Abstract
Two mammalian DNA glycosylases, methyl-CpG binding domain protein 4 (MBD4) and thymine DNA glycosylase (TDG), are involved in active DNA demethylation via the base excision repair pathway. Both MBD4 and TDG excise the mismatch base from G:X, where X is uracil, thymine, and 5-hydroxymethyluracil (5hmU). In addition, TDG excises 5mC oxidized bases i.e. when X is 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) not 5-hydroxymethylcytosine (5hmC). A MBD4 inactive mutant and substrate crystal structure clearly explains how MBD4 glycosylase discriminates substrates: 5mC are not able to be directly excised, but a deamination process from 5mC to thymine is required. On the other hand, TDG is much more complicated; in this instance, crystal structures show that TDG recognizes G:X mismatch DNA containing DNA and G:5caC containing DNA from the minor groove of DNA, which suggested that TDG might recognize 5mC oxidized product 5caC like mismatch DNA. In mutation studies, a N157D mutation results in a more 5caC specific glycosylase, and a N191A mutation inhibits 5caC activity while that when X=5fC or T remains. Here I revisit the recent MBD4 glycos ylase domain co-crystal structures with DNA, as well as TDG glycosylase domain co-crystal structures with DNA in conjunction with its mutation studies.
Keywords: DNA glycosylase, methyl-CpG binding domain 4, Thymine DNA Glycosylase, active demethylation
Cytosine modifications
In the mammalian genome, cytosine residues can be dynamically modified at the C5 position, and can exist as either unmethylated cytosine (C), methylcytosine (5mC), hydroxymethylcytosine (5hmC), fomylcytosine (5fC), or carboxylcytosine (5caC) (Fig. 1)1,2. Three DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b) methylate cytosine (C) to 5mC, and three ten-eleven translocation (TET1, TET2, and TET3) dioxygenases oxidize 5mC to 5hmC, 5fC and 5caC3,4. DNA methylation/unmethylation plays an important role in gene silencing, and DNA methylation levels vary in different tissues, at different ages, and in different genomic locations5,6. So far, there is no reported enzyme which directly removes a methyl-group from 5-methylated cytosine (5mC), but N-methylated nucleic acids (3-methylated cytosine, 3-methylated thymine and 1-methylated adenine) can be removed. Oxidization of N-methylated nucleic acids involves in a 2-step demethylation processes. N-methylated nucleic acids are oxidized by AlkB proteins, and their hydroxymethyl-groups are spontaneously released from base with the byproduct, formaldehyde7–9. However, the oxidative products of C-methylated nucleic acids are stable bases10. 5mC oxidized products, 5hmC, and 5fC, act as substrates of TET proteins, which can be converted to 5caC as the final product in vitro11,12.
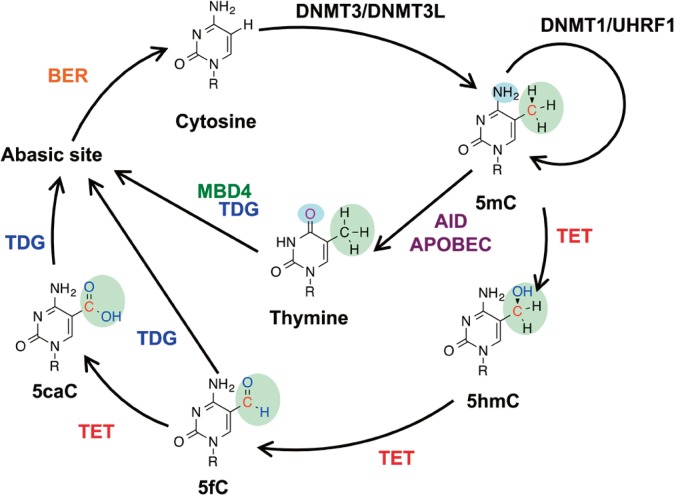
Figure 1.
Known and putative DNA demethylation pathways. DNMT3/DNMT3L complex produces new DNA methylation patterns by methylating cytosine (C) to 5-methylated cytosine (5mC), while the DNMT1/UHRF1 complex maintains previous cytosine methylation after the DNA replication. Ten-eleven translocation (TET) proteins consecutively oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). 5fC and 5caC can be removed by thymine DNA glycosylase (TDG) and replaced by cytosine via base excision repair (BER). AID (activation-induced cytidine deaminase)/APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide) enzymes deaminate 5-methylated cytosine (5mC) to thymine (T), and forms a G:T mismatch. Thymine is removed by TDG and MBD4, and replaced by cytosine via base excision repair (BER).
There are two DNA demethylation pathways, one is a replication-dependent “passive demethylation” pathway, and the other is a replication-independent “active demethylation” pathway. In mammals, two “active” demethylation pathways via base excision repair pathway have been proposed1,2,13. One is a deamination mediated demethylation pathway. DNA deaminases, like AID (activation-induced cytidine deaminase) and APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide) proteins, deaminate 5-methylcytosine (5mC) to thymine (T), and form a G:T mismatch14,15. MBD4 glycosylase and TDG glycosylase then excise the thymine paired with guanine, and generate abasic sites (AP sites). AP sites can then be repaired via base excision repair pathway. The other is a TET-mediated pathway. TET proteins consecutively oxidize 5mC and convert it to 5hmC, 5fC, and then 5caC16. Subsequently, TDG glycosylase excises 5fC and 5caC (both in vivo and in vitro)17,18 and generates AP sites. These AP sites can then be repaired via the base excision repair pathway (Fig. 1). Here I revisit the two “active” 5mC demethylation related glycosylases, MBD4 and TDG, their glycosylase domain DNA co-crystal structural studies and their mutation studies.
MBD4 glycosylase structure and substrates specificity
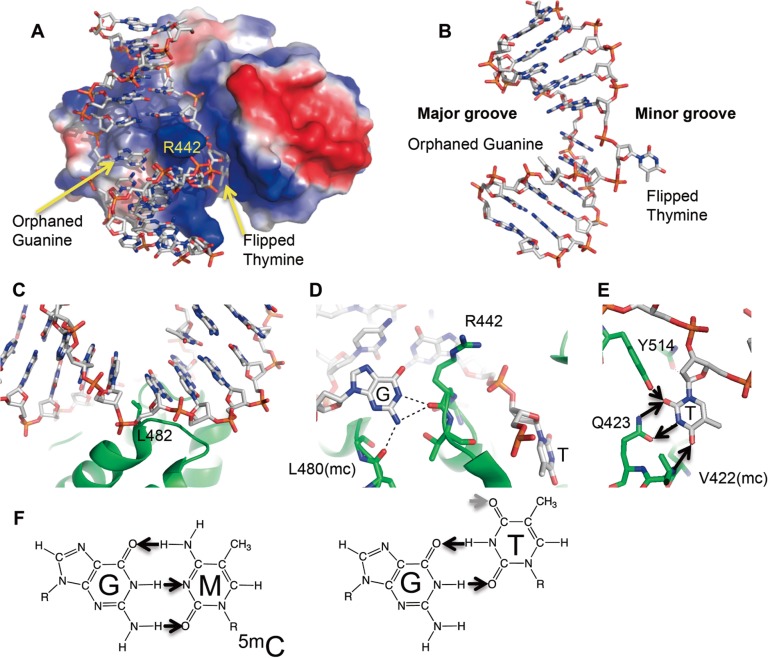
Mouse methyl-CpG binding domain 4 (MBD4) protein encodes 554 amino acids, and has two domains; the methyl-CpG DNA binding domain (MBD) domain at the amino-terminus and glycosylase domain at carboxyl-terminus19,20. The N-terminal MBD domain recognizes a G:T mismatch as well as fully methylated CpG dinucleotide19,21. The C-terminal glycosylase domain excises uracil, thymine (also known as 5-methyluracil) and 5-hydroxymethyluracil (5hmU) paired with guanine, which forms deamination linked mismatches. MBD4 catalytic inactive mutant (Asp534Asn for mouse, Asp560Ala for human) and substrate DNA crystal structures clarifies how MBD4 recognizes substrates22,23. In the mouse MBD4 inactive protein (D534N) and G:T mismatch pair substrate complex structure (Fig. 2A; PDB: 4EVV)22, in which the protein structure is highly similar to the APO structure (PDB: 1NGN)24, the G:T mismatch containing 11-bp DNA duplex was bent ∼65° (Fig. 2B), between the G:T mismatch and a C:G base pair by the Leu482 side chain (Fig. 2C). Concurrently, the thymine nucleotide in the mismatch flips out, and an arginine finger (Arg442) penetrates into the space left by the flipped thymine (Fig. 2A). The intrahelical orphaned guanine is stabilized by hydrogen bonds with the main chain carbonyl oxygen atoms of Arg442 and Leu480 (Fig. 2D). Asn441 and Leu485 sit in the minor groove of DNA, and interact weakly with the neighboring G:C pair. No interaction to the neighboring G:C pair in the major groove of DNA is observed and neighboring cytosine modification does not affect glycosylase activities25. The polar groups of the flipped thymine base along the Watson–Crick edge are all hydrogen bonded with Thr514 side chain, Gln423 side chain, and Val422 main chain (Fig. 2E). The O2 oxygen atom of thymine base accepts hydrogen bonds from the side chains of Tyr514 (hydroxyl group) and Gln423 (amino group), the N3 nitrogen atom of thymine donates a hydrogen bond to the carbonyl oxygen of Gln423 side chain and the O4 oxygen atom of thymine accepts a hydrogen bond from the main-chain amide nitrogen of Val422 (Fig. 2E). Because thymine and cytosine bases have different polar edges where MBD4 recognizes a flipped thymine base (Fig. 2E, 2F), MBD4 can discriminate against all cytosine derivatives, including 5mC, 5hmC, and 5caC. For this reason, the MBD4-mediated demethylation pathway requires a deamination process. C5 modifications of uracil are not recognized, while uracil and derivatives including 5-methyl uracil (thymine), and 5-hydroxymethyl uracil (5hmU) paired with guanine are substrates for the MBD4 glycosylase domain in vitro22,23.
Figure 2.
Active site of MBD4 (D534N) and DNA G:T mismatch substrate complex crystal structure. (A) Structure of MBD4 D534N in complex with G:T mismatch (PDB: 4EVV). The surface charge at neutral pH is displayed as blue for positive (50 kBT), red for negative (−50 kBT), and white for neutral, where kB is the Boltzmann’s constant and T is the temperature. (B) Entire 11 bp DNA structure in substrate complex with MBD4 D534N (C) Leu482 kinked DNA ∼65° (D) An arginine finger (Arg442) flips thymine and penetrates into the space left by the flipped thymine, and three hydrogen bonds are formed with the intrahelical orphaned guanine. (E) Thymine-specific interactions in MBD4. Small arrows indicate hydrogen bond donors and acceptors for a flipped thymine base (F) Hydrogen bond donors and accepters for 5mC and thymine bases in G:5mC pair, G:T mismatch.
TDG glycosylase
Human thymine DNA glycosylase (TDG) encodes 410 amino acids, and belongs to the uracil DNA glycosylase (UDG) superfamily. TDG and E. coli Mug glycosylases are highly similar (31% identity, and 43% similarity) and both can excise 5caC paired with guanine23,26. TDG excises uracil, thymine, 5-hydroxymethyl uracil, in addition to 5-formyl cytosine, 5-carboxyl cytosine and etheno-cytosine paired with guanine17,18,26,27.
An interesting substrate discrimination of TDG is that it excises 5fC, and 5caC but not 5hmC; yet it is still unknown how TDG discriminates 5-substituents (Fig. 1). Currently, all available structures of TDG in a complex with DNA reveal that TDG recognizes DNA only from the minor groove of DNA, and not from major groove of DNA (Fig. 3A)26–30. Reported mutation and crystal studies, however, have yet to explain why TDG lacks 5hmC activity, but only excises 5fC and 5caC among 5mC oxidized bases. TDG distorts various substrate/product DNA ∼18° (Fig. 3B), and there is no side chain interaction in a major groove of DNA where the modifications at the C5 position are positioned (Fig. 3C). However, the Pro198-Gly199-Ser200 (P-G-S) containing loop is located in a major groove of DNA and potentially can form interactions with C5 modifications such as in the mechanism proposed for MutM and human UNG31,32. This TDG loop can superimpose well with the corresponding loop of human UNG, but Ala-mutants in the P-G-S loop still discriminate substrate specificities. These results suggest that P-G-S loop is unlikely to play a strong role in discriminating between different 5-substituents26.
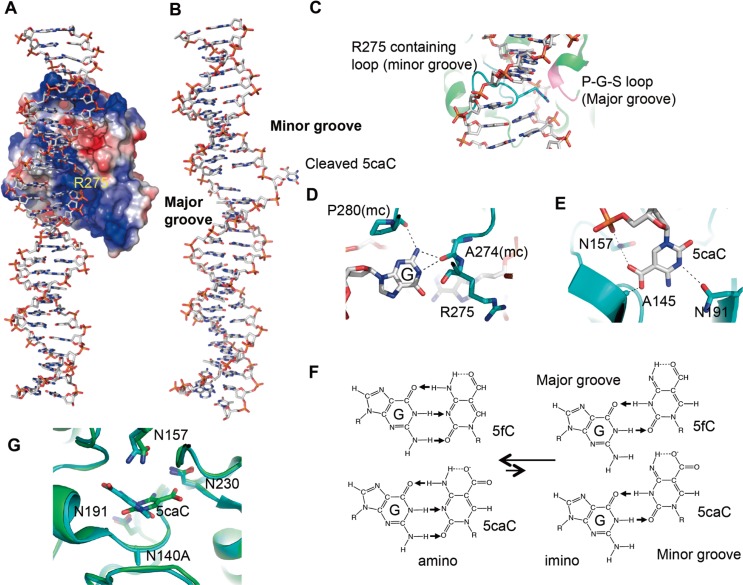
Figure 3.
Structure of TDG in complex with DNA. (A) Structure of TDG wild-type in product complex with G:5caC base pair (PDB:4JGC). The surface charge at neutral pH is displayed as blue for positive (50 kBT), red for negative (−50 kBT), and white for neutral, where kB is the Boltzmann’s constant and T is the temperature. (B) Entire 28 bp DNA structure in product complex with TDG (C) The Arg275-containing intercalation loop (cyan) and the P-G-S loop (magenta) approach the modified DNA strand from opposite directions. (D) The three hydrogen bonds formed with the intrahelical orphaned guanine (E) Proposed for amino-imino tautomerization. 5fC and 5caC exhibit an intramolecular hydrogen bond that could shift the amino/imino equilibrium toward the imino tautomeric form which would then base pair with guanine in a mismatch-like wobble pattern. (F) The interactions between 5caC base and TDG in its active site (G) Superimposition of N140A in complex with a 5caC-containing 22-bp DNA crystallized at pH 7 (colored in cyan; PDB 3UO7) and 5caC-containing 28-bp DNA crystallized at pH 4.6 (colored in green and gray) suggests a base rotation of approximately 120° around the glycoside bond. Four asparagines (Asn140Ala, Asn191, Asn 157, Asn230) are also shown.
So far, in TDG and DNA complex structures, substrate/product DNA are recognized in the minor groove of DNA by Arg275. The side chain of Arg275 penetrates into the DNA helix from the minor groove, occupying the space left by the flipped-out modified nucleotide (Fig. 3A). The intrahelical orphaned guanine is stabilized by hydrogen bonds with the main chain carbonyl oxygen atoms of Ala274 and Pro280 (Fig. 3D). Therefore, these structural studies suggest TDG potentially recognizes 5caC, 5fC as a mismatch like pair from minor groove of DNA. It was hypothesized that G:5fC /G:5caC pairs form a “wobble base pair” by amino/imino tautomeric forms33,34 (Fig. 3E), however, in which the amino tautomers of 5fC and 5caC are much more stable, the crystal structure of a 5fC containing 12-bp DNA duplex (PDB: 1VE8) shows that 5fC:G forms normal base pairs35–37. The crystal structure of the non-substrate 5hmC:G base pair containing DNA complexed with TDG would clarify how 5fC:G/5caC:G bases are recognized by TDG.
Flipped 5caC base and protein interactions are shown for the N140A TDG mutant and 23 bp double strand oligonucleotides containing 5caC:A pair crystal structure at neutral pH (PDB: 3UO7)27. The carboxyl group of the 5caC base interacts with side chains of Asn157 and Ala145 (Fig. 3F). However, N157A, N157D, and A145S mutants still have substantial activities on 5caC26. These mutation results indicate those two Asn157 and Ala145 residues are not essential for 5caC activity. Interestingly, the amino group of side chain of Asn157 forms hydrogen bonds with the carboxyl group of 5caC and the DNA phosphate backbone, and the mutant N157D makes a more 5caC specific glycosylase, by reducing the other possible substrate activity38. Another structure containing the N140A mutant and a 28 bp double strand oligo nucleotide containing 5caC, was from a crystal at a low pH (pH 4.6) (PDB: 4JGC) at which 5caC activity is enhanced. 5caC electron density was not as clear as in the neutral pH crystal, even in a higher resolution data sets (2.5 Å)28. In this crystal structure, the carboxyl group of this base was pointed outward toward Asn230, and Ser271, and not toward Asn157. N230D and S271H mutants did not affect the substrate specificity but a N230D/N157D double mutant completely inhibits the 5caC activity but not U activity28,38.
A N191A single mutation specifically inhibits the 5caC activity while 5fC activity remains. 5fC activity is pH independent, while 5caC activity is pH dependent22,37. These observations indicate that the catalytic mechanism of 5fC and 5caC are different. There are four asparagines (Asn140, Asn157, Asn191, and Asn230) in an active site and each play an individual role in substrate dependent catalysis (Fig. 3E), Asn140, and Asn157 are conserved between human TDG and E.coli Mug (eMug) (PDB: 1MWI39), but the other two are not. Asn191 of TDG position is Lys68 of eMug, and Asn230 of TDG position is Leu107 of eMug. However, eMug does have another Asn140 in its active site, equivalent to Ser271 of TDG. The catalytic mechanisms for 5caC substrates are also different between human TDG and E. coli Mug. Substrate preferences between TDG and eMug are different as well. For instance, the eMug activities on 5hmU and thymine are extremely poor, even compared to the 5caC activities of human TDG and E. coli Mug, and the human TDG activity on 5caC is pH dependent while the E. coli Mug activity on 5caC is pH independent26,40. Therefore, it is still not clear how TDG discriminates substrates and how catalysis proceeds with the various substrates of TDG.
Perspective
DNA methylation and demethylation are dynamic processes and required for embryonic development2. 5mC glycosylases which directly remove a 5mC base are found in plants41, but so far, no 5mC demethylase as well as no 5mC DNA glycosylase, has been identified in mammals. Oxidation of 5mC to 5fC/5caC by TET proteins or deamination of 5mC to T by the deaminases, AID or APOBEC, are the initial step for active DNA demethylation. Afterward, TDG and/or MBD4 glycosylases remove oxidative 5mC and/or deaminated 5mC. Structural and biochemical studies have showed that oxidation or deamination of 5mC is required for removal of the 5mC base, following by the base excision repair pathway which is required to replace the resultant abasic site with cytosine. Therefore, targeting demethylated 5mC locations seem to be controlled by TET proteins, and 5hmC to 5fC oxidization might be a key conversion for DNA demethylation. 5fC is potentially a mutagenic in mammalian cells33, DNA polymerase, and RNA polymerase also discriminate between C/5mC/5hmC and 5fC/5caC. DNA polymerases misincorporate a small amount of adenine using a 5fC template in vitro42,43. RNA polymerase II polymerization rates for GTP incorporation using 5fC/5caC containing DNA are significantly slower, and the discrimination of GTP over ATP is reduced by a factor of 30 for a 5fC template in comparison with a C template in vitro44, 5fC/5caC may introduce transition mutations during DNA replication and RNA transcription. Therefore, 5fC, and 5caC might be recognized as damaged bases, and be distinct from 5hmC among 5mC oxidized products for DNA polymerase, RNA polymerase II and TDG glycosylases. TDG could recognize G:5fC, and G:5caC as a damaged base pair from the minor groove of DNA as same manner as G:X mismatch DNA. It appears that 5hmC might be not a simple DNA demethylation intermediate product, but potentially has its own biological function(s) and is maintained by DNMT3A, DNMT3B and TET proteins after DNA replication.
References
- 1.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W, Li W, Zhou Q, Aluru N, Tang F, He C, Huang X, Liu J. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, He C. Nucleic acid modifications with epigenetic significance. Curr Opin Chem Biol. 2012;16:516–524. doi: 10.1016/j.cbpa.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi C, Jia G, Hou G, Dai Q, Zhang W, Zheng G, Jian X, Yang CG, Cui Q, He C. Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase. Nature. 2010;468:330–333. doi: 10.1038/nature09497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto H, Pais JE, Zhang X, Saleh L, Fu ZQ, Dai N, Correa IR, Jr, Zheng Y, Cheng X. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506:391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 14.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluri-potent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 16.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 20.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otani J, Arita K, Kato T, Kinoshita M, Kimura H, Suetake I, Tajima S, Ariyoshi M, Shirakawa M. Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4. J Biol Chem. 2013;288:6351–6362. doi: 10.1074/jbc.M112.431098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto H, Zhang X, Cheng X. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012;40:8276–8284. doi: 10.1093/nar/gks628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moréra S, Grin I, Vigouroux A, Couvé S, Henriot V, Saparbaev M, Ishchenko AA. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 2012;40:9917–9926. doi: 10.1093/nar/gks714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu P, Qiu C, Sohail A, Zhang X, Bhagwat AS, Cheng X. Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. J Biol Chem. 2003;278:5285–5291. doi: 10.1074/jbc.M210884200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Aacids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto H, Hong S, Bhagwat AS, Zhang X, Cheng X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012;40:10203–10214. doi: 10.1093/nar/gks845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto H, Zhang X, Cheng X. Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH. DNA Repair. 2013;12:535–540. doi: 10.1016/j.dnarep.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc Natl Acad Sci USA. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiti A, Noon MS, MacKerell AD, Jr, Pozharski E, Drohat AC. Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA. Proc Natl Acad Sci USA. 2012;109:8091–8096. doi: 10.1073/pnas.1201010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamiya H, Tsuchiya H, Karino N, Ueno Y, Matsuda A, Harashima H. Mutagenicity of 5-formylcytosine, an oxidation product of 5-methylcytosine, in DNA in mammalian cells. J Biochem. 2002;132:551–555. doi: 10.1093/oxfordjournals.jbchem.a003256. [DOI] [PubMed] [Google Scholar]
- 34.Burdzy A, Noyes KT, Valinluck V, Sowers LC. Synthesis of stable-isotope enriched 5-methylpyrimidines and their use as probes of base reactivity in DNA. Nucleic Acids Res. 2002;30:4068–4074. doi: 10.1093/nar/gkf520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irrera S, Portalone G. First X-ray diffraction and quantum chemical study of proton-acceptor and proton-donor forms of 5-carboxylcytosine, the last-discovered nucleobase. J Mol Struct. 2013;1050:140–150. [Google Scholar]
- 36.Kimura K, Ono A, Watanabe K, Takenaka A. X-Ray analyses of oligonucleotides containing 5-formylcytosine, suggesting a structural reason for codon-anticodon recognition of mitochondrial tRNA-Met; Part 1, d(CGCGAATT(5fC)GCG) 2005.
- 37.Maiti A, Michelson AZ, Armwood CJ, Lee JK, Drohat AC. Divergent mechanisms for enzymatic excision of 5-formylcytosine and 5-carboxylcytosine from DNA. J Am Chem Soc. 2013;135:15813–15822. doi: 10.1021/ja406444x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimoto H, Zhang X, Cheng X. Selective excision of 5-carboxylcytosine by a thymine DNA glycosylase mutant. J Mol Biol. 2013;425:971–976. doi: 10.1016/j.jmb.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett TE, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl LH. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 40.O’Neill RJ, Vorob’eva OV, Shahbakhti H, Zmuda E, Bhagwat AS, Baldwin GS. Mismatch uracil glycosylase from Escherichia coli: a general mismatch or a specific DNA glycosylase? J Biol Chem. 2003;278:20526–20532. doi: 10.1074/jbc.M210860200. [DOI] [PubMed] [Google Scholar]
- 41.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 42.Karino N, Ueno Y, Matsuda A. Synthesis and properties of oligonucleotides containing 5-formyl-2′-deoxycytidine: in vitro DNA polymerase reactions on DNA templates containing 5-formyl-2′-deoxycytidine. Nucleic Acids Res. 2001;29:2456–2463. doi: 10.1093/nar/29.12.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munzel M, Lischke U, Stathis D, Pfaffeneder T, Gnerlich FA, Deiml CA, Koch SC, Karaghiosoff K, Carell T. Improved synthesis and mutagenicity of oligonucleotides containing 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine. Chemistry. 2011;17:13782–13788. doi: 10.1002/chem.201102782. [DOI] [PubMed] [Google Scholar]
- 44.Kellinger MW, Song CX, Chong J, Lu XY, He C, Wang D. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]