Abstract
Skeletal myosin S1 consists of two functional segments, a catalytic-domain and a lever-arm. Since the crystal structure of ADP/Vi-bound S1 exhibits a strong intramolecular flexure between two segments, inter-conversion between bent and extended forms; i.e. “tilting of the lever-arm” has been accepted as the established molecular mechanism of skeletal muscle contraction. We utilized quick-freeze deep-etch replica electron microscopy to directly visualize the structure of in vitro actin-sliding myosin, and found the existence of a novel oppositely-bent configuration, instead of the expected ADP/Vi-bound form. We also noticed that SH1–SH2 cross-linked myosin gives an aberrant appearance similar to the above structure. Since SH1–SH2-cross-linked myosin is a well-studied analogue of the transient intermediate of the actomyosin cross-bridge cycle, we devised a new image-processing procedure to define the relative view-angles between the catalytic-domain and the lever-arm from those averaged images, and built a 3-D model of the new conformer. The lever-arm in that model was bent oppositely to the ADP/Vi-bound form, in accordance with observed actin-sliding cross-bridge structure. Introducing this conformer as the crucial intermediate that transiently appears during sliding, we propose a revised scheme of the cross-bridge cycle. In the scenario, the novel conformer keeps actin-binding in two different modes until it forms a primed configuration. The final extension of the lever-arm back to the original rigor-state constitutes the “power-stroke”. Various images observed during sliding could be easily interpreted by the new conformer. Even the enigmatic behavior of the cross-bridges reported as “loose chemo-mechanical coupling” might be adequately explained under some assumptions.
Keywords: tilting lever-arm hypothesis, in vitro motility, morphological image processing, single-particle analysis, quick-freeze deep-etch replica electron microscopy
Historical background on the study of molecular mechanism of muscle contraction
Serious pursuit in the framework of modern science on the molecular mechanism of muscle contraction was initiated by two inspiring ideas; “sliding-filament hypothesis” and “cross-bridge hypothesis”1–4 both proposed in the middle of 20th century. The former claimed that the contraction of skeletal muscle is driven by the sliding between thick and thin filaments in each sarcomere, while the latter attributes the engine of such sliding to the structural change of cross-bridges (i.e. swinging movement of myosin heads) that connect myosin molecules in the thick filaments to actin subunits in thin filaments. The sliding between two kinds of filaments was demonstrated by the microscopic observations of myofilaments before and after contraction1,2. The latter idea also seemed simple and expected to be demonstrable someday, by an adequate technique to visualize the molecular behavior. However, direct observation of the latter process has not yet been realized even today, after more than half a century of enthusiastic studies and the attempts by various means by numerous scientists.
One of the main reasons for such great difficulty, especially for skeletal muscle case, is certainly that myosin head is a highly mobile object that is too small as the target for microscopy, and no effective means is yet available to simultaneously attain sufficient spatial and time-resolution to accommodate its rapid and subtle structural change during contraction. [N.B. The movement of slower myosin-V species along actin filament was recently visualized as a movie5 using high-speed atomic-force microscopy.]
Tilting lever-arm hypothesis based on the established crystal structures of myosin
In the meantime, crystal structures of actin6 and various molecular species of myosin were successively solved in these two decades (the first myosin head structure was reported by Rayment et al.7). Among various important features of the crystal structures, the most remarkable issues might be the presence of two structural elements; the “catalytic (or motor) domain” and the “lever-arm” moieties in tandem6, and the large change in their relative angle that accompanies the binding of the nucleotides8,9. Together with the important notion that the actin-attached catalytic domain does not rotate during the power-stroke10,11, the strong flexure within a myosin head easily lead to a “titling lever-arm model” which took the place of previous simple tilting of total myosin head3,4. The message was so compelling that the novel idea readily spread, and nowadays, is widely accepted as if it were the final conclusion of extensive research history of muscle contraction over long years12–14. As a matter of fact, however, nobody showed concrete structural evidences to support the validity of such cross-bridge-cycle.
According to that hypothesis, the behavior of myosin head in the cross-bridge-cycle is presumed as follows12–14. Under nucleotide-free rigor-state, the catalytic domain of the myosin head is tightly bound to actin-filament in a configuration with its lever-arm extended. Upon binding of ATP to myosin’s catalytic site, its affinity to actin drops to ten-thousandth of that under rigor-condition to dissociate tight rigor-complex. Then, myosin instantaneously cleaves ATP to ADP and Pi, and converts itself to strongly-kinked nucleotide-bound configuration, as stated above. Though such configuration stably keeps the products of ATP-hydrolysis; ADP and Pi, reattachment of the catalytic domain with nearby actin somehow switches to accelerate the release of the products. Consequently, the catalytic domain re-binds firmly to actin and its lever-arm moiety returns to the original extended position. Since the actin-attached catalytic domain does not rotate during the power-stroke, the change in the relative angle between two modules generates the swinging movement of the lever-arm. Thus, this hypothetical cycle consists essentially of inter-conversion between the two well-documented myosin conformers, coupled with ATP-hydrolysis. In order to verify the validity of the hypothesis, the most straightforward and convincing evidence would be to directly visualize the time-course of the structural change, preferably as a movie, and confirm whether individual actin-sliding intermediates truly take nucleotide-bound kinked configuration, as postulated. Single-particle-analysis combined with quick-freeze cryo-electron microscopy15; might be proposed as the first choice to solve such problem. It is certainly a powerful solution for the analysis of large-sized supra-molecular assemblies, especially when some symmetry-based constraints could be imposed. Actually, very detailed atomic-level arguments were made for stable and regularly arranged rigor-complex16. On the other hand, the acto-S1 complex in the presence of ATP exhibited the images that were too much disordered17–20 as the target for conventional or sophisticated strategies for the structural studies like helical-reconstruction or single-particle-analysis as above. Further, the distal tail-portion of actin-attached myosin is floating free in the solution in most studies. A possibility arises that myosin head in such complex could take somewhat different configuration from that under functional states in vivo and in vitro, where myosin might bear some tension. Whatever, the most desired structural information of the intermediate remains ambiguous even now.
Advantageous features of quick-freeze deep-etch electron microscopy for direct observation of macromolecular assembly
Quick-freeze deep-etch replica electron microscopy is a unique technique that enables us to capture high-contrast snapshots of various biological events in situ21. There, target materials such as cells, organellae or supramolecular assemblies are quickly-frozen (usually by a contact with liq. He-cooled copper block) within less than a millisecond to arrest all the dynamic events. Then, the targets in the vitreous ice are exposed to a high vacuum to allow sublimation of surrounding ice (i.e. deep-etching), followed by metal evaporation and carbon backing. As long as the temperature of the mounting-stage is maintained sufficiently low, a layer of surface-bound-water, and probably, the three-dimensional architecture of the targets might be preserved frozen22 throughout the shadowing/replication process. We had consistently recognized the presence of delicate patterns on the shadowed surface of the target particles23,24, especially when the protein assemblies were pre-adsorbed onto mica-flakes25. We assumed that the distribution of the fine metal grains observed by electron microscopy could reflect the delicate surface profile of the target macromolecules, and examined if such patterns could truly be correlated with the contour of the target protein’s envelope23,24,26. Since the thickness of the accumulated metal at given position is a function of the angle between local normal-line and the direction of the evaporation source, we might be able to numerically calculate the distribution of the fine metal grains projected onto 2-D planes, according to the procedure of metal-accumulation onto rugged surface followed by its observation through transmission electron microscopy (Kimori & Katayama; unpublished data). Instead of such strict but extremely time-consuming simulation, we employed a simple light-rendering procedure to rapidly prepare a huge number of comprehensive set of artificial images by a dedicated ray-tracing software, to cover all the view-angles of the protein molecule23,24. We actually confirmed that such fuss-free method works nicely and enables us to determine the best-matched configuration and the view-angle of the model, to the image observed by freeze-replication, once the target’s atomic coordinates get available. Thus, we constructed a convenient system to select the most likely view-angle of the target proteins (complex) by a simple pattern-matching with a comprehensive set of artificial images24,26.
Search for the molecular species analogous to newly observed conformer during actin-sliding
In situ but static image data of the biological events could not give the conclusive evidence by itself, to verify underlying molecular mechanism of the dynamic phenomena. However, they could work as a strong constraint to rule out the inadequate interpretations of the observed events. In order to investigate the structure of myosin heads under various functional states, and ultimately the crucial intermediate during sliding, we took full advantage of this useful procedure combined with the mica-flake technique. We started to examine, at first, individual myosin (actually HMM; heavy-meromyosin) with or without various nucleotides27. HMM heads visualized in such way showed the images analogous to those prepared by surface-rendering of the registered X-ray data. Acto-HMM rigor-complex, most of which might be adsorbed to the mica surface through the S-2 moieties, exhibited a familiar arrowhead appearance28 that is similar to those shown by negative-staining or cryo-electron microscopy. By comparing the high magnification views of the observed particles with a series of artificial images, we confirmed that individual myosin head before addition of ATP is certainly under rigor-state having extended lever-arm (Fig.1). Taking those results as a good control, the images we would observe under sliding conditions after addition of ATP, should be reliable reflections of the real structural intermediate(s) during in vitro sliding. ATP was added to the mica slurry on which acto-HMM rigor-complex was pre-adsorbed28 and then, the total materials under functional states were quickly-frozen and replicated in the same way. What we actually observed under such condition was numerous actin-filaments, a part of which already slid out from HMM-tracks, and the rest still remaining along HMM zones on the mica substrate. The fraction of actin-bound myosin drastically decreased by the addition of ATP. We observed the structure of actin-bound HMM in such fields with higher magnification and found that it might be hard to account for the observed images by the conventional tilting-lever-arm mechanism. If well-documented actin-binding sites29 on nucleotide-bound kinked S1 is placed at the right position according to the above model, the lever-arm portion of the intermediate should stick out almost perpendicularly from the actin-filament. Contrary to the expectation, most HMM heads exhibited apparently a rounded shape that seemed to hold actin-filaments in the concave side of the curvature28 [N.B. The rounded appearance of actin-bound myosin head in the presence of ATP had been repeatedly documented both for native and chemically cross-linked acto-S1 with EDC; 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, either by negative-staining or cryo-electron-microscopy17–20]. Unless the location of actin-binding sites on the surface of the catalytic domain drastically changes during sliding, the most natural interpretation of such unexpected images is the presence of a novel configuration whose crystal structure is not yet reported. So, we started to look for candidate conformers having aberrantly flexed lever-arm by itself. After extensive search for various molecular species23,27 including those chemically-modified, we have recognized that a series of SH1–SH2 cross-linked myosin species could be the good candidates for such unusual appearance23,26, in which the lever-arm portion bent almost oppositely from that in conventional nucleotide-bound configuration.
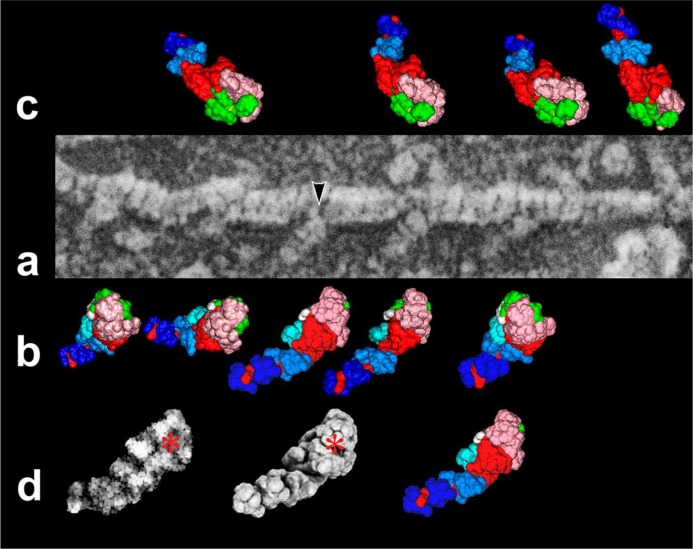
Figure 1.
a) Representative field of freeze-replicated actomyosin complex under rigor-state. All the actin-attached myosin heads show the configuration that matches rigor-form (1DFK), as defined by a comparison with simulated images. b)–c) Best-matched view for each particle along single actin filament is exhibited beside the original image. Particles on the bottom side (b) are overriding actin filament, whereas the very tip of those on the upper side (c) are partly hidden by actin. Subdomain composition of myosin S1 is color-coded as follows [upper-50 KDa, pink-tint; lower-50 K, green; N-terminal barrel, cyan; essential light-chain, sky-blue; regulatory light-chain, blue; 1st actin-contact site; yellow; 2nd actin-contact site, white; remaining part of heavy-chain including the lever-arm, red]. d) An example to examine the similarity of simulated image to replica image indicated by an arrowhead in (a). Left and center images represent the contrast-enhanced replica image and its best-matched simulation image generated by computer ray-tracing, respectively. The image texture had been pre-matched by morphological image-processing24. Note the good correlation between them. The asterisk indicates the position of the ATP-binding pocket. Subdomain constitution of the middle image was color-coded as above on the right. Here, the area of loop-2 (yellow) is much smaller than that in the actual molecule, due to the lack of atomic coordinates of that segment.
SH1 and SH2 (Cys707 and Cys697 in rabbit skeletal myosin sequence) are highly reactive thiol groups in the heart of myosin, whose modification causes prominent effects on its ATPase activity30. Though they are close to each other in the amino-acid sequence, they are pointing toward the opposite directions at both ends of an α-helix (SH1-helix), under rigor-state31. Such arrangement does not allow their approach in the real 3-D space, and chemical cross-linking could occur only when that helix is melted by the binding of nucleotides. It is also reported that the distance between the two thiols could substantially vary from zero (i.e. disulfide) up to 15 Å, indicating highly flexible nature of that portion, once disrupted32,33. Biochemical properties of such cross-linked species had been well studied and it is assumed as one of the dominant structural analogues of the transient intermediates34 in actomyosin cross-bridge-cycle.
Cohen’s team extensively studied the crystal structures of scallop myosin S1 complexed with various nucleotides, and argued on the subtle structural differences among them35–37. They also solved the atomic structure of the same S1 but pretreated with thiol-specific bifunctional reagent (p-PDM; N,N′-1,4-phenylenedimaleimide)31. In their crystal, chemical cross-linking oddly occurred between SH2 and Lys-705 (in the scallop’s sequence), instead of SH1, whose side-chain protrudes to the same face as that of SH2 under rigor-state (Fig. 2). Actually, the angle of the lever-arm in that structure did not appreciably differ from the other uncross-linked species. Nitao et al.38 later examined the reactivity of those thiols in scallop S1 and confirmed that more than 90% of the product nevertheless had SH1–SH2 cross-linking, suggesting the poor crystallizability of the inter-thiol cross-linked species. Though the crystal structure of the major product is not yet solved, we could have observed such majority whose lever-arm portion might be rotated to the other side and fixed in that position.
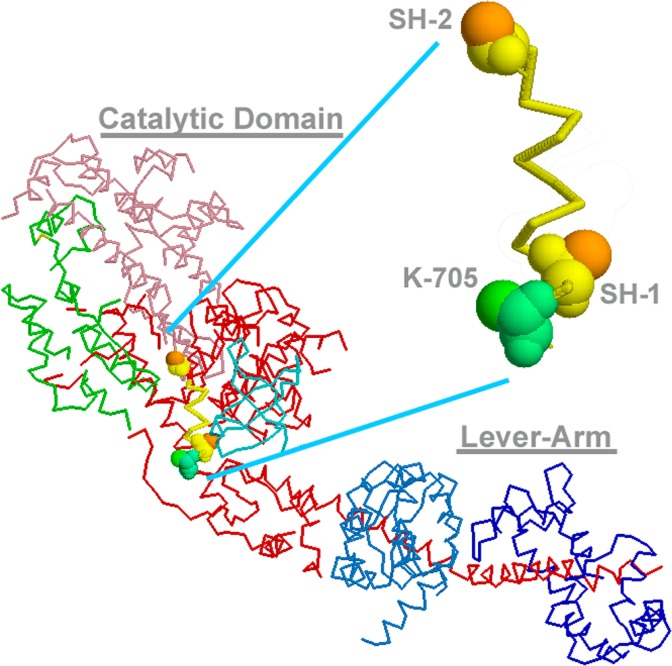
Figure 2.
Crystal structure of scallop myosin S1 under nucleotide-free rigor state (1DFK) and the 3-D spatial arrangement of SH1, SH2 (Cys-703 and Cys-693, in the scallop’s sequence) and Lys-705 along SH1-helix. SH1-helix portion is enlarged to indicate that two thiols are pointing to the opposite directions, where disulfide formation is impossible. Lys-705 is protruding to the same side as SH2 and they were actually cross-inked by p-PDM30,37 to give 1L20 structure. The lever-arm might flip to the other side, when SH1-helix is disrupted so that two thiols form disulfide.
Aberrant features of the novel conformer as compared with the other myosin structures
Since we needed, by any means, to get even the rough sketch of inter-thiol cross-linked myosin, we devised a new version of single particle analysis for freeze-replica images with the aid of morphological image-processing24. To avoid the problem which could occur by the lenient specificity of p-PDM as above, we treated HMM with DTNB; 5,5′-Dithio-bis-(2-Nitrobenzoic Acid) to form pure disulfide39; the shortest cross-link between the two thiols (SH1 and SH2), and took a number of images of rounded HMM heads. After classification and averaging, we obtained the class-averages projected to five different view-angles. We dissected the total image of each class-average into the globular catalytic domain and the rod-like lever-arm and separately determined the view-angles for each, by pattern matching with the artificial images prepared from the atomic models of those modules. Reliability of our daring method, as well as the actual values of view-angles were simultaneously ensured by a good compatibility of the relative angle between two modules in each class-average. We could also estimate the relative position of the two modules in 3-D space from the class-averages and successfully obtained coarse atomic model to mimic the novel conformer. We separately reconstructed the 3-D shape of the envelope from those projections, that covers the whole surface of the new conformer26. Though the direction of the lever-arm in the novel structure was quite apart from the other conformers, the putative actin-binding sites assigned in the tentative model (Fig. 3) well accounted for the images of the possible intermediates we observed under actin-sliding conditions.
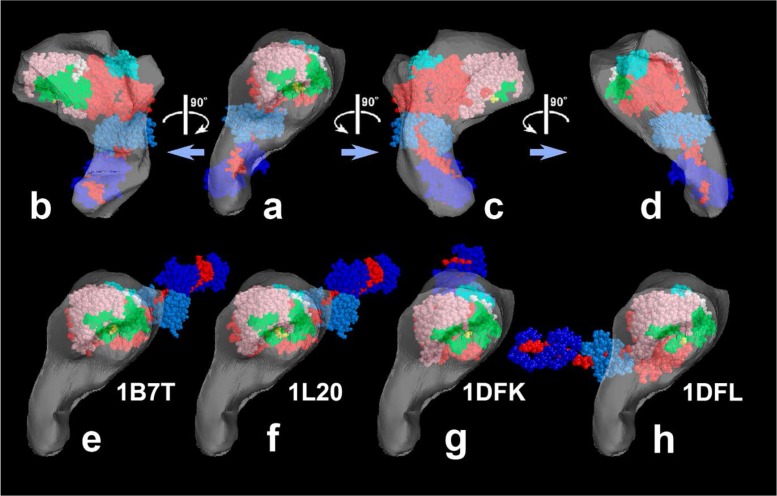
Figure 3.
Reconstructed shell to present 3-D envelope of the SH1–SH2 cross-linked myosin head [modified from Figure 7 of our paper26]. a)–d); Its tentative model was fit to the shell and viewed from four different angles. e)–h); Four known crystal structures of scallop-S1 whose catalytic domains are snugly placed in the shell of the novel structure. Note that the orientation of the lever-arm in the novel structure is directed quite differently from the others. The atomic models are, e) ADP-bound form, f) ADP-bound Lys705-Cys693 cross-linked form, g) nucleotide-free near-rigor form, h) ADP/Vi-bound form. See the legend to Figure 1 for color-coding of the subdomains.
We continued the examination on the mode of myosin attachment to actin-filament during sliding. Though the lever-arm portion was not always visible, hidden by neighboring actin-filaments or myosin head by itself, most of them seem to take the novel configuration, as judged from the appearance of the visible part. According to preliminary results26 (Katayama and Kimori, unpublished data), the mode of actin-attachment of the new conformers could be roughly classified into two populations; the first, the particles whose concave side directly faces the actin surface as if it grasps the filament, and the second, the ones 90-degrees rotated to the left around its long axis, from the first mode, and associated with actin along the side of the curved body. The first group was further classified according to the variation in the angles between long axis of the S1 body and the actin filament. All the observed images as above were easily interpreted as various views of the novel configuration we discovered, and probably correspond to respective steps of the crossbridge-cycle. Unfortunately, however, quick-freeze replica images are merely the snapshots of the contiguous events. In order to determine the sequence of such conformational states along the time-course, we absolutely need separate lines of experimental evidence.
Involvement of the new conformer in the time-course of revised cross-bridge cycle
Andreev and Reshetnyak40 published their experimental results on time-resolved chemical cross-linking during actomyosin interaction. Actomyosin complex has two pairs of contact-sites29 on each molecule’s surface, which can be covalently connected by EDC, a zero-length cross-linker. They measured the increment of the cross-linked fraction for each pair at the initial stage of the reaction and found that the rate of contact between the second sites greatly slows down if ATP was added, whereas the two reactions occur at similar rates in the absence of ATP. Coupling such invaluable information with the structural features we observed, we could deduce the sequence of the events as illustrated in Figure 426.
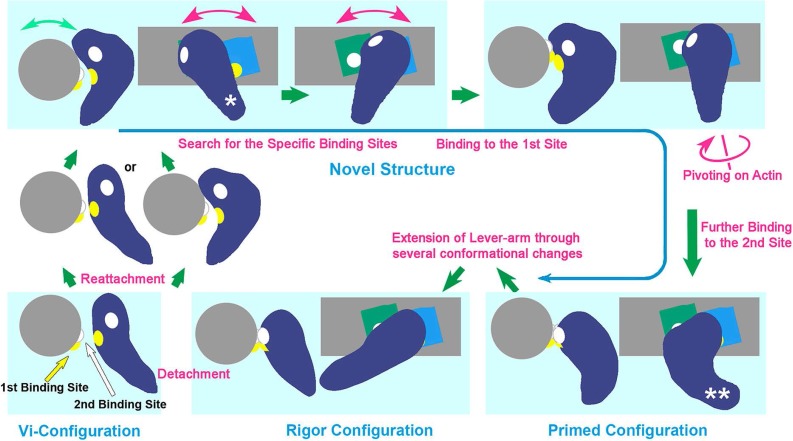
Figure 4.
Schematic drawing of revised cross-bridge-cycle of skeletal actomyosin including the new conformer (taken from Fig. 8 of the reference 26). The cycle starts from the dissociation of rigor myosin from actin by the binding of ATP, which eventually re-associates with actin through several conformational states mostly involving the new configuration. The structural states placed in light-bluish panels are presented as the side- and the barbed-end views of the actomyosin complex. Violet; myosin head, blue and green; actin-monomers along actin-filament (gray). There, the behavior of two pairs of actomyosin interaction sites (presented by yellow and white patches in the reaction sequence), are indicated according to the time-sequence of the structural change from conventionally-bent ADP/Pi-structure (similar to the ADP/Vi-bound form) right after dissociation from actin, to the final rigor structure [N.B.; The third actin-binding site is located close to the yellow patch and might concomitantly operate with the second site to start lever-arm swinging]. Hence, the new conformer plays a crucial role as the reaction intermediate, throughout those states. The configurations marked by a single and double asterisk in the drawing correspond to the views (d) and (c) in Figure 3, respectively. See text for details.
(i) By the addition of ATP to actomyosin rigor-complex, myosin hydrolyses ATP and instantaneously dissociates from actin to form the ADP/Vi (actually ADP/Pi) structure with the conventionally-kinked lever-arm. (ii) Accompanied by the reattachment of the myosin to actin, SH1 helix melts and the lever-arm portion twists to the other side converting the total myosin head into the new conformer. (iii) The myosin head in that configuration swings back and forth along the axis of the actin filament, so that the first contact-site on the inner side of the curved body kept in face to the actin filament. (iv) The interaction between the first paired sites (635–647th amino-acid residues of S1 so-called loop-2, and the N-terminal segment of actin; as indicated in yellow) occurs while the second site of myosin (567–574th amino-acid residues so-called loop-3 as indicated in white) is on the outer surface facing the opposite side (Figs. 3 and 4). (v) Then, the new conformer pivots toward the left in search for the next contact site(s), and eventually finds a counterpart (white) at the N-terminus of the neighboring actin monomer towards the barbed-end direction. (vi) Establishment of the second contacts firmly immobilizes the catalytic domain of the entire structure in the orientation similar to the rigor one, so that it takes the pre-power-stroke primed configuration. (vii) Triggered by the recovery of the disrupted SH1 helix, the lever-arm portion finally extends to return to the original rigor configuration. Since the catalytic domain had been immobilized onto actin, the last step certainly constitutes “the swing of the lever-arm” which might proceed through several other conformational steps (Katayama, unpublished data). Várkuti et al.41 reported on another interaction sites named “activation-loop”; i.e. 254–258th amino-acid residues of myosin and N-terminal segment of actin. They showed that the loop is essential to evoke actin activation of myosin ATPase. Located in between the 1st and the 2nd interaction sites of myosin, a basic residue in the 3rd loop might contribute to switch the final power-stroke41, and/or possibly, to tether total myosin head attached to the flexible acidic segments at the N-terminal of actin. [N.B. There seems to be some disagreement on the assignment of one of the chemically-cross-linked products between actin and myosin40,41. Whichever be the case, the positional relationship among them might be kept and does not much affect the conclusion of our argument.]
Considering the beginning and the ending positions of the distal end of the lever-arm during the final power-stroke that starts from the primed configuration, the resultant vector of the swinging movement is closer to parallel to the actin-filament, possibly more energy-efficient than that starting from the conventionally-kinked ATP-bound structure. This is consistent with the observation that myosin-II produces little rotational torque during sliding42,43.
Taylor and his colleagues extensively studied the 3-D molecular architecture of insect flight muscle by quick-freezing during tension-development44. They reconstructed the tomograms from freeze-substituted thin sections to examine in situ actomyosin configurations. As a summary, they presented the animated atomic models to indicate the conformational change of insect myosin-II that starts from conventional ADP/Vi-structure and ends up by rigor-like configuration. The two-stage movement and the timing of the power-stroke they showed agree well with our conclusion. On the other hand, the envelope of tension-bearing states in the tomogram seems to have a narrow and somewhat inclined neck at the origin along thick-filament and abrupt thickening toward the interface with actin. Such profile matches well with the characteristic feature of the primed configuration of the novel structure we showed (Katayama unpublished data).
Capitanio et al.45 improved their experimental systems to analyze single-molecule mechanical events, using optical tweezers. They actually examined the properties of skeletal-type myosin isoforms and found that the actomyosin interaction process can be dissected into two phases with distinct kinetic parameters. The structural change we have proposed as above might more or less reflect the functional phases they reported.
Potential involvement of the new conformer as the structural basis of “loose chemo-mechanical coupling”
Yanagida and his colleagues reported, for the first time, that the sliding distance of the unloaded actin during skeletal muscle contraction is much longer than that expected from actomyosin ATPase46 assuming “tight chemo-mechanical coupling” (i.e. one actin subunit displacement per one ATP-hydrolysis). Since then, they have persistently exhibited the “loose chemo-mechanical coupling”47,48, in which the ratio of occurrence between ATP cleavage and the actin-displacement by myosin changes according to the applied load, and deviates from one to one when it becomes lighter. As the origin of such characteristic mechanical features, they proposed “the biased Brownian-motion” of myosin along the surface of actin-filament. The experimental conditions we employed to observe actin-sliding seems to be coincidentally close to those where actin’s translocation distance is more than one unit per ATP-hydrolysis. Our replica images certainly show the existence of definite structural variants, and the differences among them could be taken as to reflect the “swinging of the lever-arm”. The energy consumption of such drastic structural change is expected to be substantially large. Thus, we speculate that if any enigmatic behavior49 of myosin responsible to loose-coupling could occur, that might be attributable to some “Brownian ratchet” mechanism50 which could predominate during the third or the fourth, seemingly more iso-energetic step(s); i.e. actin surface scanning by myosin head under weakly-bound states. If both mechanisms operate sequentially in each ATPase cycle, the actin-translocation for one ATP consumption at light load, might be substantially longer due to the displacement by the Brownian-ratchet mechanism, whereas the travel distance would approach to one actin-unit (more precisely, the span-length of the lever-arm) for the heavier load, propelled only by the powerful lever-arm swinging. The experimental results by Várkuti et al.41 sounds quite intriguing along that line. They found that the mutation of basic residue or the truncation of the activation loop greatly reduces the generated force by myosin as well as its actin-activated ATPase, whereas the velocity of actin displacement assessed by a motility assay was not much affected by the same mutation. Such experimental evidence could imply clear dissociation of the tension development process from that of actin translocation; in support of our interpretation. If such a mechanism could be true, we might predict that higher fraction of primed configuration should be observed under the conditions where actin filaments are more heavily loaded by any means. In reverse, very few or none of the myosin would take primed configuration, if the activation-loop mutants are used for the similar observation. In such case, the attachment angles of the catalytic domain to actin-filament might substantially fluctuate during actin translocation.
Epilogue
The investigation on the mechanism of biological motor started from that of the skeletal-muscle contraction. Studies along that line had been the main-stream of the biophysics for a long time. Actually, a number of cutting-edge experimental techniques such as single-molecule observation/manipulation devised for that specific purpose, not only contributed to produce various outstanding discoveries, but also rapidly spread and were applied in the neighboring fields. Since the proposal of “swinging lever-arm” hypothesis, however, the past enthusiasm as reflected by the number and the depth of the studies on skeletal-type myosin (myosin-II) seems to be substantially reduced, partly due to increasing technical limitation to further challenge the difficulty of still-persisting issues, but more likely because the majority of research scientists might get convinced of, in fact still hypothetical “swinging cross-bridge” stories. On the other hand, the discovery of novel unconventional myosin species attracted people’s research interests and the vast research activity has moved to the new materials. The movements of unconventional motor-proteins are generally slower and processive, seemingly easier to handle than faster and non-processive myosin-II. It might not be a good idea to go to the bother of dealing more difficult materials, if the underlying mechanism are evidently common to all the myosin species. However, there is no a priori way to judge whether it is so or not, unless we actually examine and get solid experimental evidence one by one, with multimodal approaches including direct visualization.
We showed the existence of a heretofore unknown configuration similar to the one realized in SH1–SH2-crosslinked myosin-II, under in vitro motility assay conditions. By including such structure as the key component of the cross-bridge cycle of the skeletal muscle, we could successfully account for the images actually observed under actin-sliding conditions. Although the above reaction scheme was proposed as a plausible interpretation of the images captured during in vitro sliding, we would presume that essentially similar events might occur in live muscle contraction.
Whatsoever, it is clear that more and more efforts are needed to truly elucidate the motor mechanism, even if the target is an extensively studied skeletal system.
Acknowledgments
A series of our long work referred in this review were carried out partly with the financial support by the System Development Program for Advanced Measurement and Analysis (Program-S) from the Japan Science and Technology Agency, and by several Grants-in-aid for Scientific Research, Grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan.
The author is greatly indebted to Drs Y. Kimori (National Institute for Basic Biology, Okazaki, Japan) and N. Baba (Kogakuin University, Hachioji, Tokyo, Japan) for their great cooperation in image-processing, Dr T. Q. P. Uyeda (National Institute of Advanced Industrial Science and Technology, Tsukuba, Ibaraki, Japan) for constructive discussion and some assistance in biochemical experiments. Dr S. Maruta and his colleagues (Soka University, Hachioji, Tokyo, Japan) cooperated in the initial part of the study and kindly prepared the protein materials for electron microscopy. Dr M. Miyata (Osaka City University, Osaka, Japan) gave his perpetual moral support.
References
- 1.Huxley AF, Niedergerke R. Structural changes in muscle during contraction: interference microscopy of living muscle fibres. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- 2.Huxley HE, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- 3.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. [PubMed] [Google Scholar]
- 4.Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 5.Kodera N, Yamamoto D, Ishikawa R, Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 6.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin: DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 7.Rayment I, Rypniewski WR, Schmidt-Bäse K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 9.Smith CA, Rayment I. X-ray structure of the Magnesium(II)·ADP·vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 10.Yanagida T. Angles of nucleotides bound to cross-bridges in glycerinated muscle fiber at various concentrations of ɛ-ATP, ɛ-ADP and ɛ-AMPPNP detected by polarized fluorescence. J Mol Biol. 1981;146:539–560. doi: 10.1016/0022-2836(81)90046-2. [DOI] [PubMed] [Google Scholar]
- 11.Cooke R, Crowder MS, Thomas DD. Orientation of spin labels attached to cross-bridges in contracting muscle fibres. Nature. 1982;300:776–778. doi: 10.1038/300776a0. [DOI] [PubMed] [Google Scholar]
- 12.Holmes KC. The swinging lever-arm hypothesis of muscle contraction. Curr Biol. 1997;7:R112–R118. doi: 10.1016/s0960-9822(06)00051-0. [DOI] [PubMed] [Google Scholar]
- 13.Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 14.Spudich JA. The myosin swinging cross-bridge model. Nat Rev Mol Cell Biol. 2001;2:387–392. doi: 10.1038/35073086. [DOI] [PubMed] [Google Scholar]
- 15.Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Biomol Struct. 2002;31:303–319. doi: 10.1146/annurev.biophys.31.082901.134202. [DOI] [PubMed] [Google Scholar]
- 16.Behrmann E, Müller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig R, Greene LE, Eisenberg E. Structure of the actin-myosin complex in the presence of ATP. Proc Natl Acad Sci USA. 1985;82:3247–3251. doi: 10.1073/pnas.82.10.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama E. The effects of various nucleotides on the structure of actin-attached myosin subfragment-1 studied by quick-freeze deep-etch electron microscopy. J Biochem (Tokyo) 1989;106:751–770. doi: 10.1093/oxfordjournals.jbchem.a122928. [DOI] [PubMed] [Google Scholar]
- 19.Pollard TD, Bhandari D, Maupin P, Wachsstock D, Weeds AG, Zot HG. Direct visualization by electron microscopy of the weakly bound intermediates in the actomyosin adenosine triphosphatase cycle. Biophys J. 1993;64:454–471. doi: 10.1016/S0006-3495(93)81387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker M, White H, Belknap B, Trinick J. Electron cryomicroscopy of acto-myosin-S1 during steady-state ATP hydrolysis. Biophys J. 1994;66:1563–1572. doi: 10.1016/S0006-3495(94)80948-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heuser JE, Reese TS, Dennis MJ, Jan Y, Jan L, Evans L. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J Cell Biol. 1979;81:275–300. doi: 10.1083/jcb.81.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross H. High resolution metal replication of freeze-dried specimens in Cryotechniques. In: Steinbrecht RA, Zierold K, editors. Biological Electron Microscopy. Springer-Verlag; Berlin, Heidelberg: 1987. pp. 205–215. [Google Scholar]
- 23.Katayama E, Ichise N, Yaeguchi N, Yoshizawa T, Maruta S, Baba N. Three-dimensional structural analysis of individual myosin heads under various functional states. Adv Exp Med Biol. 2003;538:295–304. doi: 10.1007/978-1-4419-9029-7_28. [DOI] [PubMed] [Google Scholar]
- 24.Kimori Y, Oguchi Y, Ichise N, Baba N, Katayama E. A procedure to analyze surface profiles of the protein molecules visualized by quick-freeze deep-etch replica electron microscopy. Ultramicroscopy. 2007;107:25–39. doi: 10.1016/j.ultramic.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Heuser JE. Procedure for freeze-drying molecules adsorbed to mica flakes. J Mol Biol. 1983;169:155–195. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- 26.Kimori Y, Baba N, Katayama E. Novel configuration of a myosin II transient intermediate analogue revealed by quick-freeze deep-etch replica electron microscopy. Biochem J. 2013;450:23–35. doi: 10.1042/BJ20120412. [DOI] [PubMed] [Google Scholar]
- 27.Maruta S, Uyehara Y, Aihara T, Katayama E. Interaction of myosin·ADP·fluorometal complexes with fluorescent probes and direct observation using quick-freeze deep-etch electron microscopy. J Biochem (Tokyo) 2004;136:57–64. doi: 10.1093/jb/mvh095. [DOI] [PubMed] [Google Scholar]
- 28.Katayama E. Quick-freeze deep-etch electron microscopy of the actin–heavy meromyosin complex during the in vitro motility assay. J Mol Biol. 1998;278:349–367. doi: 10.1006/jmbi.1998.1715. [DOI] [PubMed] [Google Scholar]
- 29.Milligan RA. Protein–protein interactions in the rigor actomyosin complex. Proc Natl Acad Sci USA. 1996;93:21–26. doi: 10.1073/pnas.93.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekine T, Yamaguchi M. Effect of ATP on the binding of N-ethylmaleimide to SH groups in the active site of myosin ATPase. J Biochem (Tokyo) 1963;54:196–198. [PubMed] [Google Scholar]
- 31.Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Györgyi AG, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci USA. 2002;99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells JA, Knoeber C, Sheldon MC, Werber MM, Yount RG. Cross-linking of myosin subfragment 1: nucleotide- enhanced modification by a variety of bifunctional reagents. J Biol Chem. 1980;255:11135–11140. [PubMed] [Google Scholar]
- 33.Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bobkov AA, Reisler E. Is SH1–SH2-cross-linked myosin subfragment 1 a structural analog of the weakly-bound state of myosin? Biophys J. 2000;79:460–467. doi: 10.1016/S0006-3495(00)76307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houdusse A, Szent-Györgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gourinath S, Himmel DM, Brown JH, Reshetnikova L, Szent-Györgyi AG, Cohen C. Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Å resolution: flexibility and function in the head. Structure. 2003;11:1621–1627. doi: 10.1016/j.str.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Houdusse A, Kalabokis VN, Himmel D, Szent-Györgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999;97:59–70. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 38.Nitao LK, Loo RR, O’Neall-Hennessey E, Loo JA, Szent-Györgyi AG, Reisler E. Conformation and dynamics of the SH1–SH2 helix in scallop myosin. Biochemistry. 2003;42:7663–7674. doi: 10.1021/bi027312u. [DOI] [PubMed] [Google Scholar]
- 39.Wells JA, Yount RG. Reaction of 5,5-dithiobis(2-nitro-benzoic acid) with myosin subfragment one: evidence for formation of a single protein disulfide with trapping of metal nucleotide at the active site. Biochemistry. 1980;19:1711–1717. doi: 10.1021/bi00549a030. [DOI] [PubMed] [Google Scholar]
- 40.Andreev OA, Reshetnyak YK. Mechanism of formation of actomyosin interface. J Mol Biol. 2007;365:551–554. doi: 10.1016/j.jmb.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 41.Várkuti BH, Yang Z, Kintses B, Erdélyi P, Bárdos-Nagy I, Kovács AL, Hári P, Kellermayer M, Vellai T, Málnási-Csizmadia A. A novel actin binding site of myosin required for effective muscle contraction. Nat Struct Mol Biol. 2012;19:299–306. doi: 10.1038/nsmb.2216. [DOI] [PubMed] [Google Scholar]
- 42.Sase I, Miyata H, Ishiwata S, Kinosita K., Jr Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proc Natl Acad Sci USA. 1997;94:5646–5650. doi: 10.1073/pnas.94.11.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beausang JF, Schroeder HW, 3rd, Nelson PC, Goldman YE. Twirling of actin by myosins II and V observed via polarized TIRF in a modified gliding assay. Biophys J. 2008;95:5820–5831. doi: 10.1529/biophysj.108.140319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu S, Liu J, Reedy MC, Tregear RT, Winkler H, Franzini-Armstrong C, Sasaki H, Lucaveche C, Goldman YE, Reedy MK, Taylor KA. Electron tomography of cryofixed, isometrically contracting insect flight muscle reveals novel actin–myosin interactions. PLoS ONE. 2010;5:e12643. doi: 10.1371/journal.pone.0012643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Capitanio M, Canepari M, Cacciafesta P, Lombardi V, Cicchi R, Maffei M, Pavone FS, Bottinelli R. Two independent mechanical events in the interaction cycle of skeletal muscle myosin with actin. Proc Natl Acad Sci USA. 2006;103:87–92. doi: 10.1073/pnas.0506830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanagida T, Arata T, Oosawa F. Sliding distance of actin filament induced by a myosin crossbridge during one ATP hydrolysis cycle. Nature. 1985;316:366–369. doi: 10.1038/316366a0. [DOI] [PubMed] [Google Scholar]
- 47.Harada Y, Sakurada K, Aoki T, Thomas DD, Yanagida T. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J Mol Biol. 1990;216:49–68. doi: 10.1016/S0022-2836(05)80060-9. [DOI] [PubMed] [Google Scholar]
- 48.Ishijima A, Kojima H, Funatsu T, Tokunaga M, Higuchi H, Tanaka H, Yanagida T. Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 1998;92:161–171. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 49.Cyranoski D. Swimming against the tide. Nature. 2000;408:764–766. doi: 10.1038/35048748. [DOI] [PubMed] [Google Scholar]
- 50.Vale RD, Oosawa F. Protein motors and Maxwell’s demons: Does mechanochemical transduction involve a thermal ratchet? Adv Biophys. 1990;26:97–134. doi: 10.1016/0065-227x(90)90009-i. [DOI] [PubMed] [Google Scholar]