Abstract
An ultrasensitive method for the determination of proteins is described that combines an enzyme-linked immunosorbent assay (ELISA) and a thionicotinamide-adenine dinucleotide (thio-NAD) cycling method. A sandwich method using a primary and a secondary antibody for antigens is employed in an ELISA. An androsterone derivative, 3α-hydroxysteroid, is produced by the hydrolysis of 3α-hydroxysteroid 3-phosphate with alkaline phosphatase linked to the secondary antibody. This 3α-hydroxysteroid is oxidized to a 3-ketosteroid by 3α- hydroxysteroid dehydrogenase (3α-HSD) with a cofactor thio-NAD. By the opposite reaction, the 3-ketosteroid is reduced to a 3α-hydroxysteroid by 3α-HSD with a cofactor NADH. During this cycling reaction, thio-NADH accumulates in a quadratic function-like fashion. Accumulated thio-NADH can be measured directly at an absorbance of 400 nm without any interference from other cofactors. These features enable us to detect a target protein with ultrasensitivity (10−19 mol/assay) by measuring the cumulative quantity of thio-NADH. Our ultrasensitive determination of proteins thus allows for the detection of small amounts of proteins only by the application of thio-NAD cycling reagents to the usual ELISA system.
Keywords: androsterone, enzyme cycling, 3α-hydroxysteroid dehydrogenase, insulin, thio-NAD
To diagnose and assess the progression of diseases, the quantitative examination of mRNA amounts in blood for target molecules of diseases is commonly performed1,2. The recent spread of real-time polymerase chain reaction (PCR) methods has enabled us to determine a few copies of mRNAs even in single cells3, as nucleic acids, i.e., RNA and DNA, can be amplified by PCR even when they exist in very small quantities. However, to determine the principal cause of a disease requires the identification of both quantitative and qualitative changes in proteins or peptides in the body. That is, the exact determination of proteins is required for the diagnosis, as opposed to just the quantitative determination of mRNAs. For this purpose, we must develop a novel determination method for trace amounts of proteins, because proteins consisting of 20 kinds of amino acids cannot be amplified.
Generally, the concentrations of the majority of proteins that are used for the diagnosis and judgments regarding the disease’s progression of cancer, neurological disorders and early stage of infection are thought to circulate in the range of 10−12 to 10−16 M4–7. When we use a 100 μL sample isolated from the body (such as blood) for an assay, we must detect concentrations of 10−16 to 10−20 mol/assay. On the other hand, because the usual immunoassays measure proteins at concentrations of 10−12 M8, many attempts have been made to develop methods capable of measuring very low concentrations of proteins using nanoparticle-based methods, flow-based methods, immuno-PCR techniques, digital ELISA and so on. The limits of detection for these methods have been reported to be 10−13 to 10−18 M9–18. Even though the assay volumes are different in each method, and thus the comparison among these data is not straightforward, we need to develop the determination method with higher, or at least comparative, sensitivity for the limit of detection.
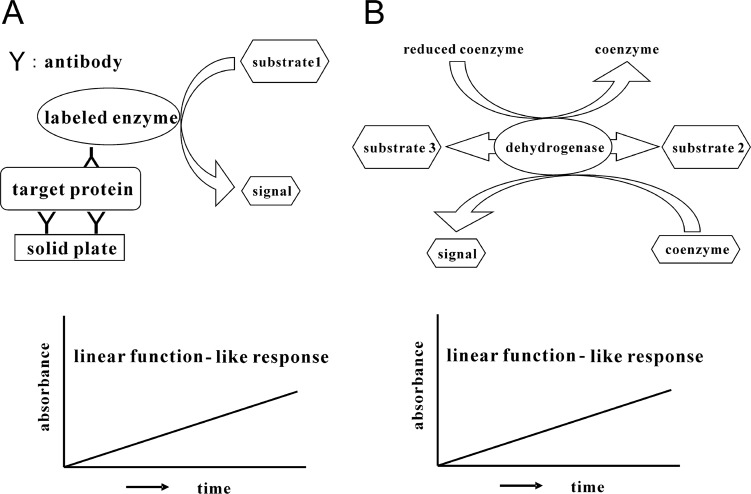
For the exact quantification of trace amounts of proteins, a suitable method may be a ‘sandwich’ enzyme-linked immunosorbent assay (ELISA)19. ELISA is an easy, rapid, specific and highly sensitive detection method and thus has been widely used as a diagnostic tool in medicine and for quality-control checks in various industries. In this assay, an enzyme linked to a secondary antibody converts its substrate to another form. Most commonly, this produces a color change in the substrate, that is, a detectable signal. We usually detect this visible signal that indicates the quantity of antigen, i.e., proteins, with a microplate reader (Fig. 1A). This detectable signal changes linearly with time.
Figure 1.
A. ELISA (sandwich method). B. Enzyme cycling method. Signals in both methods can be obtained in a linear function against measuring time.
There is, however, another assay for the determination of trace amounts of substrates by amplification techniques as a result of the continuous reaction of enzyme function. This is referred to as an enzyme cycling method20,21. In general, amplification is achieved by two enzyme reaction systems in which each enzyme independently and cooperatively acts on the same substrate in a di erent way. In addition, there is another substrate cycling reaction conducted by a single dehydrogenase such as 3α-hydroxysteroid dehydrogenase (3α-HSD, EC. 1.1.1.50; Fig. 1B)22–28. In this cycling reaction, 3α-HSD catalyzes a substrate cycling between 3α-hydroxysteroid and its corresponding 3-ketosteroid in the presence of an excess amount of NADH and thionicotinamide-adenine dinucleotide (thio-NAD), because 3α-HSD utilizes both NADH and thio-NAD as cofactors29. In each turn of the cycle, one molecule of thio-NAD is reduced to thio-NADH, which can be measured directly by an increase in the absorbance at 400 nm (11,900 M−1 cm−1), e.g., 405 nm with a commercially available microplate reader, without any interference from other cofactors such as thio-NAD, NAD and NADH, the absorbance maximums of which are all under 340 nm. These features make it possible to determine the amount of 3α-hydroxysteroids with high sensitivity by measuring the cumulative quantity of thio-NADH. This detectable signal also changes linearly with time.
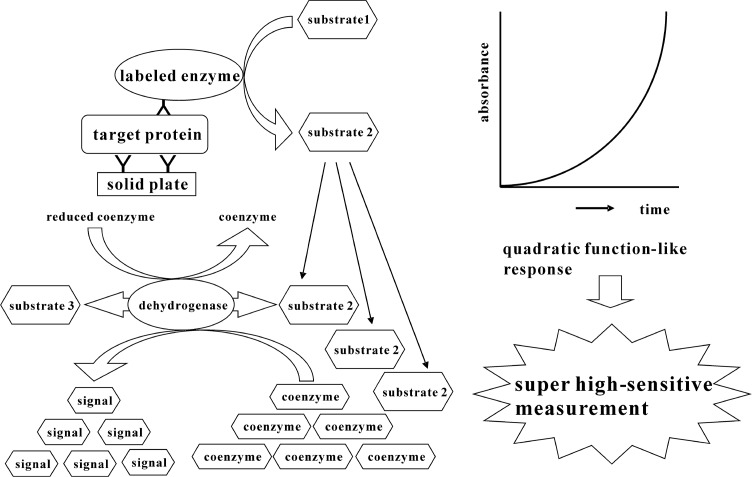
In the present study, we propose the combination of a sandwich ELISA and a thio-NAD cycling method for an ultra-sensitive determination of trace amounts of proteins (Fig. 2). InasandwichELISA, we use 17β-methoxy-5β-androstan-3α-ol 3-phosphate as a synthetic substrate for alkaline phosphatase (ALP, EC. 3.1.3.1) linked to a secondary antibody, because 17β-methoxy-5β-androstan-3α-ol 3-phosphate is easily hydrolyzed by ALP to 17β-methoxy-5β-androstan-3α-ol, which shows highly efficient thio-NAD cycling. The claim for our novel method is that proteins themselves cannot be amplified, but a detectable signal for proteins can be amplified by means of a quadratic function-like response. We here describe how human insulin can be determined with ultra-sensitivity using our new method.
Figure 2.
Ultrasensitive determination of proteins by combination of ELISA and enzyme cycling method. Signals can be obtained as a quadratic function-like response occurring over time.
Materials and Methods
Chemicals and equipment
The primary antibody was monoclonal mouse anti-human insulin 7F8 from HyTest (Turku, Finland). The secondary antibody was monoclonal mouse anti-human insulin D4B8 from HyTest (Turku, Finland). Alkaline phosphatase (ALP) was purchased from Roche (origin from calf intestine, recombinant by Pichia pastoris; Mannheim, Germany). Sulfo-EMCS was purchased from Dojindo (Kumamoto, Japan). Human insulin was purchased from MP Biomedicals (recombinant by yeast; MP Bio Japan, Tokyo, Japan). Blocking One-P was purchased from Nacalai Tesque (Kyoto, Japan). Thio-NAD and NADH were purchased from Roche (Mannheim, Germany). 3α-hydroxysteroid dehydrogenase (3α-HSD) was purchased from Kikkoman Biochemifa (origin from Comamonas testosteroni, recombinant by E. coli; Tokyo, Japan) and purified by BL (Numazu, Japan). 5β-Androsterone was purchased from Steraloids (Newport, RI, USA). 17β-methoxy-5β-androstan-3α-ol 3-phosphate was synthesized according to Iwai et al. (unpublished data). Briefly, 17β-methoxy-5β-androstan-3α-ol 3-phosphate was synthesized from 5β-androsterone (5β-androstan-3α-ol-17-one) via 5 steps including (1) protection of 3α-hydroxyl group by converting into 3-(2′-tetrahydropyranyl) ether, (2) reduction of 17-keto group to 17β-hydroxyl group with NaBH4, (3) methylation of 17β-hydroxyl group with CH3I/NaH, (4) elimination of 3-(2′-tetrahydropyranyl) group with HCl, and (5) phosphorylation of 3α-hydroxyl group with POCl3 to give 17β-methoxy-5β-androstan-3α-ol 3-phosphate. Para-Nitrophenylphosphate (p-NPP) was purchased from KPL (Gaithersburg, MD, USA). Absorption measurements were made with a Corona Electric MTP-500 microplate reader (Hitachinaka, Japan) thermostated at 37°C.
Enzyme-linked secondary antibody
The secondary antibody was digested to F(ab′)2 by pepsin, and reduced to Fab′ by 2-mercaptoethylamine. A maleimide terminal was then introduced into ALP by sulfo-EMCS. Finally, a SH group of Fab′ and a maleimide terminal of ALP were joined.
Ultrasensitive determination
-
Coat a primary antibody.
Dilute the primary antibody with 50 mM Na2CO3 (pH 9.6) to a concentration of 20 μg/mL. Add 50 μL of the antibody into each well of 96-well microplates. Incubate for 1 h at room temperature.
-
Wash microplates.
Wash the microplates 3 times with TBS including 0.05% Tween 20.
-
Block nonspecific binding sites.
Dilute Blocking One-P 5 times with distilled water. Block nonspecific binding sites by filling wells this solution at 150 μL/well. Incubate for 45 min at room temperature.
-
Wash microplates.
Wash the microplates 3 times with TBS including 0.05% Tween 20. Repeat this procedure 3 times.
-
Add an antigen.
Dilute insulin with TBS including 5% Blocking One-P to 0.1–1.0 pg/mL. Add 50 μL of this antigen solution to each well. Shake the microplates for 1 h at room temperature.
-
Wash microplates.
Wash the microplates 3 times with TBS including 0.05% Tween 20. Repeat this procedure 3 times.
-
Add an enzyme-linked secondary antibody.
Dilute an enzyme-linked secondary antibody with TBS including 5% Blocking One-P and 0.05% Tween 20 to 10 pmol/mL. Add 50 μL of this antibody solution to each well. Incubate overnight at 4°C.
-
Wash microplates.
Wash the microplates 3 times with TBS including 0.05% Tween 20. Repeat this procedure 3 times.
-
Add a thio-NAD cycling solution.
Dissolve 1 mM NADH, 1.5 mM thio-NAD, 0.25 mM 17β-methoxy-5β-androstan-3α-ol 3-phosphate and 5 U/mL 3α-HSD into 0.1 mM Tris-HCL (pH 9.0). This is referred to as a thio-NAD cycling solution. Add 50 μL of this thio-NAD cycling solution to each well.
-
Measure absorbance.
Measure the absorbance at 405 nm with a microplate reader every 5 min for 1 h at 37°C.
Limit of detection and coefficient of variation
The experimental data were obtained by subtracting the mean value of blank signals from each of the corresponding measured data. The limit of detection was estimated from the mean of the blank, the standard deviation of the blank and a confidence factor of 3. The coefficient of variation calculated from 3 data points was obtained for 0.1 pg/mL human insulin.
Results and Discussion
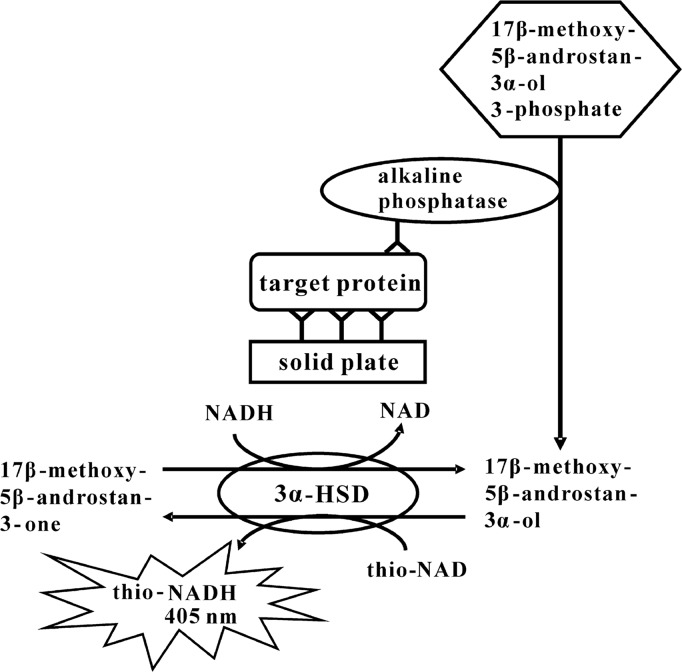
We developed an ultrasensitive ELISA for the determination of human insulin based on 3α-HSD-catalyzed enzyme cycling (Fig. 3). In an ELISA, we employed a sandwich method using the primary and the secondary antibody for human insulin. ALP-linked to the second antibody hydrolyzed 17β-methoxy-5β-androstan-3α-ol 3-phosphate to 17β-methoxy-5β-androstan-3α-ol. 17β-methoxy-5β-androstan-3α-ol was then oxidized to 17β-methoxy-5β-androstan-3-one under a catalytic reaction of 3α-HSD with a cofactor thio-NAD. By the opposite reaction, 17β-methoxy-5β-androstan-3-one was reduced to 17β-methoxy-5β-androstan-3α-ol with a cofactor NADH. During this cycling reaction, thio-NADH accumulated in a quadratic function-like fashion. More specifically, because the thio-NADH signaling intensity depends on the number of thio-NADH molecules accumulated by the enzyme reactions, the thio-NADH signaling intensity = . Here a is the turnover rate of ALP per min; b is the cycling rate of 3α-HSD per min; n=min of measuring time. The amount of insulin was calculated from the increase in absorbance at 405 nm.
Figure 3.
Our ultrasensitive determination of proteins using alkaline phosphatase, androsterone derivative, 3α-HSD and its coenzymes in combination with ELISA and the thio-NAD cycling method.
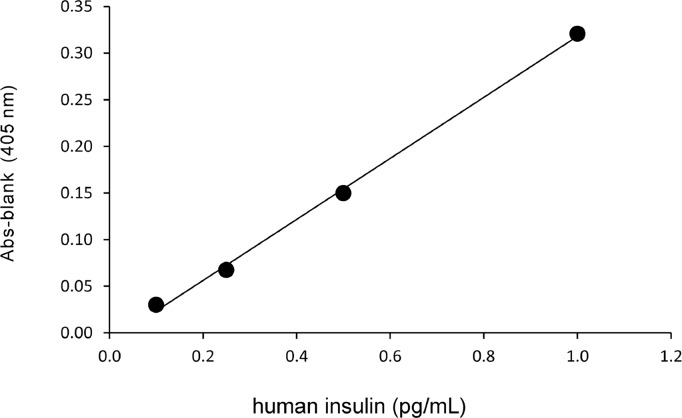
Our ultrasensitive method gave a linear calibration curve (y = 0.33x – 0.009, R2 = 1.00) for human insulin in the range of 0.1–1.0 pg/mL (Fig. 4). This curve was obtained from the absorbance of thio-NADH at a cycling reaction of 60 min. The limit of detection was 0.0047 pg/assay (i.e., 8.0×10−19 mol/assay), and the coefficient of variation was 4% for 0.1 pg/mL. Here we note that the molecular mass of human insulin is 5.8 kDa.
Figure 4.
Linear calibration curve for human insulin in our ultrasensitive determination. The absorbance of thio-NADH was obtained at the cycling reaction of 60 min. y =0.33x–0.009, R2=1.00.
For comparison with a conventional ELISA, p-NPP was used as a chromogenic substrate for ALP. p-NPP has been widely used in an ELISA. Under their influence, the decay to p-nitrophenol is catalyzed and can also be measured with a 405 nm spectrophotometer. For the use of p-NPP, the limit of detection was 0.28 pg/assay (i.e., 4.9×10−17 mol/assay). These results showed that our ultrasensitive determination is at least 2 orders of magnitude more sensitive than conventional ELISA.
Although we only noted the results of the thio-NAD cycling reaction, the combination of 3α-hydroxysteroid 3-phosphate, 3α-HSD, thio-NAD and NADH has been found to detect ALP at 10−20 mol/assay (Iwai et al., unpublished data). That is, our method also has the potential to reach limits of detection of 10−20 mol/assay. Ishikawa and Hashida30,31 have achieved the limit of detection of 10−20 mol/assay for HIV-1 p24 by their own method, the ‘immune complex transfer enzyme immunoassay’. This method, however, requires time and effort for the detection compared with our method. If their method would be combined to ours, we might obtain further sensitivity.
We can apply our ultrasensitive method to the measurement of insulin in human blood. Because our method is based on an ELISA, the procedure includes the terms of ‘washing microplates’. Thus the debris contaminated in blood will be washed out. The study using blood will be reported in the near future. Further, we can use another enzyme, which catalyzes the formation of 3α-hydroxys-teroids or 3-ketosteroids as products, instead of ALP in our ultrasensitive measurement. For example, β-galactosidase can give 3α-hydroxysteroid by using β-galactoside of 3α-hydroxysteroid as its synthetic substrate.
In conclusion, our ultrasensitive determination of proteins by a combination of ELISA and the thio-NAD cycling method is a powerful tool for early disease detection because of its ultrasensitivity. Our method is very convenient because it only requires adding an enzyme cycling solution to a usual ELISA. Our next target is to reduce the procedure time.
Acknowledgments
This study was supported by a grant of the Development of Systems and Technology for Advanced Measurement and Analysis from JST, a grant of the Regional Innovation Strategy Support Program 2009 from MEXT, a grant of the New Regional Consortium Research and Development Project from the METI and the Hokkaido Bureau of Economy, Trade, and Industry, and KAKENHI (Nos. 24657055 and 25291074) from JSPS to E.I.
Footnotes
Conflict of Interest
H.K., M.K., T.Y., A.I., T.M. and E.I. declare that they have no conflict of interest. S.W. is an employee of BL Co. Ltd.; and M.M. and K.N. are the employees of TAUNS Co. Ltd.
Author Contributions
S.W., T.M. and E.I. directed the entire project. H.K., M.K., M.M., K.N. and A.I. performed the experiments. T.Y. synthesized the androsterone derivatives. S.W., K.N., T.Y., T.M. and E.I. co-wrote the manuscript.
References
- 1.Llombart V, Garcia-Berrocoso T, Bustamante A, Fernandez-Cadenas I, Montaner J. Cardioembolic stroke diagnosis using blood biomarkers. Curr Cardiol Rev. 2013;9:340–352. doi: 10.2174/1573403X10666140214122633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganepola GA, Nizin J, Rutledge JR, Chang DH. Use of blood-based biomarkers for early diagnosis and surveillance of colorectal cancer. World J, Gastrointest, Oncol. 2014;15:83–97. doi: 10.4251/wjgo.v6.i4.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagatsuma A, Sadamoto H, Kitahashi T, Lukowiak K, Urano A, Ito E. Determination of the exact copy numbers of particular mRNAs in a single cell by quantitative real-time RT-PCR. J Exp Biol. 2005;208:2389–2398. doi: 10.1242/jeb.01625. [DOI] [PubMed] [Google Scholar]
- 4.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 5.Barletta JM, Edelman DC, Constantine NT. Lowering the detection limits of HIV-1 viral load using real-time immuno-PCR for HIV-1 p24 antigen. Am J Clin Pathol. 2004;122:20–27. doi: 10.1309/529T-2WDN-EB6X-8VUN. [DOI] [PubMed] [Google Scholar]
- 6.Galasko D. Biomarkers for Alzheimer’s disease—clinical needs and application. J Alzheimers Dis. 2005;8:339–346. doi: 10.3233/jad-2005-8403. [DOI] [PubMed] [Google Scholar]
- 7.de Jong D, Kremer BPH, Olde Rikkert MGM, Verbeek MM. Current state and future directions of neurochemical biomarkers for Alzheimer’s disease. Clin Chem Lab Med. 2007;45:1421–1434. doi: 10.1515/CCLM.2007.320. [DOI] [PubMed] [Google Scholar]
- 8.Giljohann DA, Mirkin CA. Drivers of biodiagnostic development. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Y, Wei QQ, Park HK, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 10.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 11.Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Label-free, single-molecule detection with optical microcavities. Science. 2007;317:783–787. doi: 10.1126/science.1145002. [DOI] [PubMed] [Google Scholar]
- 12.Todd J, Freese B, Lu A, Held D, Morey J, Livingston R, Goix P. Ultrasensitive flow-based immunoassays using single-molecule counting. Clin Chem. 2007;53:1990–1995. doi: 10.1373/clinchem.2007.091181. [DOI] [PubMed] [Google Scholar]
- 13.Adler M, Wacker R, Niemeyer CM. Sensitivity by combination: immuno-PCR and related technologies. Analyst (Lond) 2008;133:702–718. doi: 10.1039/b718587c. [DOI] [PubMed] [Google Scholar]
- 14.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaster RS, Hall DA, Nielsen CH, Osterfeld SJ, Yu H, Mach KE, Wilson RJ, Murmann B, Liao JC, Gambhir SS, Wang SX. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nat Med. 2009;15:1327–1332. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee JS, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, Mirkin CA. Nanoparticle-based bio-barcode assay redefines “undetectable” PSA and biochemical recurrence after radical prostatectomy. Proc Natl Acad Sci USA. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SH, Iwai S, Araki S, Sakakihara S, Iino R, Noji H. Large-scale femtoliter droplet array for digital counting of single biomolecules. Lab Chip. 2012;12:4986–4991. doi: 10.1039/c2lc40632b. [DOI] [PubMed] [Google Scholar]
- 19.Crowther JR. The ELISA Guidebook. 2nd ed. Springer; New York: 2009. [Google Scholar]
- 20.Lowry OH, Passonneau JV, Schulz DW, Rock MK. The measurement of pyridine nucleotides by enzymatic cycling. J Biol Chem. 1961;236:2746–2755. [PubMed] [Google Scholar]
- 21.Kato T, Berger SJ, Carter JA, Lowry OH. An enzymatic cycling method for nicotinamide-adenine dinucleotide with malic and alcohol dehydrogenases. Anal Biochem. 1973;53:86–97. doi: 10.1016/0003-2697(73)90409-0. [DOI] [PubMed] [Google Scholar]
- 22.Mashige F, Imai K, Osuga T. A simple and sensitive assay of total serum bile acids. Clin Chim Acta. 1976;70:79–86. doi: 10.1016/0009-8981(76)90007-3. [DOI] [PubMed] [Google Scholar]
- 23.Mashige F, Tanaka N, Maki A, Kamei S, Yamanaka M. Direct spectrophotometry of total bile acids in serum. Clin Chem. 1981;27:1352–1356. [PubMed] [Google Scholar]
- 24.Yoshimura T, Kurosawa T, Ikegami S, Tohma M. Sub-strate specificity of 3α-hydroxysteroid dehydrogenase for the oxidation of fetal bile acids. Bunseki Kagaku. 1995;44:865–869. [Google Scholar]
- 25.Ueda S, Oda M, Imamura S, Ohnishi M. Kinetic study of the enzymatic cycling reaction conducted with 3α- hydroxysteroid dehydrogenase in the presence of excessive thio-NAD+ and NADH. Anal Biochem. 2004;332:84–89. doi: 10.1016/j.ab.2004.04.035. [DOI] [PubMed] [Google Scholar]
- 26.Zhang GH, Cong AR, Xu GB, Li CB, Yang RF, Xia TA. An enzymatic cycling method for the determination of serum total bile acids with recombinant 3α-hydroxysteroid dehydrogenase. Biochem Biophys Res Commun. 2005;326:87–92. doi: 10.1016/j.bbrc.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka T, Ueda S, Matsumoto H, Kawakami M. An ultrasensitive enzymatic method for measuring mevalonic acid in serum. J Lipid Res. 2012;53:1987–1892. doi: 10.1194/jlr.D028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwai A, Yoshimura T, Wada K, Watabe S, Sakamoto Y, Ito E, Miura T. Spectrophotometric method for the assay of steroid 5α-reductase activity of rat liver and prostate microsomes. Anal Sci. 2013;29:455–459. doi: 10.2116/analsci.29.455. [DOI] [PubMed] [Google Scholar]
- 29.Skålhegg BA. 3α-hydroxysteroid dehydrogenase from Pseudomonas testosteroni: kinetic properties with NAD and its thionicotinamide analogue. Eur J Biochem. 1975;50:603–609. doi: 10.1111/j.1432-1033.1975.tb09901.x. [DOI] [PubMed] [Google Scholar]
- 30.Hashida S, Ishikawa S, Hashinaka K, Nishikata I, Saito A, Takamizawa A, Shinagawa H, Ishikawa E. Optimal conditions of immune complex transfer enzyme immunoassay for p24 antigen of HIV-1. J Clin Lab Anal. 1998;12:115–120. doi: 10.1002/(SICI)1098-2825(1998)12:2<115::AID-JCLA7>3.0.CO;2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikawa E, Ishikawa S, Hashida S, Hashinaka K. Potential of the immune complex transfer enzyme immunoassay for antigens and antibodies to improve the sensitivity and its limitations. J Clin Lab Anal. 1998;12:154–161. doi: 10.1002/(SICI)1098-2825(1998)12:3<154::AID-JCLA5>3.0.CO;2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]