholar]
- 12.Mezei M. Adaptive umbrella sampling: Self-consistent determination of the non-Boltzmann bias. J Comp Phys. 1987;68:237–248. [Google Scholar]
- 13.Higo J, Ikebe J, Kamiya N, Nakamura H. Enhanced and effective conformational sampling of protein molecular systems for their free energy landscapes. Biophys Rev. 2012;4:27–44. doi: 10.1007/s12551-011-0063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]