Abstract
CRM1 (also known as exportin 1 or Xpo1) is the most versatile nuclear export receptor (exportin) that carries a broad range of proteins and ribonucleoproteins from the nucleus to the cytoplasm through the nuclear pore complex. The majority of the export substrates of CRM1 contain a short peptide sequence, so-called leucine-rich nuclear export signal (NES), which typically harbor four or five characteristically spaced hydrophobic residues. The transport directionality is determined by the small GTPase Ran and Ran-binding proteins that control the binding and dissociation of cargo. Here we review recent structural studies that advanced understanding of how NES is specifically recognized by CRM1 in the nucleus, and how NES is rapidly dissociated from CRM1 in the cytoplasm.
Keywords: CRM1, Ran, nuclear export signal, nuclear pore, RanBP1
In eukaryotic cells, the transport of macromolecules into and out of the nucleus is a fundamental and essential cellular activity that regulates many physiological functions including gene expression, signal transduction and cell growth. The nuclear transport occurs through the nuclear pore complex (NPC), embedded in the nuclear envelope and built from approximately 30 different proteins collectively termed nucleoporins (Nups). In the fully assembled NPC, each Nup exists in multiple copies (8, 16, or 32) with a total of approximately 500 Nups per NPC. NPCs pose barriers that prevent passive diffusion of inert objects > 5nm in diameter1 and yet accommodate active transport of large macromolecules2. Most of the active transport pathways through the NPCs are mediated by multiple families of soluble transport receptors. The largest class of the nuclear transport receptors is the karyopherin-β superfamily of proteins. There are more than 20 karyopherin-βs in human cells, and 14 in budding yeast. The karyopherin-βs can be classified into two types, importins and exportins, depending on the directionality of transport. Importins carry cargoes to the nucleus, whereas exportins carry cargoes to the cytoplasm.
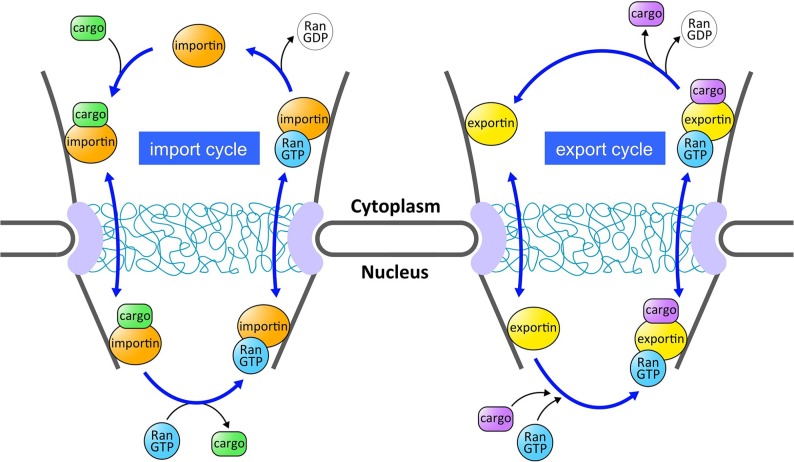
Nuclear transport pathways mediated by the karyopherin-βs are regulated by the small GTPase protein Ran2. Like other Ras-family GTPases, Ran cycles between GDP- and GTP-bound states3. The low intrinsic rates of nucleotide exchange and hydrolysis on Ran are stimulated by specific factors2. Ran GTPase activity is stimulated by the cytoplasmic protein RanGAP, whereas nucleotide exchange is stimulated by the Ran guanine nucleotide exchange factor (GEF) in the nucleus. Because of the compartmentalized localization of GAP and GEF, cytoplasmic Ran is primarily in the GDP-bound state whereas nucleoplasmic Ran is kept primarily in the GTP-bound state. This RanGDP-RanGTP gradient from the cytoplasm to the nucleus is an important determinant of the directionality of nuclear transport2 (Fig. 1). In general, importins bind cargo in the cytoplasm, and release it in the nucleus upon RanGTP binding. By contrast, exportins bind cargo in the nucleus in complex with RanGTP, and the ternary export complex (exportin-cargo-RanGTP complex) is disassembled in the cytoplasm, where Ran GTPase is activated.
Figure 1.
Nuclear import and export cycles mediated by carrier proteins (importins and exportins) belonging to the karyopherin-β superfamily. The directionality of the transport is regulated by the RanGDP-RanGTP gradient from the cytoplasm to the nucleus. Importin binds to its cargo in the cytoplasm and crosses the NPC. Once in the nucleus, RanGTP induces dissociation of cargo from importin. Exportin forms a ternary complex with its cargo and RanGTP in the nucleus, and this ternary export complex is disassembled in the cytoplasm, where Ran GTPase is activated. Although not shown in this figure, RanGDP is recycled back to the nucleus by the Ran-specific nuclear import factor NTF2, and then regenerated with GTP to participate in another transport cycle.
In this review, we focus on recent advances in structural characterization of a nuclear export pathway mediated by CRM1 (for a more comprehensive review of structural biology of various nuclear transport pathways, the readers are referred to many excellent reviews4–7). CRM1 (Chromosome Region Maintenance 1) is the most versatile exportin that carries a plethora of cargo macromolecules from the nucleus to the cytoplasm8–11. The majority of the export substrates of CRM1 contain a short peptide sequence, so-called leucine-rich nuclear export signal (NES). The amino acid sequences of experimentally verified NESs have strong preferences for the Φ1-X3-Φ2-X2-Φ3-X-Φ4 pattern, where Φn represents Leu, Val, Ile, Phe or Met and X can be any amino acid but tends to be negatively charged12. The NES-containing cargo (NES-cargo) cannot passively diffuse through the NPC, and it is only when the cargo is complexed with CRM1 that the cargo can be translocated across the NPC. The translocation across the NPC appears to be a reversible facilitated diffusive process mediated by weak interactions between CRM1 and Nups, and it is crucially important that the loading of NES-cargo occurs in the nucleus and the unloading occurs in the cytoplasm, in order for the transport to be unidirectional. We describe below recent structural studies that are revealing how the loading and unloading of cargoes are regulated in a compartment-specific manner.
Mechanism of NES recognition
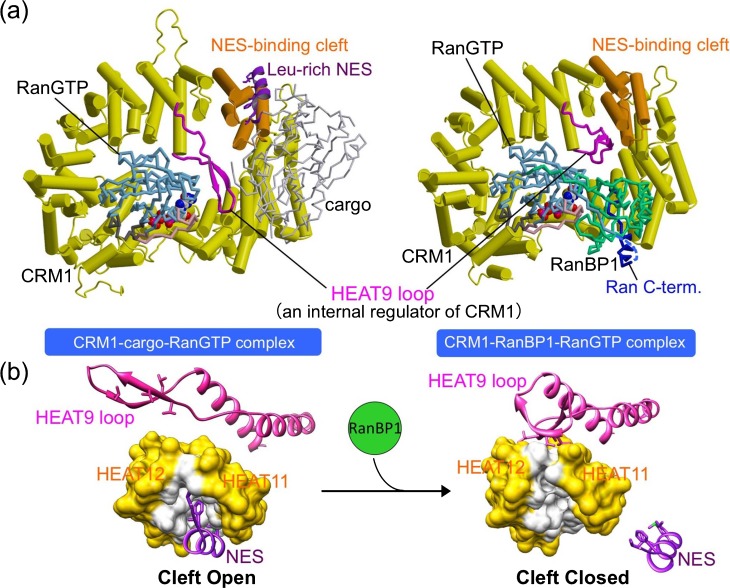
The structures of CRM1-cargo complexes with and without RanGTP have recently been elucidated by X-ray crystallography (Fig. 2a)13–15. CRM1 is a ring-shaped molecule constructed from a tandem array of 21 HEAT repeats. Each of the HEAT repeats consists of two antiparallel α-helices connected by loops of varying length. In general, tandem stacking of HEAT repeats produces a super-helical structure, which has a large surface-to-volume ratio and so has the ability to function as a coordinating scaffold to interact with a broad range of different substrates16. Another important property of the HEAT repeat proteins is that they have conformational flexibility that can be utilized for the regulation of the interactions with their substrates17. HEAT repeat proteins typically wrap around its target proteins inside the super-helical structure to form a stable complex, but the outer convex surface can also be used for protein-protein interactions. The structures of CRM1 bound to three different NESs invariably showed that the hydrophobic side chains of NESs fit into five hydrophobic pockets within a narrow groove formed on the outer convex surface of CRM1 between HEAT repeats 11 and 12. In the CRM1-cargo complexes, the NESs of snurportin and PKI adopt combined α-helix-loop conformations, whereas the NES of Rev adopts an entirely loop conformation. Interestingly, the conformation of the NES-binding cleft in the binary CRM1-RanGTP complex is essentially identical to that of the various ternary CRM1-cargo-RanGTP complexes14. This implies that diverse NESs adapt structurally to fit into a structurally invariant binding site. The binding site of Leptomycin B (LMB) is located in this hydrophobic NES-binding cleft, explaining why LMB is a potent inhibitor of CRM1-mediated nuclear export9,18. The fact that the cargo-binding site is located on the outer surface of CRM1 is probably important for CRM1 to carry a broad range of cargoes that vary greatly in size and shape. RanGTP, on the other hand, binds to the inner surface of CRM1 at four distinct binding surfaces. CRM1 directly binds to both switch I and switch II loops of Ran, and this is possible only when both switch loops adopt the GTP-bound conformation. This accounts for the ability of CRM1 to discriminate between GTP- and GDP-bound Ran.
Figure 2.
Structural basis for cargo binding and release in the CRM1-mediated nuclear export. (a) Crystal structures of CRM1-cargo (snurportin)-RanGTP complex (PDB code, 3GJX) and CRM1-RanBP1-RanGTP complex (PDB code, 3M1I). CRM1 is colored in yellow, except that the HEAT9 loop and HEAT repeats 11 and 12 (the NES-binding site) are highlighted in magenta and orange, respectively. Ran is colored in cyan, with its switch I, switch II and the C-terminal extension highlighted in pink, gray and blue, respectively. The cargo (snurportin) is colored in lightgray, with its NES at the N-terminus highlighted in purple. RanBP1 is colored in green. (b) The movement of the HEAT9 loop, induced by RanBP1 binding, drives closure of the NES-binding cleft. HEAT repeats 11 and 12 are shown in surface representation. The residues that directly interact with NES are white, whereas the other residues are yellow. The N-terminal NES of snurportin is shown in purple.
An allosteric mechanism to accelerate NES dissociation
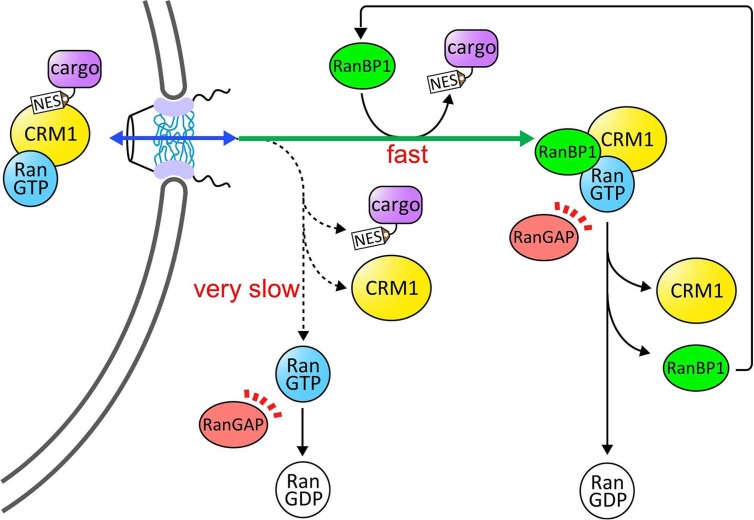
The ternary CRM1-cargo-RanGTP complex is disassembled in the cytoplasm by the combined action of the cytoplasmic proteins RanBP1 (or RanBP2) and RanGAP. The current view of the disassembly mechanism is illustrated in Figure 3. In essence, recent biochemical studies suggested that the major pathway of the disassembly reaction occurs as follows19. First, RanBP1 binds to the CRM1-cargo-RanGTP complex, causing rapid release of cargo and forming a ternary CRM1-RanBP1-RanGTP complex (the rapid release of NES accelerated by RanBP1 occurs much faster than the spontaneous dissociation of NES from CRM1 and RanGTP19). RanGAP then acts on the CRM1-RanBP1-RanGTP complex and promotes rapid hydrolysis of GTP, which causes dissociation of RanBP1 from Ran. The free RanBP1 can then participate in the next round of disassembly. Thus, it has been proposed that RanBP1 functions catalytically in the disassembly reaction.
Figure 3.
Disassembly pathways of the CRM1 nuclear export complex in the cytoplasm. Upon translocation across the nuclear pore complex, CRM1-cargo-RanGTP complex would encounter Ran-binding domains (RanBDs) of either freely diffusing RanBP1 as depicted in this figure RanBP2 that is a major component of the cytoplasmic fibril of the nuclear pore complex. RanBD rapidly dissociates cargo from CRM1 and RanGTP by an active displacement mechanism. RanGAP-mediated GTP hydrolysis terminates the disassembly reaction. Alternatively, the dis assembly reaction could proceed without using RanBDs, but RanBD-accelerated dissociation of cargo from CRM1 and RanGTP is much faster than the spontaneous dissociation of cargo, and so the disassembly pathway using RanBDs is most likely the major pathway of disassembly in the cytoplasm.
Recently, a high-resolution crystal structure of yeast CRM1-RanBP1-RanGTP complex, an intermediate complex in the disassembly reaction, has been determined (Fig. 2a)19. Importantly, the NES-binding cleft of CRM1 in this disassembly intermediate adopts a closed conformation (Fig. 2b), suggesting that RanBP1, which binds to a site located away from the NES-binding site, displaces NES by inducing conformational changes of the NES-binding cleft from the open structure to the closed structure. Comparison of the CRM1-RanBP1-RanGTP complex with the CRM1-cargo-RanGTP complex shows that a long β-hairpin loop of CRM1 (referred to as HEAT9 loop because this loop is a linker connecting the two α-helices of HEAT repeat 9) plays an important role in the long-range allosteric communication between the RanBP1- and NES-binding sites. In the NES-bound state, the HEAT9 loop is located away the NES-binding site and reaches across the CRM1 ring to interact with the switch I loop of Ran. In the RanBP1-bound state, RanBP1 interacts extensively with RanGTP, recruiting the C-terminal tail of Ran. Because of this, the HEAT9 loop can no longer bind Ran in this complex, and in turn moves to the inner surface of HEAT repeats 11 and 12 (immediately behind the NES-binding cleft), driving rotations and translations of the α-helices constituting the NES-binding cleft. This results in closure of the hydrophobic cleft to dissociate NES (Fig. 2b). Thus, the movement of the HEAT9 loop is the key for allosteric communication in CRM1. In addition to the movement of the HEAT9 loop, RanBP1-binding is also associated with a change in the superhelical path followed by HEAT repeats 12–19 of CRM1, which might contribute to stabilize the closed state of the NES-binding cleft. However, mutational analyses of the HEAT9 loop provided strong evidence that it is the direct interaction between the HEAT9 loop and the inner surface behind the NES-binding site that is vital for the release of cargo19.
How RanGAP acts on the CRM1-RanBP1-RanGTP complex remains an unresolved issue, but in the CRM1-RanBP1-RanGTP complex, the surface area of Ran directly contacting CRM1 is significantly reduced compared to CRM1-cargo-RanGTP complexes. This indicates that the binding of RanBP1 would facilitate dissociation of RanGTP from CRM1 and formation of RanGAP-RanGTP-RanBP1 complex. The crystal structure of the RanGAP-RanGTP-RanBP1 transition state complex suggests that RanGAP activates the GTPase by orientating catalytic glutamine in the switch II loop of Ran for activation of a water molecule that attacks the γ-phosphate of GTP20. The RanGAP-mediated GTP hydrolysis makes the disassembly reaction irreversible, and also provides the driving force for the active transport across the NPC, allowing accumulation of cargo against concentration gradient.
An autoinhibition hypothesis
In the CRM1-RanBP1-RanGTP complex, the closed conformation of the NES-binding cleft is stabilized by intra-molecular interactions in CRM1, namely the interactions between the HEAT9 loop and the inner surface of HEAT repeats 11 and 12. This indicates that the HEAT9 loop has an autoinhibitory function to inhibit cargo-binding in the absence of RanGTP, and that RanBP1 exploits the auto-inhibitory function of the HEAT9 loop to accelerate NES release19. Mutational analyses provided strong support for this autoinhibition hypothesis19. However, this is probably not the complete story of autoinhibition. Interestingly, in the binary CRM1-snurportin complex, the C-terminal α-helix of CRM1 adopts dramatically different conformation compared to the ternary export complex, and lies across the central cavity of CRM1 ring with its C-terminus located close to the NES-binding site13. This indicates that the C-terminus of CRM1 might regulate the affinity of NES in a RanGTP-sensitive manner. Indeed, mutagenesis studies showed that the C-terminus of CRM1 is required for inhibition of cargo binding in the absence of RanGTP21,22. The structure determination of unliganded CRM1 at high resolution is eagerly awaited to elucidate the precise mechanism of autoinhibition that renders CRM1 incapable of NES-binding in the absence of RanGTP.
Outlook
The determination of X-ray crystal structures of the CRM1 nuclear export complex and its disassembly intermediate was a significant advance and provided rich insights into the mechanism of cargo binding and release, which is crucial for transport directionality. Nevertheless, the inferences drawn from comparison of the crystal structures need to be corroborated by experimental observation of dynamic motions of CRM1 relevant to allosteric regulation. We also need to have complete description of the thermodynamics and kinetics of the nuclear export reaction. Another major remaining challenge in this field is the elucidation of the NPC passage mechanism, that is, the precise mechanism of how nuclear transport receptors such as CRM1 overcome the permeability barrier at the NPC. Recent advances in structural characterization of the NPC23 and development of experimental systems to analyze the formation of the passive permeability barrier at the NPC24 have been impressive, and form a firm foundation to decipher the enigmatic mechanism of NPC passage.
Acknowledgments
We thank our colleagues in Nagoya, especially Natsumi Saito, Natsuki Shirai, Junya Kobayashi and Hidemi Hirano, for valuable discussion. This work was supported in part by the Sumitomo Foundation and JSPS/MEXT KAKENHI (18687010, 21770109 and 23770110). MK was supported by JSPS Research Fellowship.
References
- 1.Mohr D, Frey S, Fischer T, Güttler T, Görlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 3.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 4.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 5.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr Opin Struct Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SJ, Jiko C, Yamashita E, Tsukihara T. Selective nuclear export mechanism of small RNAs. Curr Opin Struct Biol. 2011;21:101–108. doi: 10.1016/j.sbi.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Güttler T, Görlich D. Ran-dependent nuclear export mediators: a structural perspective. EMBO J. 2011;30:3457–3474. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 9.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 11.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 12.Xu D, Farmer A, Collett G, Grishin NV, Chook YM. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol Biol Cell. 2012;23:3677–3693. doi: 10.1091/mbc.E12-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong X, Biswas A, Süel KE, Jackson LK, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458:1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monecke T, Güttler T, Neumann P, Dickmanns A, Görlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324:1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 15.Güttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, Ficner R, Sattler M, Görlich D. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol. 2010;17:1367–1376. doi: 10.1038/nsmb.1931. [DOI] [PubMed] [Google Scholar]
- 16.Andrade MA, Petosa C, O’Donoghue SI, Müller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 17.Conti E, Müller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyama M, Matsuura Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 2010;29:2002–2013. doi: 10.1038/emboj.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seewald MJ, Körner C, Wittinghofer A, Vetter IR. RanGAP mediates GTP hydrolysis without an arginine finger. Nature. 2002;415:662–666. doi: 10.1038/415662a. [DOI] [PubMed] [Google Scholar]
- 21.Dong X, Biswas A, Chook YM. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol. 2009;16:558–560. doi: 10.1038/nsmb.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox AM, Ciziene D, McLaughlin SH, Stewart M. Electrostatic interactions involving the extreme C terminus of nuclear export factor CRM1 modulate its affinity for cargo. J Biol Chem. 2011;286:29325–29335. doi: 10.1074/jbc.M111.245092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 24.Hulsmann BB, Labokha AA, Gorlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]