Abstract
Comparison and classification of protein structures are fundamental means to understand protein functions. Due to the computational difficulty and the ever-increasing amount of structural data, however, it is in general not feasible to perform exhaustive all-against-all structure comparisons necessary for comprehensive classifications. To efficiently handle such situations, we have previously proposed a method, now called GIRAF. We herein describe further improvements in the GIRAF protein structure search and alignment method. The GIRAF method achieves extremely efficient search of similar structures of ligand binding sites of proteins by exploiting database indexing of structural features of local coordinate frames. In addition, it produces refined atom-wise alignments by iterative applications of the Hungarian method to the bipartite graph defined for a pair of superimposed structures. By combining the refined alignments based on different local coordinate frames, it is made possible to align structures involving domain movements. We provide detailed accounts for the database design, the search and alignment algorithms as well as some benchmark results.
Keywords: protein structure comparison, relational database, Hungarian algorithm, protein-ligand interaction
Comparison and classification of protein structures are important steps towards understanding the principles of protein structures and functions. Accordingly, there have been many methods for comparing protein structures1–8 (see Eidhammer et al.9 for review) and there are standard databases of protein taxonomy such as SCOP10 and CATH11. When studying protein functions, it is important to notice that protein functions may not necessarily be intrinsic properties of proteins since proteins need to interact with other molecules ranging from single atom ions to macromolecules in order to execute their functions. Therefore, structural studies of protein functions must be based on the mode of interactions, and there are already a number of comparative studies of interaction sites of proteins12–39.
A classification study of protein structures naturally requires comparing a large number of structures. At present, there are nearly 80,000 entries in the Protein Data Bank (PDB)40,41, and the amount of data is still increasing due to the increased efficiency of structure determination as well as to technological development for solving large complex structures. When ligand interaction sites including those for small molecules, proteins and nucleic acids are considered, the number is even higher by an order of magnitude. Since structure comparison is a computationally difficult problem9,42, it is often assumed necessary to reduce the data size by making a “representative” set of structures by removing redundancy (usually based on homologous relationships and experimental qualities) even when efficient (approximate) algorithms are employed. However, when atomic details are to be examined, the structural diversity of closely homologous or even identical proteins may not be negligible. This is especially true when one is interested in different interaction states of a protein bound with different types of ligands. Therefore, it becomes necessary to take into account all the available structures without removing any redundancy to achieve a detailed classification of interaction site structures.
In order to compare the ever-increasing amount of interaction site structures in the PDB, we have previously developed an extremely efficient method that exploits a relational database (RDB) management system and its indexing mechanism29. This method was later named GIRAF (Geometric Indexing and Refined Alignment Finder) and, with some further improvements, has been applied to comprehensive structural classifications of interaction sites for small molecules33 and proteins34 as well as the combinations of various types of binding sites39. This article is a detailed account of the most recent implementation of the GIRAF method. The main improvements include the simplified and integrated RDB schema for facilitating functional annotations combined with structure similarity search, improved geometric indexing (GI) search by using a rich set of structural features associated with each backbone-based local coordinate frame, and flexible alignment that can handle domain movements. With these improvements, it is made possible to search through a database containing hundreds of thousands of interaction site structures in several seconds to a few minutes on a modern desktop computer with multi-core CPUs.
Materials and Methods
Since the first presentation29, the GIRAF method has been subject to many significant improvements, which include the design of its back-end database, structural features for geometric indexing, and the alignment algorithm.
Before proceeding to the details of the method, let us introduce some technical terms. A template structure is a binding site structure that is stored in the GIRAF database. The GIRAF database is a relational database in which structural data extracted from the PDB are stored and indexed. A query structure is a protein subunit structure, or a part thereof, for which similar templates in the GIRAF database are to be searched.
In the following, most of the parameters were chosen in such a way that some anecdotal examples12,15,29 were covered.
Database design
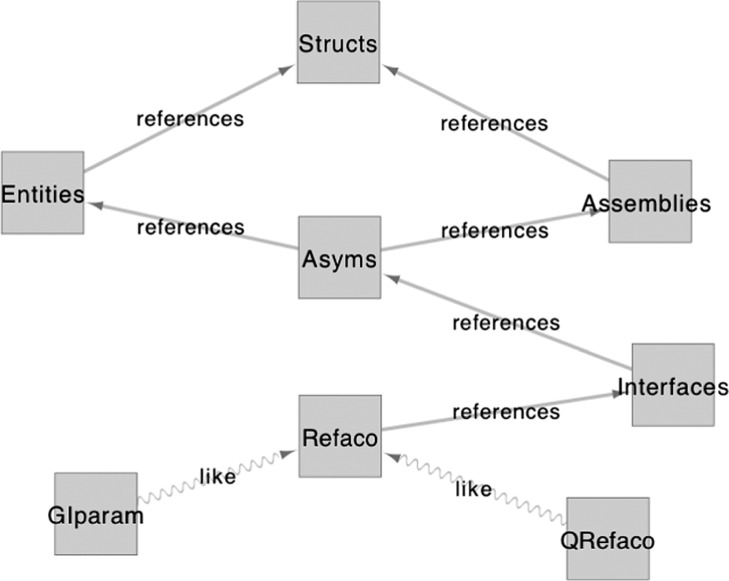
GIRAF relies on a relational database (RDB) for storing a large amount of structural data as well as for fast look-up of similar structures. At the same time, the RDB stores relevant annotations of PDB entries. The overall database structure is depicted in Figure 1.
Figure 1.
Relational tables in the GIRAF database. A simplified view of the GIRAF database schema. Each rectangle represents a relational table, possibly connected to another table via foreign key reference (edges labeled with “references”). The table structure of the GIparam and Qrefaco tables are copied from that of the Refaco table. The Qrefaco table is created temporarily for each query. The diagram was created using Cytoscape45.
The source of data is PDBML files43,44. The tables “Structs” and “Entities” store the annotations of PDB entries and those of individual molecular entities, respectively, extracted from PDBML files. Biological units are generated when available in the pdbx_struct_assembly category of the PDBML files the contents of which are stored in the “Assemblies” table. The chain ID’s (label_asym_id) are renamed by appending the identifier(s) of symmetry operators (pdbx_struct_assembly_gen.oper_expression=pdbx_struct_oper_list.id), and they are stored in the “Asyms” table with a reference (entity_id) to the corresponding entity in the “Entities” table as well as a reference (assembly_id) to the “Assemblies” table. These 4 tables (Structs, Entities, Assemblies, Asyms) provide the basic annotations of the molecules in PDB entries.
A ligand binding site is defined as a set of at least 10 atoms that are in contact with some ligand within 5 Å. This size (10 atoms) was set because at least 10 aligned atom pairs are required for an alignment to be statistically significant29. Ligands are defined as one of non-polymer molecules (as annotated in the entity category of PDBML files), proteins (those annotated as “polypeptide(L)” in the entity_poly category with at least 25 amino acid residues), or nucleic acids (annotated as “polydeoxyribonucleotide,” “polyribonucleotide” or “polydeoxyribonucleotide/poly-ribonucleotide hybrid” in the entity_poly category). Other polymer molecules such as polypeptide with less than 25 residues and polysaccharides are treated separately. The non-polymer molecules, short peptides, and “other” polymers are collectively referred to as “small molecules” and are treated together in the following. The information on binding sites are identified by the pair of chain ID’s of the receptor and the ligand (corresponding to the Asyms table) in addition to the PDB ID and biological assembly ID, and is stored in the table “Interfaces” along with their atomic coordinates in a compressed binary format.
The fast search capability of GIRAF is made possible by looking up similar local structural features defined on various affine frames for each receptor subunit. The affine frames associated with the structural features as well as discretized local atomic coordinates are stored in the “Refaco” table (see also Appendix). The details of structural features are explained in the next subsection.
Pre-processing
Affine frames
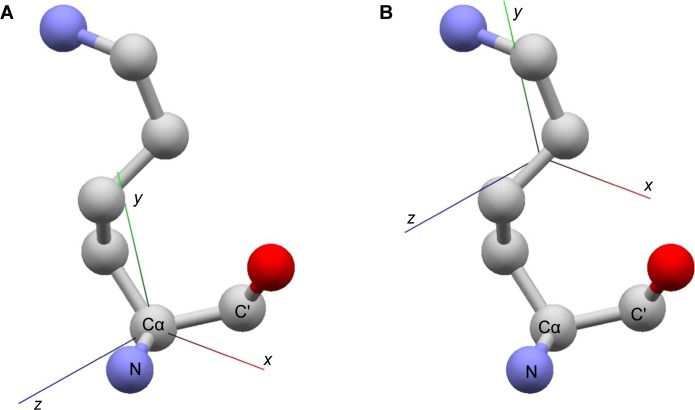
An affine frame is a tuple of 4 3-dimensional vectors (O, x̂, ŷ, ẑ) which defines the origin, and x, y and z axes of a local coordinate system of a given structure. In the present case, an affine frame is defined for each amino acid residue where the origin, O, is the center of mass of side-chain atoms (or Cα atom for glycines), and the basis vectors are based on backbone N, Cα and C′ atoms (Fig. 2). Thus, for an atom whose PDB coordinate is s, its local coordinate s′ with respect to an affine frame f = (O, x̂, ŷ, ẑ) is given as s′ = f(s) where the action of f is defined as
| (1) |
with the dot “·” indicating the dot product.
Figure 2.
Affine frame. (A) For a given amino acid residue, the directions of x, y and z axes are determined by the backbone atoms N, Cα, and C′. (B) The origin of the affine (local coordinate) frame is set to the center of mass of side-chain atoms (Cα for glycines).
We discard affine frames for which no atom of the residue belongs to any binding sites in order to reduce the computational cost.
Structural features of affine frames
Each affine frame is characterized by a set of structural features. These features are subsequently indexed and serves for fast retrieval of potentially similar binding site structures. The fundamental assumption is that if two binding sites are structurally similar, they should share affine frames with similar features. There are in total 44 features which are grouped into two categories: local coordinates of 5-residue fragments and atomic compositions. Atomic compositions are defined in 4 overlapping hemispheres of 10 Å radius centered at the origin of the affine frame, separated by the y–z or z–x planes.
After all affine frames with structural features have been stored in the GIRAF database, the standard deviations of the values of structural features are calculated and stored in a table (“GIparam”, c.f., Fig. 1). These standard deviations multiplied by constant factors are used as thresholds of structural similarity in the geometric indexing search (see below).
Discretized local atomic coordinates
Based on each affine frame, each atomic coordinate of the corresponding interface, if its distance from the origin of the frame is less than 15 Å, is transformed to the local coordinate system and discretized into a lattice point in a lattice of 1 Å×1 Å×1 Å voxel size, which is also characterized by its atom type. (Also see Appendix).
Geometric indexing (GI) search
The goal of geometric indexing search is to find template structures that are potentially similar to a query structure. Conceptually, this is done in two steps. The first is to find pairs of affine frames in the query and templates with similar structural features; the second is to estimate the similarity between the query and template structures by counting the number of overlapping atoms in the respective local coordinate systems defined by the affine frames found in the first step. Thus found potentially similar structures are subject to alignment refinement described in the next subsection. Practically, the two steps can be achieved at once by a single SQL query.
A query structure for a GIRAF search can be either a single binding site or a whole subunit structure. In either case, the affine frames and their associated structural features are extracted in the same manner as template structures. The affine frames with structural features of the query structure, as well as its discretized local atomic coordinates, are stored in a temporary table (“Qrefaco”, c.f., Fig. 1) in the GIRAF database, which has the same structure as the Refaco table. For the discretized coordinate of each atom in the query structure, the nearest and next-nearest neighbor lattice points are also filled with that atom so that structural deviation between the query and template can be taken into account.
To find potentially similar template structures, an SQL query is constructed that returns a list of potentially similar structures (see Appendix for the details).
The criteria for filtering potentially similar structures are given as follows. We define the GI score, SGI, by
| (2) |
where cnt(fq, ft) is the number of overlapping atom pairs (in discretized representation), Nq and Nt are the number of lattice points in the query and template, respectively, that are specified for the affine frames (see the previous subsection). Then the cutoff condition is given as
| (3) |
where Smin is set to 50 by default. This means that the overlapping atom pairs needs to exceed 50% of the atoms in the smaller site. In addition, we also require that cnt(fq, ft) ≥ 10.
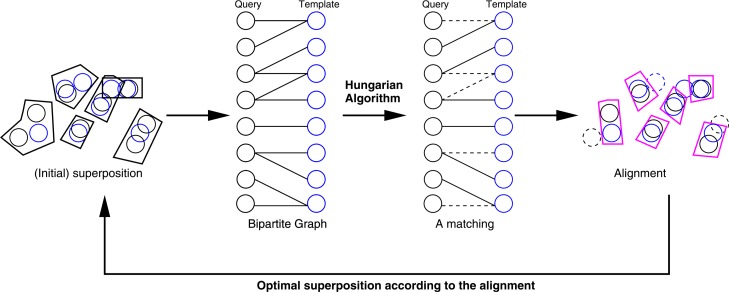
Iterative refinement (IR) of alignment
After the GI search step, we have a list of potentially similar structures. The next step is to obtain optimal alignments of these structures. This is done by iterative applications of superposition46 and bipartite graph matching47,48 as described previouly29 (Fig. 3). Bipartite graph matching has been applied to protein structure alignment previouly by others8,20. However, Taylor8 applied it in a different problem setting (matching pairs of secondary structure elements), and Shulman-Peleg et al.20 did not apply iterative refinement.
Figure 3.
Iterative refinement of alignment.
The Hungarian algorithm (or, more specifically, the Kuhn-Munkres algorithm) for bipartite graph matching was implemented using the functional scheduled binomial heap structure49 to increase performance48, and runs in O(|V|(|E|+|V|log|V|)) worst case time where |V| and |E| are the numbers of nodes and edges, respectively, in a bipartite graph.
After a refined alignment is obtained, we compute the score (GIRAF score) of the alignment given by
| (4) |
where the sum of the edge weights, Σ′(a,b) w(a,b), is computed over aligned atom pairs. The GIRAF score is a generalization of the GI score (Eq. 2): in place of the number of overlapping atoms, the sum of edge weights are used, and the maximum possible GIRAF score is 100 for exactly overlapping binding sites with no deviation. This definition of GIRAF score differs from the previous version29 in that the right hand side of Eq. (4) is not multiplied by the number of aligned atom pairs. We find the present definition more convenient for detecting similarities especially for small binding sites.
Significant matches are selected based on the following criterion:
| (5) |
where C is a constant set to 95 by default, and r(x) is a function of the number of aligned atom pairs Nali, and is defined as:
| (6) |
where the constants N0 and W are set to 10. This criterion imposes a constraint that smaller alignments (Nali ≈10) should score much higher than larger alignments, and effectively removes small binding sites that are liable to be partially aligned with larger binding sites by chance. For example, when C=95, an alignment with Nali= 10 must have a score of at least 95 whereas alignments with Nali >30 need to have a score of only ≈29.
In addition to this score, the number of aligned atom pairs is also used for judging whether an alignment is significant.
It should be noted that the refined alignments obtained from different pairs of affine frames can be exactly identical. In this sense, some affine frame pairs obtained in the GI search are redundant. Since the IR procedure is relatively time-consuming, we remove the redundancy as follows. Let (fq, ft) be a pair of query and template affine frames with their respective origins being O(fq) and O(ft). For another pair of affine frames, (gq, gt), with the same template interface but with a lower GI score, if the distance between the transformed origins ‖gq(O(fq)) – gt(O(ft))‖ is smaller than a certain cutoff (1.5 Å in the present implementation), then the frame pairs (fq, ft) and (gq, gt) are judged to be redundant and the latter of them is discarded. Although it is also necessary in theory to check the distance between fq(O(gq)) and ft(O(gt)) in order to determine the redundancy, we found that omitting it did not affect the result in practice. We apply this procedure to all pairs of the frames for each template interface, and all the redundant frame pairs with lower GI score (SGI) are discarded before the IR procedure is applied.
Flexible alignment
For a given pair of query and template structures, there may be a number of (suboptimal) alignments based on different pairs of affine frames. These alignments are “rigid” in the sense that they are based on rigid body transformation. We can simply select the alignment with the highest IR score to obtain the best rigid alignment for a given query-template pair.
Alternatively, we can integrate the rigid alignments for a given query-template pair into a bipartite graph in the following manner. Let there be N rigid alignments M1, ..., MN with IR score greater than a certain cutoff value. Each alignments is regarded as a matching of a bipartite graph Mm= ((Vq, Vt), Em), m = 1, ..., N, where Vq and Vt are the sets of nodes consisting of query and template atoms, respectively, and Em is the set of weighted edges representing the one-to-one correspondence of the alignment m.
Sort the rigid matchings Mm in the descending order of their number of aligned atom pairs and GIRAF score (in this order of priority).
Let the integrated graph B initially be an empty graph with no vertices or edges. Set m=1.
- Classify each edge e in Mm into one of the following three types:
- “common” if e (disregarding the weight) is already in B;
- “competing” if only one of the vertices of e is already in B;
- “additional” if neither of the vertices of e is in B.
If there are at least two common edges, add competing and additional edges to the graph B.
If there are less than two common edges, but there exist some competing edges, if all the competing edges are consistent with B (in the sense explained below), then add all the competing and additional edges to B.
Set m:=m+1 and go to step 3 unless m=N.
Thus, an integrated bipartite graph B is constructed. By applying the Hungarian algorithm to B, an optimal alignment integrating multiple rigid alignments is obtained. The GIRAF score is computed as in the case for the rigid alignment (Eq. 4).
In step 5, we need to check if the competing edges of Mm are consistent with the already partially defined B. This is done as follows. Let us pick one competing edge e=(vq, vt) where vq and vt are the vertices corresponding to the query and template atoms, respectively, that comprise the edge. Thus, if vq∈Vq(B), then vt ∉Vt(B) by construction. The set W={w|(vq, w) ∈E(B)} consists of template atoms that are connected to the query atom vq in B. If, for all w ∈ W, the distance between vt and w is less than a certain cutoff D, then the edge e is defined to be consistent with B. In the case when vt∈Vt(B), then apply the same argument by swapping query and template. We set D=5 Å in the following. The motivation behind this consistency check is that addition of a new matching to an existing one should not involve a large translation of the local coordinate frames.
It should be noted that the construction of an integrated alignment is not based on a unique rigid-body transformation, but on a set of rigid-body transformations. Thus, it is in general not possible to describe the proximity of all atom pairs in a single affine frame of the 3-dimensional Euclidean space. The integrated bipartite graph should be regarded as being embedded in a manifold spanned by the underlying rigid alignments. In this sense, the integrated alignment is flexible.
One caveat in making flexible alignment is that at least one pair of affine frames should produce a sufficiently large rigid alignment. Otherwise, the combination of a set of fragmentary rigid alignments would result in an inconsistent flexible alignment in which small pieces of rigid alignments are interlaced more or less randomly. The procedure for constructing the integrated bipartite graph described above addresses this problem by limiting the rigid alignments to those with a small number of conflicts with better rigid alignments. In addition, we also impose a constraint that each rigid body alignment to be integrated must have at least 10 pairs of aligned atoms.
Pre-processing of query structures
When a query is a pre-defined interface (binding site), the set of atoms and affine frames are prepared in the same manner as the template structures in the GIRAF database.
When a query is a whole subunit, a special treatment is required for selecting relevant sets of atoms and affine frames. If all the atoms and affine frames found in the query subunit are directly compared to a template structure, it is likely to result in a meaningless alignment. For example, a template, which is an interface to some ligand and therefore is found on a surface of some protein subunit, may be aligned to a deeply buried region of the query structure simply because there are more atoms in the protein core. This is a limitation of the present method which does not “align” empty spaces or cavities. Therefore, we first select atoms that are near to the surface of the query subunit. We define exposed atoms to be those having non-zero solvent accessible surface area (ASA). Exposed atoms and atoms that are within 2 Å from some exposed atoms are selected as near-surface atoms. For selecting affine frames, we calculate residue-wise ASA and those residues having residue-wise ASA less than the mean of that subunit are discarded. ASA was calculated numerically based on pentakis dodecahedron tessellation of a shere (960 triangles) with atomic radii defined by Ooi et al.50 using an in-house OCaml library routine. Affine frames are defined for the remaining, relatively exposed, residues. The set of near-surface atoms and affine frames based on relatively exposed residues are used to search the GIRAF database.
Implementation
PostgreSQL (http://www.postgresql.org) was used as the back-end relational database management system for GIRAF. The PostgreSQL system was customized to enable multi-column indexing of the 44 structural features of affine frames. All the programs were written in the OCaml language (http://caml.inria.fr/, see also Jambon et al.17 for the benefit of using OCaml). The computation for the GI search can be distributed to P processes by splitting the set of query affine frames (i.e., the QRefaco table) into P groups. The IR process can be also distributed to P processes by splitting the list of potential hits found in the GI search into P groups. The parallelization was implemented by using the Berkeley (BSD) socket application programming interface.
Benchmarks were performed on a PC cluster machine running the Linux operating system with Intel Xeon X5460 processor (3.16 GHz, 8 cores) and 8 gigabytes (GB) of random access memory (RAM).
The source code of GIRAF is available at http://pdbj.org/giraf/source/distr.tar.gz.
Data set
We used all the 77,985 PDB entries as of December 21, 2011. Entries with only Cα atom coordinates, those without any protein molecules, or those without ligands were discarded. In total, 114,724 biological assemblies were generated according to the annotations in the PDBML files. The number of interfaces and affine frames are listed in Table 1. When all the PDB entries have been loaded, the GIRAF database consumed approximately 37 GB of hard disk space including indexes.
Table 1.
Database statistics
| type | interfaces | affine frames (disk spacea) | # atoms (S.D.)b |
|---|---|---|---|
| small | 531,491 | 4,843,757 (5 GB) | 35.5 (30.3) |
| protein | 418,344 | 11,921,888 (14 GB) | 142.0 (120.5) |
| nucleic acids | 23,372 | 682,644 (862 MB) | 147.3 (139.4) |
The hard disk space occupied by the corresponding Refaco table.
The average number of atoms and its standard deviation of the interfaces.
Results
Execution time
To measure the efficiency of the GIRAF method, we randomly extracted 1,000 interfaces for small molecules, proteins and nucleic acids, and measured execution time of each GIRAF similarity search. The GIRAF search was executed using 8 CPU cores in parallel and the execution time includes “dead” times for synchronization between the CPU cores. Flexible alignment was enabled and template structures with GIRAF scores (Eq. 4) satisfying the cut-off criterion (Eq. 5) were selected at the final stage.
Depending on the interface type (small, protein or nucleic acid), the mean execution time varies significantly from a few seconds to a few minutes (Table 2). The median values indicate most searches are executed rather quickly. In general, interfaces for nucleic acids and proteins tend to require short and long execution times, respectively. This difference is apparently due to the size of the Refaco tables (Table 1): whereas the table for nucleic acid interfaces (862 MB in size) can be totally loaded into the 8GB RAM, such memory caching is difficult or even impossible for the small molecule (5 GB) or protein interfaces (14 GB). To confirm this, we have also performed the same benchmark by using a computer (Intel Xeon X5560, 8 cores, 2.8 GHz) with a RAM sufficiently large (64 GB) for holding all the Refaco table data. Then the average execution time for small molecule, protein and nucleic acid interfaces were 3.43 (S.D. 3.57; median 2.21), 31.3 (S.D. 33.5; median 20.9) and 2.68 (S.D. 5.26; median 1.20) seconds, respectively. Note that the change in speed does not vary significantly for the nucleic acid interfaces, but it is significant for the other interface types.
Table 2.
Total execution time and number of hits in GIRAF search
| type | Execution time (sec.) | Number of hits | |||
|---|---|---|---|---|---|
|
|
|
||||
| mean | S.D. | median | GIa | IRb | |
| small | 18.7 | 25.3 | 9.61 | 971 (1832) | 178 (317) |
| protein | 145 | 126 | 109 | 5608 (12003) | 565 (1192) |
| nucleic acids | 6.0 | 7.3 | 3.7 | 909 (1332) | 63 (67) |
The average number of template structures selected after the GI search (standard deviation in the parentheses).
The average number of template structures selected after the final stage (standard deviation in the parentheses).
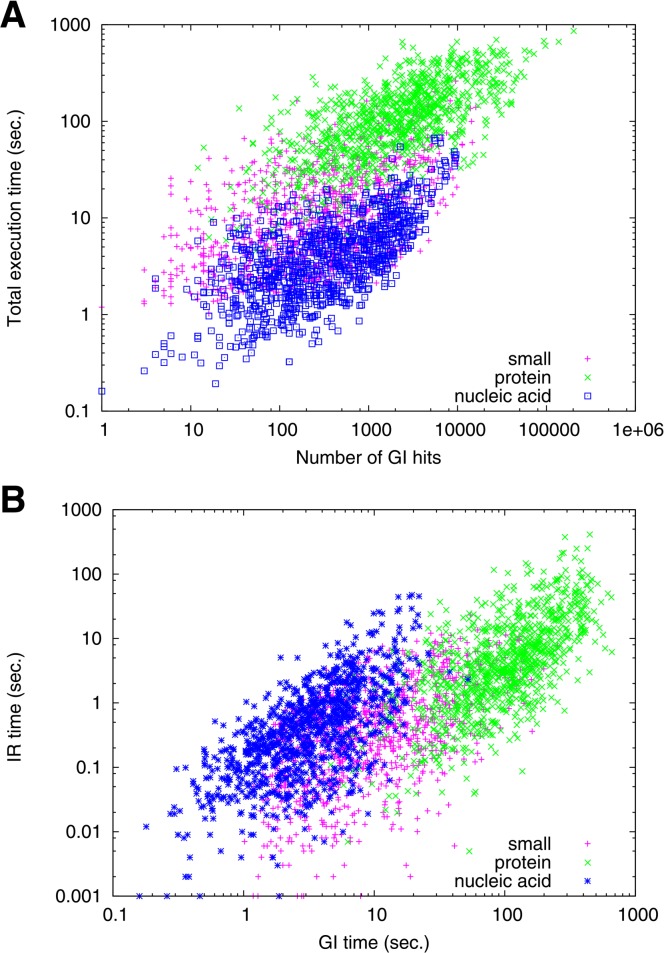
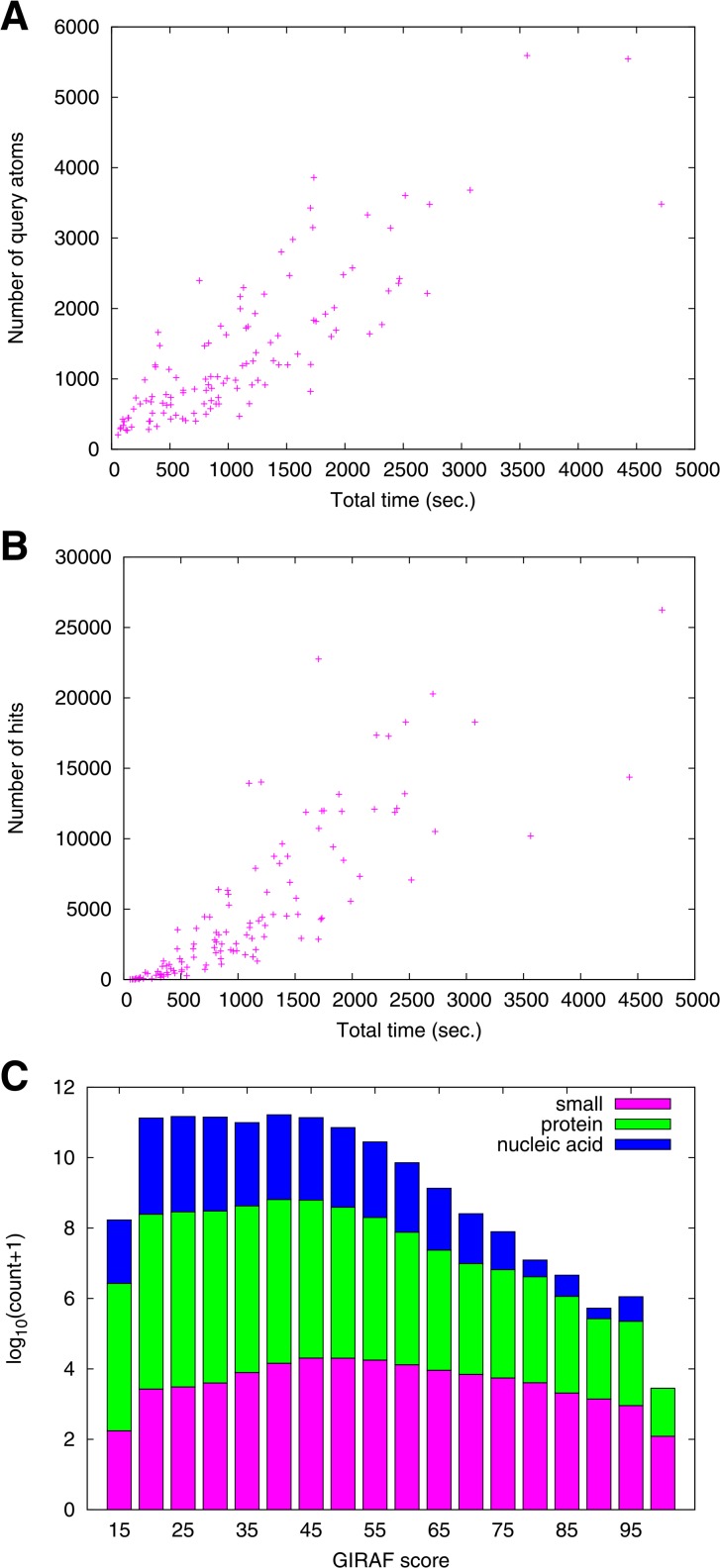
For a given interface type, the execution time seems to be mainly determined by the number of similar interfaces found in the GI search step (Fig. 4A).
Figure 4.
Execution time. (A) Correlation between the number of GI hits and the total execution time (in seconds). (B) Execution times of geometric indexing search (GI time) and of iterative refinement (IR time) in seconds. Colors are magenta for small molecule, green for protei and blue for nucleic acid interfaces.
By breaking up the total execution time into GI search and IR procedure, we can observe the behavior of GIRAF search process in more detail (Fig. 4B). The execution time for GI search is in accordance with the number of template structures (c.f., Table 1). That is, there are the least number of interfaces for nucleic acid binding and its GI search time is in general lower than other interface types. The time required for IR procedure seems to be influenced by the interface size (i.e., the number of interface atoms). Small molecule interfaces require less time for the IR procedure in general than interfaces of other types; interfaces for proteins and nucleic acids are in general large so that it takes more time to refine their alignments.
Effects of iterative refinement
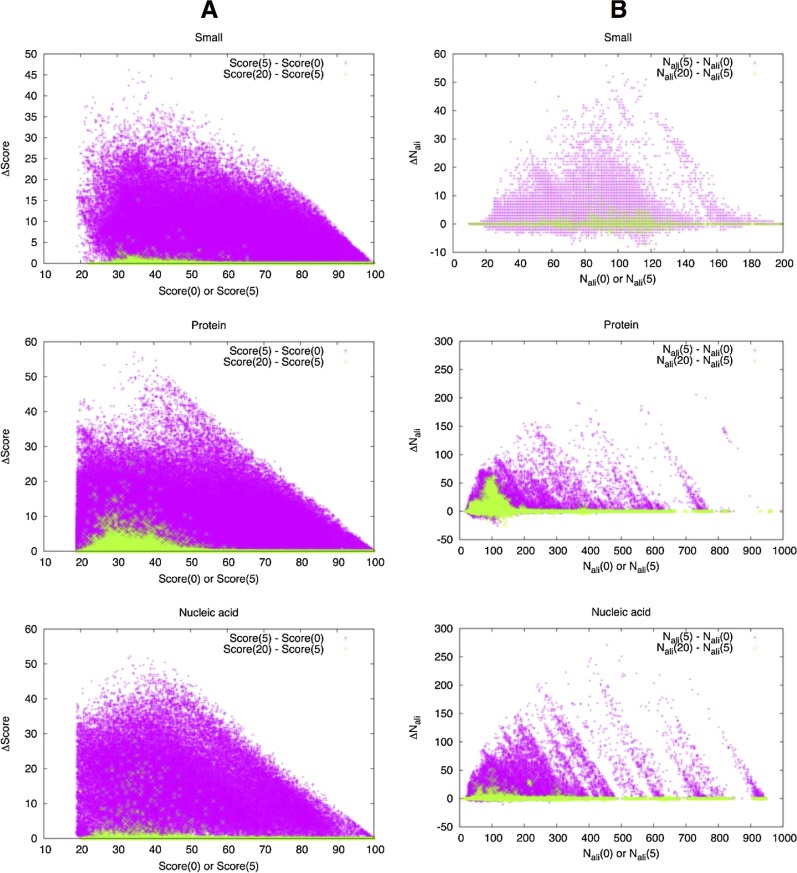
To examine the effectiveness of iterative refinement of alignment, we used the same set of 1,000 interfaces for each interface type, and performed GIRAF searches by varying the number of iterations of refinement from 0 to 5 and to 20 (Fig. 5). In this benchmark, we have used only rigid alignments.
Figure 5.
Iterative refinement. (A) GIRAF scores with 0 [Score(0)] or 5 [Score(5)] iterations for alignment refinement are compared with the difference from GIRAF scores with 5 [Score(5)–Score(0)] or 20 [Score(20)–Score(5)] iterations, respectively. (B) Similar to (A), the number of aligned atom pairs (Nali) are compared. Colors are magenta for the difference between 5 and 0 iterations, green for that between 20 and 5 iterations.
Comparison of the cases with no iteration and with 5 iterations of refinement clearly shows that the distribution of the rigid GIRAF scores indeed improve for all the interface types. A general trend is that alignments with relatively low GIRAF scores tend to improve more significantly after iterative refinement (Fig. 5A). Further increase in the number of iterations does not lead to marked differences in case of small molecule binding sites. However, for binding sites for proteins and nucleic acids, there are cases where an increased number of iterations is effective, possibly due to the large size of these binding sites.
The number of aligned atom pairs exhibits a similar trend as the GIRAF score (Fig. 5B). After 5 iterations of refinement, many alignments are augmented by a significant number of additional atom pairs. As in the case for the GIRAF score, further iterations result in further improvement not for most small molecule binding sites, but for protein and nucleic acid binding sites.
It is interesting to note that although the GIRAF score always improves after iterative refinement (by construction), the number of aligned atom pairs may decrease in some cases. This indicates that removing some largely deviating atoms in the superimposed structures may improve the overall GIRAF score.
Flexible alignment
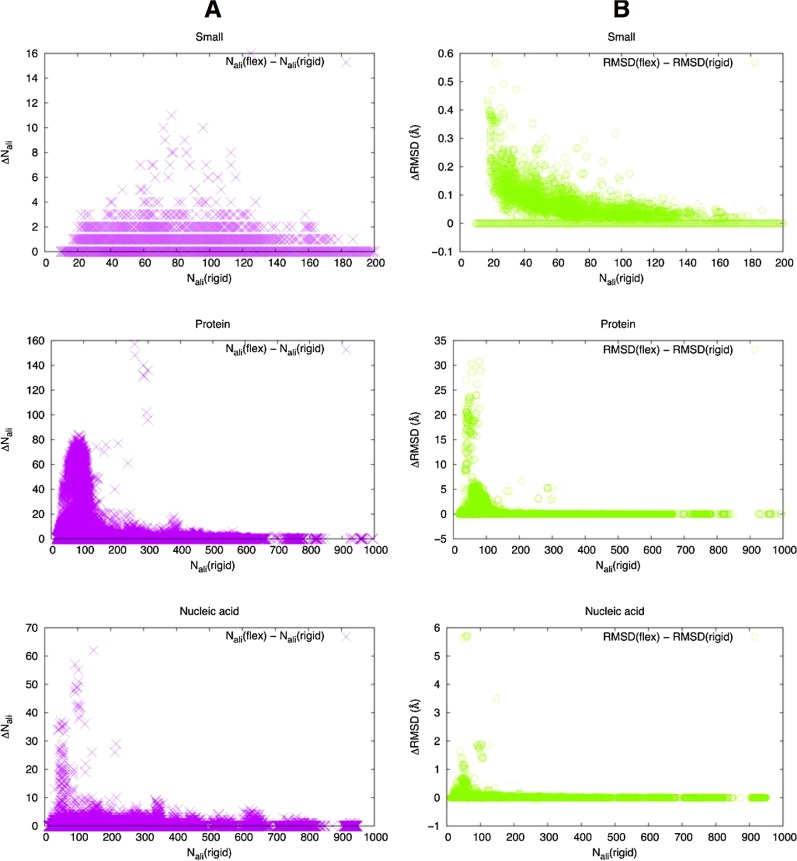
The objective of flexible alignment is to align larger regions of structures that cannot be superimposed by rigid body transformation. This is achieved by patching together smaller regions each of which can be superimposed by rigid body transformation. Thus, one measure for the effectiveness of flexible alignment is the number of aligned atom pairs. It is also expected that the root mean square deviation (RMSD) of superimposed structures be larger for flexible alignments than for rigid alignments. Therefore, we have compared the difference in the number of aligned atom pairs as well as the difference in RMSD in flexible and rigid alignments using the same data set as above (Fig. 6). We applied up to 20 iterations of refinement in this benchmark.
Figure 6.
Flexible alignment. (A) Change in the number of aligned atom pairs between rigid and flexible alignments. (B) Change in RMSD (Å) between rigid and flexible alignments.
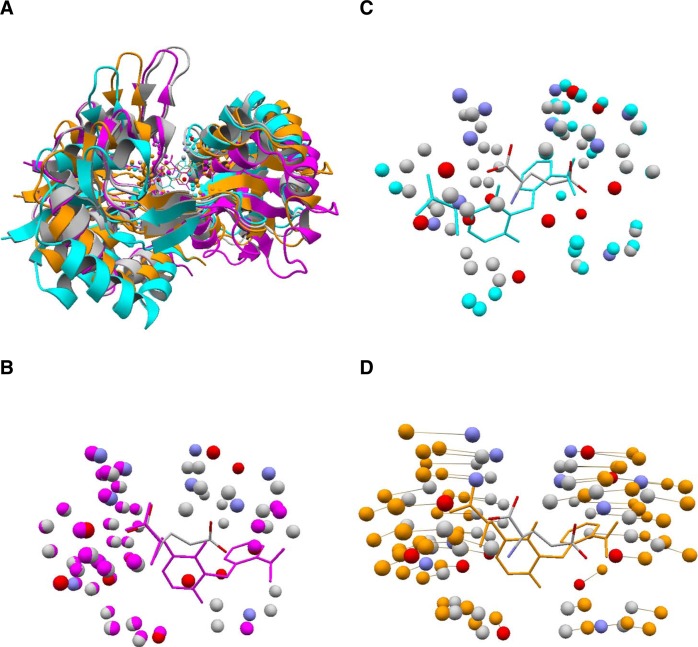
Not many small molecule interfaces exhibit large changes due to flexible alignment. This is expected because most small molecule interfaces are themselves small so that they cannot satisfy the conditions for flexible alignments. Nevertheless, flexible alignment is helpful for aligning structures with different conformational states as seen in the case of glutamate receptor from Rattus norvegicus (Fig. 7). In this example, a GluR6 structure (grey/CPK) is solved with a bound glutamate51 and the other, GluR5 (orange/magenta/cyan) with an antagonist UBP30252. The latter is superimposed to the former based on different alignments. Two suboptimal rigid alignments were found: one of which was surrounding the backbone carboxyl group of glutamate (Fig. 7B), the other around the side-chain carboxyl group of glutamate (Fig. 7C). Due to the difference in the ligand size, the two alignments are not compatible with each other within either rigid frame. A flexible alignment was obtained (Fig. 7D) by integrating these two alignments. As noted above, two structures may not be superimposed well based on an integrated alignment because they cannot be described by a single coordinate frame. We can still observe, however, that the integrated alignment (orange in Fig. 7A) is a sort of an average of the two rigid alignments.
Figure 7.
An example of flexible alignment for small molecule interfaces. The UBP302 binding site of GluR5 from Norway rat (colored orange, magenta, cyan; PDB 2F3552) is superimposed to the glutamate binding site of GluR6 from the same species (colored grey and CPK; PDB 2XXR51). (A) Superposition of folds based on the alignments of ligand binding sites. The colors correspond to those in B–D. (B) An optimal rigid alignment where mainly the atoms around the backbone carboxyl group of glutamate and one of two carboxyl group of UBP302 are aligned. 37 atom pairs are aligned with RMSD of 0.57 Å. (C) A suboptimal rigid alignment where mainly the atoms around the side-chain carboxyl group of glutamate and the other carboxyl group of UBP302 are aligned. 31 atom pairs are aligned with RMSD of 1.05 Å. (D) The resulting flexible alignment which integrates those in (B) and (C). Corresponding atom pairs are connected with a line. In total, 58 atom pairs are aligned with RMSD of 2.43 Å. Note the two structures (orange and CPK) cannot be closely superimposed based on this alignment.
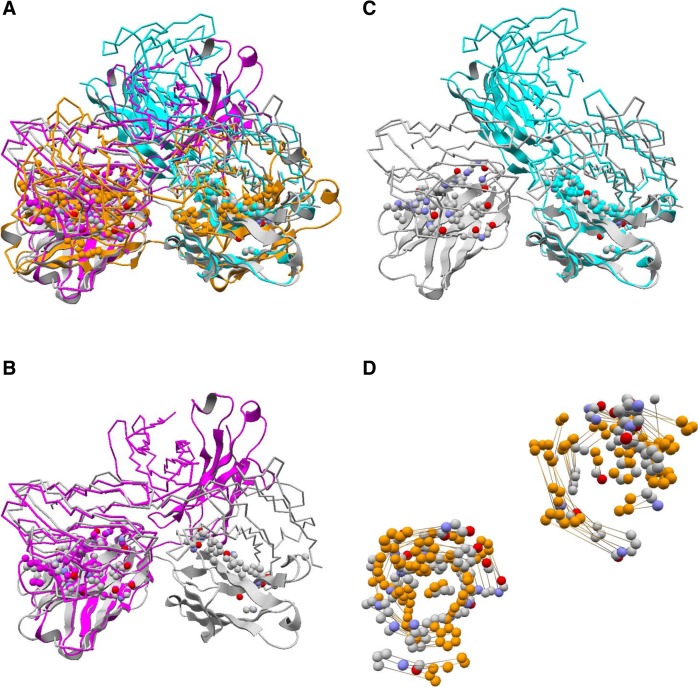
In contrast, many protein interfaces do show marked changes due to flexible alignments. In some cases, more than 100 atom pairs were added in flexible alignments compared to the rigid ones, and RMSD can change up to 10 Å or more. The extreme cases where RMSD increases by more than 20 Å are usually alignments between a very long stretch of a coiled coil interface and a kinked helix bundle. Many of the interfaces with more moderate changes (Nali < 100 and RMSD < 6 Å) are immunoglobulin chains with two domains (constant and variable), the relative orientation of which may widely vary depending on complexes (Fig. 8). A similar trend is observed for nucleic acid binding interfaces although the change in RMSD tends to be smaller.
Figure 8.
An example of flexible alignment for protein interfaces. Two immunoglobulin light chains (cartoon representation) are superimposed (PDB 2FL553 colored orange, magenta, cyan; 3L7F54 colored grey) where their “ligands” are immunoglobulin heavy chains (backbone representation). (A) Superposition of folds based on the alignments of protein binding sites. The colors correspond to those in B–D. (B) An optimal rigid alignment where mainly the interface atoms in N-terminal variable domains are aligned. 89 atom pairs are aligned with RMSD of 0.76 Å. (C) A suboptimal rigid alignment where mainly the interface atoms in the C-terminal constant domains are aligned. 75 atom pairs are aligned with RMSD of 0.91 Å. (D) The resulting flexible alignment which integrates those in (B) and (C). Corresponding atom pairs are connected with a line. In total, 164 atom pairs are aligned with RMSD of 6.8 Å.
Whole subunit queries
Up to the previous subsection, the queries for GIRAF searches were individual (known) interfaces. In this subsection, we demonstrate the results of GIRAF searches with whole subunits as queries. As noted in Materials and Methods, near-surface atoms were extracted and searched against the GIRAF database. For benchmarking, we randomly selected 120 SCOP10 fold representatives; 20 folds from each of 6 SCOP classes (all-α, all-β, α/β, α+β, “membrane and cell surface proteins and peptides”, and “small proteins”). All types of interfaces were searched against a given query subunit.
There were on average ∼5,000 hits per query, ranging from 2 to 26,244. It was observed that the total execution time (including pre-processing of the query subunit) is roughly correlated with the number of atoms in the query structure (correlation coefficient 0.84; Fig. 9A). The most time-consuming query (PDB 2HYD A chain, multi-drug ABC transporter55; 3483 near-surface atoms and 266 affine frames) required approximately 80 minutes for finding 26,244 matching templates. It appears that many of the matches are due to its transmembrane helices which are one of the most common patterns in protein-protein interfaces, often found irrespective of homologous relationships34. As was the case for isolated interfaces (Fig. 4A), the search time is roughly proportional to the number of hits (correlation coefficient 0.83; Fig. 9B). The distributions of GIRAF scores are qualitatively the same for all the ligand types (Fig. 9C) although the number of aligned atom pairs does differ significantly reflecting the interface sizes (not shown).
Figure 9.
Results of whole subunit queries. (A) Total execution time (seconds) vs. number of query atoms. The execution time includes pre-processing of the query in addition to GI search and IR procedure. (B) Total execution time vs. number of hits (matching templates) with GIRAF score of at least 40. (C) Histogram of GIRAF score where the scores are dissected into bins of width 5. Colors of the bars are magenta for small molecule, green for protein and blue for nucleic acid interfaces that were found by GIRAF searches.
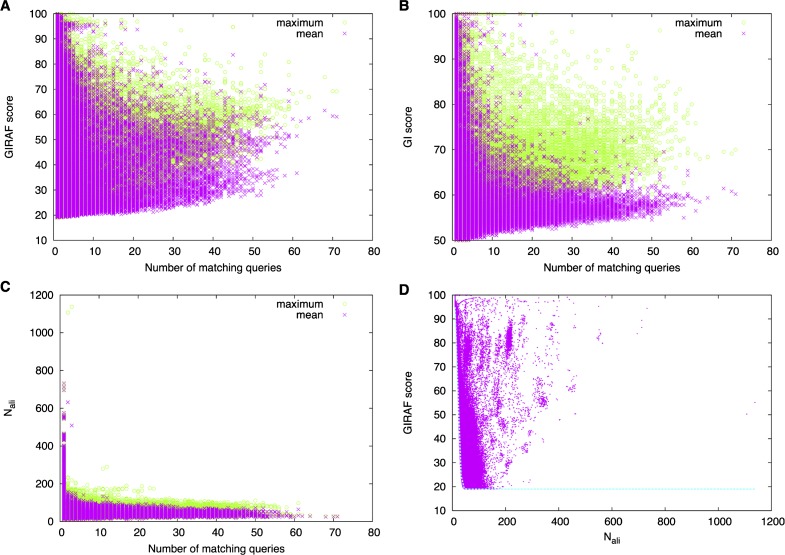
It has been previously noticed that some binding sites are frequently found in many unrelated queries29. Such binding sites are potential false positives. To examine the behavior of such binding sites, we examined the number of matching queries for each interface across the set of queries (Fig. 10). In total, 132,574 template interfaces were matched to some query out of which 71,184 matched with more than one query. Some interfaces were found to have matched with over 70 out of the 120 query structures. Those interfaces with high number of matching queries may have relatively high maximum GIRAF scores (Fig. 10A) or GI scores (Fig. 10B) for some particular query although the mean scores are lower. Nevertheless, the number of aligned atom pairs tends to be low for these frequently found binding sites (Fig. 10C). In fact, when we discard those interfaces with the mean number of aligned atom pairs less than 15 or 30, the number of interfaces with more than one matching query drops to 70,826 or to 49,513, respectively. Although these interfaces are often found as false positives, they may be true positives for some query. In such cases, the GIRAF scores are expected to be higher compared to the average, and it is reflected in the distribution of the maximum scores of each interface (Fig. 10A–C, green points).
Figure 10.
Mean (magenta crosses) and maximum (green circles) GIRAF score (A), GI score (B), or number of aligned atom pairs (C) of matching templates (binding sites in the GIRAF database). The number of matching queries indicates the number of queries to which a given template was matched. (D) Distribution of GIRAF score against the number of aligned atom pairs (Nali). The line in cyan indicates the threshold.
When the relationship between the number of aligned atom pairs (Nali) and GIRAF score is examined, we find that many hits are concentrated at the region of low Nali and low GIRAF score (Fig. 10D), many of which are deemed potential false positives. On the other hand, there are only high scoring hits in the region of large Nali. Therefore, increasing the value of W in the threshold function r(x) in Eq. (5) may remove many of the potential false positives.
What are these potential false positives? As we have already noted in a previous work29, they are interfaces residing at very common and regular structural motifs such as sides of α helices or faces of β sheets. For example, the most frequently found interface was the resveratrol (PDB compound ID: STL) binding site in ATP synthase gamma chain (PDB 2JIZ56), in which the ligand is located at and covering a side of an α helix. Perhaps, it may be useful to filter out these potential false positives by reweighting the scores based on a statistics derived from a representative set of query subunits.
Discussion
There are typically two approaches for aligning a pair of protein structures. The one which we call the “coordinate-based” method is based on direct superposition of atomic coordinates. A typical example of such methods is the geometric hashing method3,57, and GIRAF is also a coordinate-based method. The other, “distance-based” method, is based on comparison of distance matrices as employed in DALI5 or other methods based on clique detection detection15,30. Apart from various heuristics introduced in particular methods, these two approaches are expected to yield similar results (or even identical results in a limiting case)58,59, and both involve a computationally intensive combinatorial problem for finding alignments9. An advantage of the coordinate-based method is that the coordinate transformation of template structures can be done independently of that of a query so that it can be performed at once in advance. This implies that the half of the comparison problem is done even before a query is given. However, it is also a disadvantage in that it results in a large size of the pre-processed data. For example, a naive implementation of the geometric hashing method, which requires every atom as a key of a hash table, is prohibitive with the current data size of the PDB. The distance-based method, based on frame-independent variables (i.e., distances), does not require a large data size compared to the coordinate-based method, but since preprocessing cannot be applied, efficient search for a large data set seems more difficult.
There are mainly two approaches for handling a large data set. One is to reduce the data set by selecting some representatives by removing the redundancy in the data based on, for example, homologous relationships between proteins. Although this approach has been employed for many years in many studies, it is inadequate if one is interested in the diversity of structures and interaction states37. The other approach is to filter out irrelevant templates based on some structural features that can be compared without explicitly matching or aligning structures. Recently, several such methods have been proposed20,35,38, and GIRAF is one of them. An efficient all-against-all comparison is still difficult even with clever filtering techniques, and there are very few studies that truly treat the entire PDB data33,34,38,39. By using their SketchSort method, Ito et al.38 conducted an all-against-all comparison of 1.2 million known and predicted (small molecule) ligand binding sites. While the structural features in GIRAF are defined for each affine frame locally, those of SketchSort are defined globally. Thus, in the latter, the number of feature sets is equal to the number of interfaces, while in GIRAF there are in general much more feature sets (affine frames) than interfaces (Table 1). The advantage of locally defined feature sets is that they make it possible to match partially similar structures, which is especially important for obtaining flexible alignments. Moreover, the feature set of SketchSort does not define a coordinate frame so that the final alignments are obtained by using an external program, TM-align60.
Compared to the previous versions of GIRAF, there are many changes introduced in the present implementation. First of all, we no longer use the Delaunay tessellation of atomic coordinates for defining affine frames. In the original version, some stringent condition was imposed on the Delaunay tetrahedra to achieve fast look-up, but this sacrificed sensitivity. That is, some clear similarities have been missed. This was mainly because some tetrahedra were discarded although they were important for identifying the similarities as well as because the Delaunay tessellation is not robust against small changes in structures. Based on backbone conformation, the present version produces more stable results. Second, more structural features are included for even faster look-up of potentially similar interfaces. These features were absent in the original version29, but some of them (namely, atomic compositions) have been already used and proved effective in later studies33,34,39. Third, the GI search is now entirely executed in the RDB system. This greatly simplifies the implementation and reduces the traffic between the RDB system and the client program, resulting in slightly faster execution. Lastly, the flexible alignment is a novel feature of the current version. It was shown effective especially for large interfaces with structural changes.
Finally, we discuss practical advantages of using a RDB system. The large amount of structural data easily exceeds the size of RAM in conventional computers. Therefore, structural data usually must be stored and searched in secondary storage, namely, hard disks. Efficient search using hash tables or indexes on hard disks involves complicated programming. By using a RDB system which is optimized for large data stored on hard disks, it is a trivial matter to exploit index-based searches. If the RAM is sufficiently large, the operating system will cache the data and very fast search is possible. Even if the RAM is not large enough as was the case in the benchmarks described above, RDB systems work equally well and the GIRAF program runs normally and relatively efficiently owing to the geometric indexing. Furthermore, the GIRAF back-end database contains annotations extracted from the PDBML files so that the result of a GIRAF search can be immediately combined with these annotations as well as annotations from other sources to provide a useful view for examining the biological implications of structural similarities.
In summary, we have developed a method to efficiently search and flexibly align similar interfaces in protein structures by exploiting a RDB system and using novel algorithms. We hope that exhaustive analyses of protein structures using this method serve as a basis for further understanding protein functions from the atomic resolution.
Acknowledgments
A.R.K. thanks Prof. Motoko Kotani for valuable discussion on mathematical aspects of structure comparison. This work was supported by a JSPS Grant-in-Aid for Young Scientists B (22770149) to A.R.K. and a JSPS Grant-in-Aid for Scientific Research B (23370071) to H.N. and A.R.K., and by National Bioscience Database Center, the Japan Science and Technology Agency.
Appendix. Database table and SQL query for geometric indexing search
In this appendix, we describe the details of the database table and SQL query for geometric indexing (GI) search.
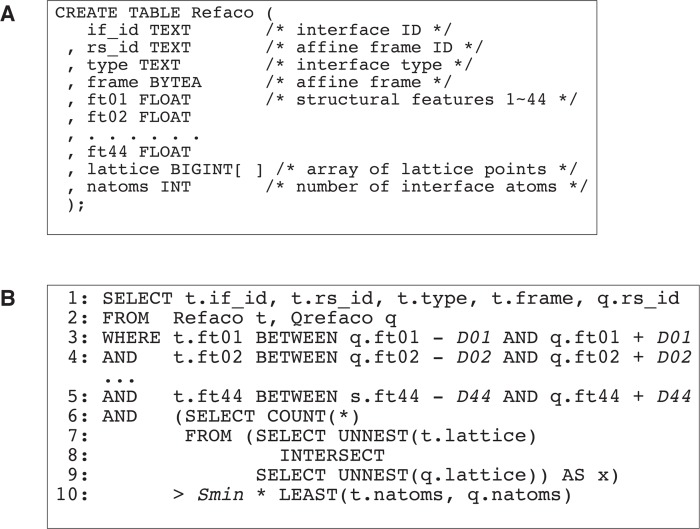
The Refaco table Among the tables in the GIRAF database (Fig. 1), the information necessary for the GI search is stored in the Refaco table (Fig. 11A). In addition to the identifiers for each interface (if_id) and affine frame (rs_id), it contains interface type (type), structural features (ft01, ..., ft44), and the number of atoms contained in the lattice points (natoms). The affine frame itself, that is, a set of vectors specifying the origin and axes of the local coordinate system, is saved in the frame column in a binary format so that it can be directly converted to the data structure of the GIRAF client program. The lattice points together with atom types are encoded as 8-byte integers and are stored in the lattice column of an array type in the Refaco table (Fig.11A).
Figure 11.
Pseudo SQL codes. (A) A pseudo SQL code defining the Refaco table (c.f. Fig. 1). (B) The SQL query for geometric indexing search. The tables containing structurally featured affine frames and discretized atomic coordinates of templates and the query are joined (line 2). The identifiers of the interfaces (t.if_id) and affine frames (t.rs_id), interface type (t.type) and the affine frames (t.frame) of templates and the identifier of the matching affine frames of the query (q.rs_id) are returned (line 1) if the structural features of the templates are sufficiently similar to those of the query (lines 3–5) and the number of overlapping atom pairs (lines 6–9) is greater than a threshold (line 10).
The Qrefaco table (Fig. 1) that stores the affine frames of a query has the same structure as the Refaco table with the value of the interface identifier being arbitrary.
SQL query for GI search An SQL query for the GI search is constructed by joining the Refaco and Qrefaco tables with tolerance parameters extracted from the GIparam table (Fig. 11B). This query returns the identifier of the template interfaces (t.if_id in Fig. 11B), the identifier of the template affine frames (t.rs_id), template affine frames themselves (t.frame), and the identifier of query affine frames (q.rs_id). In the WHERE clause, parameters extracted from the GIparam table are used to define the permitted range of structural features (Fig. 11B, lines 3–5). For the Cα coordinate xj of i-th atom (relative to the central residue defining the affine frame), the half width of the range is defined as
| (7) |
where Sc is a scaling factor set to 1.0 by default, and σ(x′i) is the standard deviation of the local coordinate x′j in the GIRAF database. The half width is defined for each of x′, y′, and z′ (local) coordinate of each Cα atoms of the five residue segment. For the atomic composition ca of atom type a in each of the four regions, the half width is
| (8) |
where Sa is a scaling factor which is set to 1.2 by default, and σ(ca) is the standard deviation of the composition in the database.
To count the number of overlapping atoms, the arrays containing the discretized atomic coordinates are unnested and the cardinality of their intersection is computed (Fig. 11B, lines 6–9). If the count is greater than a certain thresh-old value, the result is selected (Fig. 11B, line 10).
References
- 1.Taylor WR, Orengo CA. Protein structure alignment. J Mol Biol. 1989;208:1–22. doi: 10.1016/0022-2836(89)90084-3. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell EM, Artymiuk PJ, Rice DW, Willett P. Use of techniques derived from graph theory to compare secondary structure motifs in proteins. J Mol Biol. 1990;212:151–166. doi: 10.1016/0022-2836(90)90312-A. [DOI] [PubMed] [Google Scholar]
- 3.Nussinov R, Wolfson HJ. Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc Natl Acad Sci USA. 1991;88:10495–10499. doi: 10.1073/pnas.88.23.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov NN, Takahashi K, Go N. Common spatial arrangements of backbone fragments in homologous and non-homologous proteins. J Mol Biol. 1992;225:5–9. doi: 10.1016/0022-2836(92)91021-g. [DOI] [PubMed] [Google Scholar]
- 5.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 6.Mizuguchi K, Go N. Comparison of spatial arrangements of secondary structural elements in proteins. Protein Eng. 1995;8:353–362. doi: 10.1093/protein/8.4.353. [DOI] [PubMed] [Google Scholar]
- 7.Kawabata T, Nishikawa K. Protein tertiary structure comparison using the markov transition model of evolution. Proteins. 2000;41:108–122. [PubMed] [Google Scholar]
- 8.Taylor WR. Protein structure comparison using bipartite graph matching and its application to protein structure classification. Mol Cell Proteomics. 2002;1:334–339. doi: 10.1074/mcp.t200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Eidhammer I, Jonassen I, Taylor WR. Protein bioinformatics. Wiley & Sons; Chichester, England: 2004. [Google Scholar]
- 10.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 11.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi N, Go N. ATP binding proteins with different folds share a common ATP-binding structural motif. Nat Struct Biol. 1997;4:6–7. doi: 10.1038/nsb0197-6. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi N, Go N. A method to search for similar protein local structures at ligand-binding sites and its application to adenine recognition. Eur Biophys J. 1997;26:135–144. doi: 10.1007/s002490050065. [DOI] [PubMed] [Google Scholar]
- 14.Russell RB. Detection of protein three-dimensional side-chain patterns: New examples of convergent evolution. J Mol Biol. 1998;279:1211–1227. doi: 10.1006/jmbi.1998.1844. [DOI] [PubMed] [Google Scholar]
- 15.Kinoshita K, Sadanami K, Kidera A, Go N. Structural motif of phosphate-binding site common to various protein superfamilies: all-against-all structural comparison of protein-mononucleotide complexes. Protein Eng. 1999;12:11–14. doi: 10.1093/protein/12.1.11. [DOI] [PubMed] [Google Scholar]
- 16.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jambon M, Imberty A, Deléage G, Geourjon C. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins. 2003;52:137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- 18.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J Mol Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brakoulias A, Jackson RM. Towards a structural classification of phosphate binding sites in protein-nucleotide complexes: an automated all-against-all structural comparison using geometric matching. Proteins. 2004;56:250–260. doi: 10.1002/prot.20123. [DOI] [PubMed] [Google Scholar]
- 22.Keskin O, Tsai CJ, Wolfson H, Nussinov R. A new, structurally nonredundant, diverse data set of protein-protein interfaces and its implications. Protein Sci. 2004;13:1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henschel A, Kim WK, Schroeder M. Equivalent binding sites reveal convergently evolved interaction motifs. Bio-informatics. 2006;22:550–555. doi: 10.1093/bioinformatics/bti782. [DOI] [PubMed] [Google Scholar]
- 24.Kim WK, Henschel A, Winter C, Schroeder M. The many faces of protein-protein interactions: A compendium of interface geometry. PLoS Comput Biol. 2006;2:e124. doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gold ND, Jackson RM. Fold independent structural comparisons of protein-ligand binding sites for exploring functional relationships. J Mol Biol. 2006;355:1112–1124. doi: 10.1016/j.jmb.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 26.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: a structural classification of protein-protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aung Z, Tan S-H, Ng S-K, Tan K-L. PPiClust: efficient clustering of 3D protein-protein interaction interfaces. J Bioinform Comput Biol. 2008;6:415–433. doi: 10.1142/s0219720008003485. [DOI] [PubMed] [Google Scholar]
- 28.Minai R, Matsuo Y, Onuki H, Hirota H. Method for comparing the structures of protein ligand-binding sites and application for predicting protein-drug interactions. Proteins. 2008;72:367–381. doi: 10.1002/prot.21933. [DOI] [PubMed] [Google Scholar]
- 29.Kinjo AR, Nakamura H. Similarity search for local protein structures at atomic resolution by exploiting a database management system. BIOPHYSICS. 2007;3:75–84. doi: 10.2142/biophysics.3.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie L, Bourne PE. Detecting evolutionary relationships across existing fold space, using sequence order-independent profile-profile alignments. Proc Natl Acad Sci USA. 2008;105:5441–5446. doi: 10.1073/pnas.0704422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuncbag N, Gursoy A, Guney E, Nussinov R, Keskin O. Architectures and functional coverage of protein-protein interfaces. J Mol Biol. 2008;381:785–802. doi: 10.1016/j.jmb.2008.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teyra J, Paszkowski-Rogacz M, Anders G, Pisabarro MT. SCOWLP classification: structural comparison and analysis of protein binding regions. BMC Bioinformatics. 2008;9:9. doi: 10.1186/1471-2105-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinjo AR, Nakamura H. Comprehensive structural classification of ligand binding motifs in proteins. Structure. 2009;17:234–246. doi: 10.1016/j.str.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Kinjo AR, Nakamura H. Geometric similarities of protein-protein interfaces at atomic resolution are only observed within homologous families: an exhaustive structural classification study. J Mol Biol. 2010;399:526–540. doi: 10.1016/j.jmb.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 35.Moll M, Bryant DH, E KL. The LabelHash algorithm for substructure matching. BMC Bioinformatics. 2010;11:555. doi: 10.1186/1471-2105-11-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasahara K, Kinoshita K, Takagi T. Ligand-binding site prediction of proteins based on known fragment-fragment interactions. Bioinformatics. 2010;26:1493–1499. doi: 10.1093/bioinformatics/btq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dasgupta B, Nakamura H, Kinjo AR. Distinct roles of overlapping and non-overlapping regions of hub protein interfaces in recognition of multiple partners. J Mol Biol. 2011;411:713–727. doi: 10.1016/j.jmb.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 38.Ito J, Tabei Y, Shimizu K, Tomii K, Tsuda K. PDB-scale analysis of known and putative ligand binding sites with structural sketches. Proteins. 2012;80:747–763. doi: 10.1002/prot.23232. [DOI] [PubMed] [Google Scholar]
- 39.Kinjo AR, Nakamura H. Composite structural motifs of binding sites for delineating biological functions of proteins. PLoS One. 2012;7:e31437. doi: 10.1371/journal.pone.0031437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinjo AR, Suzuki H, Yamashita R, Ikegawa Y, Kudo T, Igarashi R, Kengaku Y, Cho H, Standley DM, Nakagawa A, Nakamura H. Protein data bank japan (PDBj): Maintaining a structural data archive and resource description framework format. Nucleic Acids Res. 2012;40:D453–D460. doi: 10.1093/nar/gkr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lathrop RH. The protein threading problem with sequence amino acid interaction preferences is NP-complete. Protein Eng. 1994;7:1059–1068. doi: 10.1093/protein/7.9.1059. [DOI] [PubMed] [Google Scholar]
- 43.Westbrook J, Ito N, Nakamura H, Henrick K, Berman HM. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 44.Kinjo AR, Yamashita R, Nakamura H. PDBj Mine: Design and implementation of relational database interface for Protein Data Bank Japan. Database. 2010;2010:baq021. doi: 10.1093/database/baq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond R. A note on the rotational superposition problem. Acta Cryst A. 1988;44:211–216. [Google Scholar]
- 47.Lawler E. Combinatorial Optimization: Networks and Matroids. Dover; New York, U.S.A.: 2001. Originally published in 1976. [Google Scholar]
- 48.Gupta A, Ying L. On algorithms for finding maximum matchings in bipartite graphs. (Technical Report RC 21576 (97320), IBM 1999).
- 49.Okasaki C. Purely functional data structures. Cambridge University Press; Cambridge, U. K.: 1999. [Google Scholar]
- 50.Ooi T, Oobatake M, Némethy G, Scheraga HA. Accessible surface areas as a measure of the thermodynamic parameters of hydration of peptides. Proc Natl Acad Sci USA. 1987;84:3086–3090. doi: 10.1073/pnas.84.10.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayeem N, Mayans O, Green T. Conformational flexibility of the ligand-binding domain dimer in kainate receptor gating and desensitization. J Neurosci. 2011;31:2916–2924. doi: 10.1523/JNEUROSCI.4771-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu X, Wentworth P, Kyle RA, Lerner RA, Wilson IA. Cofactor-containing antibodies: Crystal structure of the original yellow antibody. Proc Natl Acad Sci USA. 2006;103:3581–3585. doi: 10.1073/pnas.0600251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fransson J, Teplyakov A, Raghunathan G, Chi E, Cordier W, Dinh T, Feng Y, Giles-Komar J, Gilliland G, Lollo B, Malia TJ, Nishioka W, Obmolova G, Zhao S, Zhao Y, Swanson RV, Almagro JC. Human framework adaptation of a mouse anti-human IL-13 antibody. J Mol Biol. 2010;398:214–231. doi: 10.1016/j.jmb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Dawson RJ, Locher KP. Structure of a bacterial multi-drug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 56.Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. Mechanism of inhibition of bovine F1-ATPase by resver-atrol and related polyphenols. Proc Natl Acad Sci USA. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolfson HJ, Rigoutsos I. Geometric hashing: An overview. IEEE Comput Sci Eng. 1997;4:10–21. [Google Scholar]
- 58.Burago D, Burago Y, Ivanov S. A course in metric geometry, volume33 of Graduate Studies in Mathematics. American Mathematical Society; Providence, Rhode Island, U.S.A: 2001. [Google Scholar]
- 59.Mémoli F. In: On the use of Gromov-Hausdorff distances for shape comparison. in Eurographics Symposium on Point-based Graphics. Botsch M, Pajarola R, editors. The Eurographics Association; 2007. 2007. [Google Scholar]
- 60.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]