Abstract
Saccharomyces cerevisiae and related species, the main workhorses of wine fermentation, have been exposed to stressful conditions for millennia, potentially resulting in adaptive differentiation. As a result, wine yeasts have recently attracted considerable interest for studying the evolutionary effects of domestication. The widespread use of whole-genome sequencing during the last decade has provided new insights into the biodiversity, population structure, phylogeography and evolutionary history of wine yeasts. Comparisons between S. cerevisiae isolates from various origins have indicated that a variety of mechanisms, including heterozygosity, nucleotide and structural variations, introgressions, horizontal gene transfer and hybridization, contribute to the genetic and phenotypic diversity of S. cerevisiae. This review will summarize the current knowledge on the diversity and evolutionary history of wine yeasts, focusing on the domestication fingerprints identified in these strains.
Keywords: evolutionary history, comparative genomics, wine fermentation, domestication, horizontal transfer, hybrids
This review summarizes current knowledge and recent advances on the diversity and evolutionary history of Saccharomyces cerevisiae wine yeasts, focusing on the domestication fingerprints identified in these strains.
Graphical Abstract Figure.

This review summarizes current knowledge and recent advances on the diversity and evolutionary history of Saccharomyces cerevisiae wine yeasts, focusing on the domestication fingerprints identified in these strains.
INTRODUCTION
Although Saccharomyces cerevisiae is a well-studied model that has aided our understanding of eukaryotic cellular processes and was the first eukaryotic genome to be completely sequenced (Goffeau et al. 1996), this yeast has only recently been established as a model for studies in ecological and evolutionary genetics (Landry et al. 2006). Some authors have defended that its closest relative S. paradoxus is a better model for ecology and evolutionary biology because it is not associated with humans (Replansky et al. 2008). However, S. cerevisiae is not domesticated as a whole and population genetics analysis of both domesticated and a growing number of wild isolates is continuously offering new insights into the ecological distribution, population structure and biogeography of this species. In this context, S. cerevisiae wine yeasts have attracted considerable interest in recent years.
Wine has a long and rich history, dating back thousands of years, closely associated with the history of agriculture. The earliest archaeological evidence for the production of a fermented beverage made of a mixture of rice, honey and fruit is in China dated to 7000 BC (McGovern et al. 2004). The first chemical evidence of the presence of wine is dated to 5400–5000 BC, from the Neolithic period site called Hajji Firuz in Iran, where the presence of combined calcium salt from tartaric acid and terebinth resin was identified in a pottery jar (McGovern, Donald and Glusker 1996). Evidence that S. cerevisiae was probably responsible for wine fermentation in Egypt by at least 3150 BC was provided (Cavalieri et al. 2003). Wine fermentation technologies expanded from Mesopotamia towards Europe and subsequently spread to the New World (McGovern 2003). Alcoholic fermentation is not only an efficient method for the preservation of the quality and safety of beverages and foods, but wine is also a widespread drug and medicine of antiquity, reflecting the analgesic, disinfectant and preservative properties of this alcoholic beverage. Over time, wine has influenced geography, economics, archeology, history, mythologies and religions, arts and traditions, law and medicine. Today, this beverage has a unique place in most societies, with tremendous economic and cultural value.
It was not until 1860 that Louis Pasteur discovered the origin of fermentation and the implication of yeast (Pasteur 1860). In the early 1880s, Emile Christian Hansen, from the Carlsberg laboratory in Denmark, developed the first pure yeast culture and wort inoculation was performed some years later. In 1890, Müller-Thurgau performed the first inoculation of a grape must with a pure yeast culture. Surprisingly, this practice was effectively used in oenology only in the 1970s, almost one century later. After the 1970s, these practices were generalized and currently most wine fermentations worldwide use selected starter yeasts primarily belonging to S. cerevisiae. These practices have improved the control and reliability of the fermentation process, limiting microbiological alterations and have largely contributed to increased wine quality in recent decades. Consistently, most pioneering research in the genetics, genomics, physiology and evolutionary biology of wine yeasts has been developed in S. cerevisiae and to a lesser extent on other Saccharomyces species and hybrids prominent in the wine industry. This review will focus on the most recent advances on the history, diversity and evolution of Saccharomyces wine yeasts.
SACCHAROMYCES AND THE MAKE-ACCUMULATE-CONSUME STRATEGY
The fermentation of grape musts can spontaneously occur through the activity of various microorganisms naturally present on grape berries. More than 40 yeast species have been identified from grape must (reviewed in Jolly, Varela and Pretorius 2014), with the most frequent yeast being from the genera Hanseniaspora (Kloeckera), Candida, Pichia, Rhodotorula, Debaryomyces, Metschnikowia, Kluyveromyces, Schizosaccharomyces, Torulaspora, Zygosaccharomyces and Dekkera. A sequential succession of these yeast species is observed during the early phase of spontaneous fermentation, followed by the disappearance of the majority of them, even if certain non-Saccharomyces yeasts persist longer (Fleet, Lafon-Lafourcade and Ribereau-Gayon 1984). This phenomenon might reflect several factors, such as their low fermentative capacity, low tolerance to oxygen limitation and high concentrations of SO2 and ethanol. Ethanol-tolerant species, such as Zygosaccharomyces bailii (Martorell et al. 2007), have been identified throughout fermentation. Non-Saccharomyces species might contribute positively or negatively to the organoleptic characteristics of wines (Fleet 1993; Jolly, Varela and Pretorius 2014). Nevertheless, even during spontaneous fermentation, S. cerevisiae dominate fermentation and is the primary species responsible for the conversion of sugars to ethanol and CO2, reflecting the combination of several ‘winning’ traits, including rapid sugar degradation, ethanol production, accumulation and tolerance, and anaerobic propagation.
One of the most remarkable characteristics of S. cerevisiae and closely related species is their ability to produce and accumulate ethanol, referred to as the Crabtree effect, even under aerobic conditions. The long-term Crabtree effect has been explained as a limited respiratory capacity reflecting the repression of respiratory genes (Postma et al. 1989; Alexander and Jeffries 1990). However, the immediate occurrence of alcoholic fermentation after the addition of sugar to sugar-limited and respiratory cultures, called the short-term Crabtree effect, has been attributed to an overflow in sugar metabolism (Pronk, Steensma and Van Dijken 1996; Vemuri et al. 2007). It has recently been suggested that overflow metabolism is the fundamental mechanism behind both long- and short-term Crabtree effect, which originated approximately 125–150 million years ago (Mya) in the Saccharomyces lineage (Hagman and Piškur 2015). Overflow metabolism was first acquired, providing a general strategy to increase energy production rates and enabling rapid glucose consumption. The glucose-mediated repression of respiration would have been acquired as a second step to further increase overflow and ethanol production, thereby inhibiting the growth of other microbes. This characteristic is primarily confined among S. cerevisiae and closely related species that diverged after whole-genome duplication, less than 100 Mya (Hagman and Piškur 2015).
Saccharomyces and Dekkera are characterized by a ‘make-accumulate-consume’ strategy, as these yeasts also efficiently catabolize ethanol. This strategy could confer an advantage to these species in nature, as these yeasts rapidly consume a high quantity of sugars, transforming these carbohydrates into ethanol, which inhibits the growth of other species, and subsequently consuming ethanol after establishing competitive dominance within the ecological niche (Thomson et al. 2005; Piškur et al. 2006; Rozpędowska et al. 2011; Dashko et al. 2014).
The make-accumulate-consume strategy emerged after the split between the S. cerevisiae and Kluyveromyces lactis lineages approximately 100 Mya, suggesting that this process reflected the appearance of modern plants with fruits, which occurred >125 Mya, far earlier than the human domestication of yeast (Thomson et al. 2005; reviewed in Piškur et al. 2006). Comparative genomics approaches revealed that at least two mechanisms might be involved in the acquisition of this capacity. Thomson et al. (2005) reconstructed an ancestral Saccharomyces alcohol dehydrogenase gene (ADH) and showed that the pre-duplication enzyme was optimized to produce (not consume) ethanol. The make-accumulate-consume strategy emerged with the duplication of ADH1 and ADH2, which occurred after whole-genome duplication approximately 100 Mya (Wolfe and Shields 1997; Kellis, Birren and Lander 2004). The duplication of other genes controlling the flux from hexose to ethanol might have also contributed to the emergence of this strategy (Thomson et al. 2005; Conant and Wolfe 2007). Another mechanism to achieve ethanol accumulation involves the global rewiring of the transcriptional network after whole-genome duplication in the S. cerevisiae lineage, resulting in the massive loss of regulatory elements from genes involved in respiration (Ihmels et al. 2005). Interestingly, the Dekkera bruxellensis lineage also lost these specific elements, contributing to the observed Crabtree effect (Rozpędowska et al. 2011).
WINE FERMENTATION: A CHALLENGING ENVIRONMENT
Wine fermentation is a fluctuating environment that exposes yeast to a variety of stresses, including high osmolarity, reflecting increased sugar concentrations, high sulfite levels, anaerobiosis, acid stress, nutrient (nitrogen, lipids and vitamins) depletion and ethanol toxicity. A typical wine fermentation (Fig. 1) comprises a lag phase, which lasts for several hours, a short growth phase of 24–36 h, followed by a stationary phase, during which most of the sugar (between 50 and 80%) is fermented. During this phase, yeast activity continually decreases, although the viability levels remain high, generally over 90%, until the sugar is exhausted. The most desirable traits of wine yeasts include the rapid and complete degradation of sugars into ethanol and CO2 to provide metabolites and aroma compounds that positively impact the sensory balance of wine, without producing undesirable compounds (Pretorius 2000; Dequin 2001). Numerous fermentative by-products (glycerol, carboxylic acids, aldehydes, higher alcohols, esters, carbonyl compounds, sulfur compounds, etc.) are derived from the degradation of sugars, amino acids and fatty acids, and yeasts can also transform grape precursors to release varietal aromas (monoterpenes and thiols) (Swiegers et al. 2005).
Figure 1.
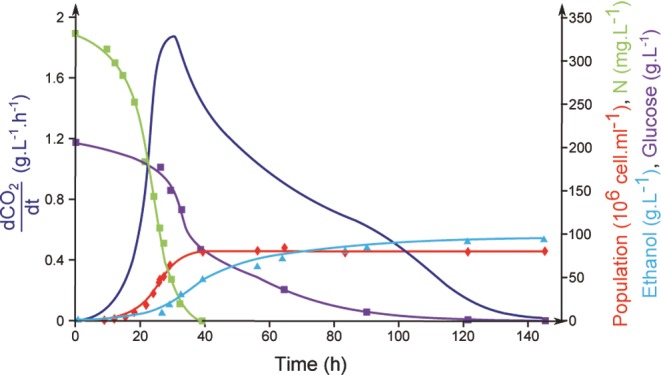
Main phases of wine fermentation. Evolution of the main fermentation parameters during wine fermentation on a synthetic medium containing 200 g L−1 glucose/fructose and 330 mg L−1 assimilable nitrogen, with the commercial wine strain EC1118 at 24°C. Dark blue: fermentation rate; light blue: ethanol; red: cell number; green: nitrogen; and purple: sugars.
The outcome of fermentation depends on many factors, particularly the amount and quality of nutrients available in grape musts. The primary constituents of grapes must include sugars (glucose and fructose in equimolar amounts, present in high concentrations, 180–300 g L−1), organic acids (tartaric and malic), mineral cations (especially potassium), nitrogen compounds and lipids (phytosterols). As yeasts preferably metabolize glucose rather than fructose, fructose is the major sugar present during the late stages of wine fermentation. Wine yeasts must ferment this non-preferred sugar after long periods of starvation and in the presence of large amounts of ethanol. These stressful conditions might alter yeast activity, frequently resulting in sluggish or stuck fermentations (Bisson 1999; Bauer and Pretorius 2000). The ability of wine yeasts to ferment fructose is therefore critically important for the maintenance of a high fermentation rate at the end of alcoholic fermentation.
Nitrogen is an important nutrient present in limited amounts in grape musts, and the availability of this nutrient is directly associated with biomass production, which governs the fermentation rate and production of volatile compounds. Consequently, nitrogen deficiency is the most prevalent cause of stuck and sluggish fermentations (Bisson 1999; Bell and Henschke 2005; Sablayrolles 2008). Yeast assimilable nitrogen (YAN) primarily comprises ammonium ions and amino acids (Henschke and Jiranek 1993). However, other nitrogen sources, e.g. oligopeptides, polypeptides, proteins, amides, biogenic amines and nucleic acids, might constitute substantial nitrogen resources (Ough, Huang and Stevens 1991; Henschke and Jiranek 1993). It is generally considered that 140 mg L−1 of YAN is necessary to complete the fermentation of 200 g L−1 sugar, whereas approximately 300 mg L−1 of YAN is required to optimize fermentation kinetics (Sablayrolles 2008).
Lipids are also key nutrients in alcoholic fermentation. The synthesis of sterols and unsaturated fatty acids requires oxygen. Limited amounts of unsaturated fatty acids or sterols negatively affect viability at the end of fermentation (Alexandre, Rousseaux and Charpentier 1994; Luparia et al. 2004). The strong clarification of grape musts leads to lipid limitation typically associated with a loss of yeast cell viability. Recent studies have shown that the nitrogen status of grape musts is a strong determinant of the outcome of alcoholic fermentations under conditions of lipid limitation (Tesnière et al. 2013). Other important nutrients are vitamins such as thiamin, which is rapidly consumed by yeasts. Thus, indigenous yeasts might cause thiamin depletion in grape musts, with negative consequences for the inoculation efficiency and fermentation kinetics (Bataillon et al. 1996). In addition, various inhibitors, including mid-length chain fatty acids, killer toxins or SO2 which is frequently used in grape juice as antimicrobial or antioxidant, might affect yeasts during wine fermentation. However, ethanol is widely recognized as one of the causes of stuck or sluggish alcoholic fermentation (Bisson 1999). Ethanol exerts a biological effect on growth and fermentation efficiency, reflecting an increase in membrane permeability and changes in membrane fluidity (Alexandre, Rousseaux, Charpentier 1994). Wine yeasts are also exposed to a wide range of temperatures, as red wine fermentation is generally conducted at high temperatures (l8–25°C) and white wine fermentation typically occurs at cooler temperatures (l0–15°C).
Thus, for millennia, winemaking conditions have exposed yeasts to a combination of stresses (osmotic, acidic, nutrient starvation, ethanol toxicity) acting individually or synergistically, potentially leading to adaptive differentiation.
DIVERSITY AND EVOLUTIONARY HISTORY OF S. CEREVISIAE WINE STRAINS
Genetics and life style
Saccharomyces cerevisiae has a haplo-diploid life cycle and a life history predominantly involving clonal reproduction. The frequency of outcrossing remains a matter of debate. The outcrossing rate was initially considered a rare event, occurring once every 50 000 divisions (Ruderfer et al. 2006). However, a recent study, based on different methods, estimated a considerably higher rate of approximately 1 per 100 mitotic divisions (Kelly et al. 2012). Outcross matings have been estimated at 2% and 36–52% for oaks and domesticated isolates (Magwene et al. 2011), and 8–20% for vines and wines strains (Gayevskiy and Goddard 2012). Wine yeasts also have a low level of linkage disequilibrium (falls to one half of is maximal value in approximately 2.5 kb) compared with other isolates, likely reflecting a higher frequency of outcrossing events (Schacherer et al. 2009). These data suggest that human-associated environments might create greater opportunities to bring diverse strains into proximity (Goddard et al. 2010) or for spore dispersal through insect vectors (Reuter, Bell and Greig 2007). Indeed, Reuter, Bell and Greig (2007) suggested that outcrossing would be more effective after the partial digestion of asci by Drosophila.
Most S. cerevisiae strains isolated from the environment, including vineyards, are diploid cells (Cubillos et al. 2009). Wine isolates are primarily homothallic (Mortimer 2000), producing haploid spores that switch mating types and undergo self-diploidization. The pioneering studies of Robert Mortimer showed that homothallic isolates are generally heterozygous at one or more loci, and the frequency of heterozygosity was negatively correlated with the viability of the spores produced from the vineyard isolates. This heterozygosity in homothallic isolates has been attributed to both mutations that occur during the mitotic growth of homozygous diploid isolates (Mortimer et al. 1994; Johnston, Baccari and Mortimer 2000) and the outcrossing of homothallic isolates (McCusker 2006). This finding led Mortimer to propose the ‘Genome Renewal Hypothesis’ (Mortimer et al. 1994; Mortimer 2000), suggesting that recessive, deleterious heterozygous mutations accumulate during mitotic growth and are subsequently eliminated through rare sexual cycles involving meiosis, followed by mating-type switching and autodiploidization. Thus, deleterious alleles would be lost, and beneficial alleles would be fixed, thereby facilitating adaptation in yeast. Recent data obtained through the whole-genome sequencing of diploid isolates revealed a more extensive level of heterozygosity than initially considered, particularly in domesticated isolates (Magwene et al. 2011). The number of heterozygous single nucleotide polymorphisms observed in the genome of commercial wine yeasts ranged from 1000 to more than 18 000 (Novo et al. 2009; Borneman et al. 2011). The high levels of heterozygosity reflect population admixture due to human domestication, coupled with selfing during rare sexual cycles, and these effects may facilitate rapid adaptation to novel environments by increasing the genetic and phenotypic diversity in the population deriving from a single isolate (Magwene 2014). Thus, sexual reproduction and outcrossing are rare but important features in the Saccharomyces life cycle.
Wine yeast strains have a unique origin
In the last decade, numerous studies, based on multigene sequencing (Fay and Benavides 2005; Stefanini et al. 2012; Wang et al. 2012), microsatellite analysis (Legras et al. 2007), tiling array hybridization (Schacherer et al. 2009), low coverage whole-genome sequencing (Liti et al. 2009) or restriction-site-associated sequencing (Rad-seq) (Cromie et al. 2013) have provided deep insight into the population structure and evolutionary history of S. cerevisiae. Domesticated strains of S. cerevisiae, particularly those used for the production of sake and wine, were derived from the natural population through independent domestication events (Fay and Benavides 2005; Legras et al. 2007). Legras et al. (2007) showed that 95% of strains associated with wine belong to the same cluster, suggesting a unique origin of wine yeasts, followed by the expansion of populations through human activities.
From a set of S. cerevisiae isolates with worldwide origin, five distinct lineages were revealed based on their technological and geographic origin (West African, Malaysian, North American, Sake and European/wine), and many strains with mosaic genomes resulting from crosses between these well-defined lineages were identified (Liti et al. 2009). Using another set of strains, Schacherer et al. 2009 identified three distinct lineages (wine, sake and laboratory), which reflect different ecological origins. A recent genomic survey of a higher number of strains suggested a model of geographic differentiation, followed by human-associated admixture, primarily occurring between European and Asian populations and more recently between European and North American populations (Cromie et al. 2013). In addition to global-scale pictures, several studies have investigated the structure and gene flow at ecological levels. Analyses of vineyard isolates have provided evidence for region-specific subpopulations (Gayevskiy and Goddard 2012; Schuller et al. 2012), consistent with previous observations (Legras et al. 2007). Evidence of gene flow across small distances between populations inhabiting vineyards and distinct oak tree populations has also been reported, suggesting some degree of connectivity between populations (Hyma and Fay 2013; Knight and Goddard 2015).
These studies showed that S. cerevisiae as a whole is not domesticated and that the population structure of this species, at least partially, reflects different ecological niches. Specifically, wine strains form a distinct phylogenetic group, with low diversity (Fay and Benavides 2005; Legras et al. 2007; Liti et al. 2009; Schacherer et al. 2009; Cromie et al. 2013). The diversity between wine strains has been estimated as 1 to 1.4 substitutions per kb and 5 to 6 substitutions per kb between wine and other S. cerevisiae strains from other origins (Fay and Benavides 2005; Liti et al. 2009; Schacherer et al. 2009).
A microsatellite-based study suggested that wine yeast strains could originate from Mesopotamia (Legras et al. 2007; Sicard and Legras 2011). Two migration routes could have led the yeasts into Europe: the first route through the Mediterranean Sea to Italy, France and Spain, and in France from the Mediterranean coast to Burgundy through the Rhone valley, and the second route through the Danube valley. Yeast strains could have also been transferred via comigration with grape varieties (Sicard and Legras 2011). The identification of three Chinese wild isolates belonging to the Wine/European lineage by Wang et al. (2012) led these authors to raise the possibility that Wine/European strains have an Asian origin, in line with previous archaeological evidence for fermented beverage in China dated to 9000 years ago (McGovern et al. 2004). However, these isolates were sampled from orchard soil and grape and might not be truly natural. The opposite hypothesis stipulating that the wine yeast could have migrated to Asia is also plausible.
Phenotypic divergence between wine and non-wine strains
There is strong evidence of marked phenotypic divergence between wine and non-wine strains. Wine yeasts strains demonstrate better resistance to copper (Fay et al. 2004; Warringer et al. 2011) and sulfites (Park and Bakalinsky 2000; Pérez-Ortín et al. 2002), two chemical compounds used in vineyards and during winemaking. Several wine yeasts strains have also demonstrated an ability to utilize xylose as a carbon source (Wenger, Schwartz and Sherlock 2010) or different types of ditripeptides as nitrogen sources (Homann et al. 2005). Liti et al. (2009) reported a correlation between strain genetic diversity and growth in different environments or in the presence of drugs. The Wine/European lineages and mosaics showed rapid growth compared with other lineages, and this feature could be advantageous for fermentation processes in which many of these strains are used. There are few large-scale studies comparing the performance and metabolite production of strains under relevant conditions for fermentation. Divergence in life-trait strategies has been reported between industrial strains, referred to as extreme ‘grasshoppers’, which reproduce slowly, display a small carrying capacity and have a large cell size compared with natural and laboratory isolates, referred to as ‘ants’, which reproduce rapidly, display a large carrying capacity and have a small cell size (Spor et al. 2009).
The comparison of 72 strains of various origins in wine fermentation conditions revealed substantial variations in fermentative properties and growth and metabolite traits. Wine strains and other strains isolated from sugar-rich environments (fruits) had a better fermentative capacity under oenological conditions than natural strains isolated from sugar-poor environments (Camarasa et al. 2011) (Fig. 2). Another study, based on a restricted number of strains and on sensorial analysis, suggested that wine yeasts produce higher fruity aromas in wine compared with strains from other origins (Hyma et al. 2011). The analysis of a higher number of strains representing each group and under conditions simulating different habitats is needed to obtain a better picture of the adaptation of these strains to specific conditions.
Figure 2.
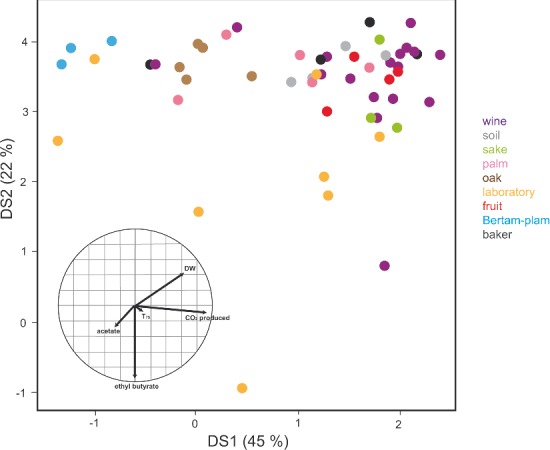
Ability of S. cerevisiae from various origins to ferment large amounts of sugar under winemaking conditions. Linear discriminant analysis (LDA) of the population based on dry weight, 75% sugar fermentation, CO2 production, acetate and ethyl butyrate production of 53 strains during fermentation in a synthetic must (adapted from Camarasa et al. 2011).
Saccharomyces cerevisiae traits might be a consequence of genetic drift rather than selection (Warringer et al. 2011). For example, the West-African population is phenotypically unique, with an extreme abundance of low-performance alleles. However, domestication traits likely reflect both genetic drift and human selection of specific traits (Warringer et al. 2011). In the following section, we will describe compelling evidence and cases of adaptive evolution in wine yeasts.
EVIDENCE FOR HUMAN-ENFORCED ADAPTIVE EVOLUTION
Yeasts use several mechanisms to respond to environmental challenges and evolve corresponding adaptive functions. Adaptation can be achieved through small-scale nucleotide changes (base insertions, deletions or substitutions), which alter protein structure, protein interactions or gene expression, large-scale genome rearrangements (chromosome duplications, translocations and aneuploidy), which alter gene expression through the modification of the genomic context, or copy number variations (CNV), which might alter the gene dosage. In addition, introgression and horizontal transfer could exert powerful evolutionary forces by generating novelties that cannot be introduced using other nucleotide changes or structural rearrangements.
These mechanisms have been shown to contribute to the adaptation of wine yeast genomes (Pretorius 2000; Barrio et al. 2006; Blondin et al. 2009; Dequin and Casaregola 2011).
Hallmarks of domestication in Flor strains
An interesting case of traits acquired after human domestication has been reported in flor yeast. Flor strains form a biofilm on the surface of wine after fermentation and develop oxidative metabolism in the presence of a high concentration of ethanol and a low level of fermentable sugar, i.e. fructose (Alexandre 2013). Because flor yeasts continuously grow on the surface of wine during the sherry wine making, the life style of these microorganisms is completely different from that of fermentative S. cerevisiae wine yeasts, which makes these strains an interesting model to study evolution. The acquisition of two mutations in the promoter and coding regions of the FLO11 gene encoding a GPI-anchored cell surface glycoprotein (flocculin) required for pseudohyphal formation, invasive growth, flocculation and biofilm formation (Guo et al. 2000; Fidalgo et al. 2006) results in increased FLO11 expression and enhanced cell adhesion. A study in a fructophilic wine yeast strain (Guillaume et al. 2007) identified a natural allelic variant of HXT3 encoding a major glucose transporter during wine fermentation, which enhances fructose fermentation. This allele is frequently found in flor strains (Coi A, Dequin S, Legras JL, unpublished data), which might be related to a better capacity of these strains to use fructose. Interestingly, a population study of flor yeasts using microsatellite analyses recently showed that these strains form a unique group, closely related to wine yeasts (Legras, Erny and Charpentier 2014). Comparative genomics between these two close but distinct groups with contrasting life styles offers promising perspectives to identify traces of selection in this group.
Adaptation of wine yeasts to chemicals used in vineyards
The existence of gross chromosomal rearrangements (GCR), i.e. translocations, deletions and amplifications of chromosomal regions, was proposed in the 1990s based on the high level of chromosome polymorphism observed in wine yeasts (Vezinhet, Blondin and Hallet 1990; Yamamoto et al. 1991; Bidenne et al. 1992; Codón, Benítez and Korhola 1998). These GCR events are mediated through ectopic recombination between repeated Ty sequences or other repeated sequences (Rachidi, Barre and Blondin 1999). Several chromosomal translocations have been identified in wine yeast genomes, particularly in telomeric regions, consistent with the idea that peripheral regions are highly plastic (Borneman et al. 2008, 2011; Novo et al. 2009). In most cases, there is no evidence that these rearrangements contribute to yeast fitness.
A well-documented example of chromosomal rearrangement with an adaptive advantage is the reciprocal translocation between chromosome VIII and XVI, which is widespread among wine yeasts. This translocation generated a dominant allele of the sulfite pump, SSU1-R1, which is expressed at much higher levels than SSU1 (Goto-yamamoto et al. 1998) and confers a high level of sulfite resistance (Goto-Yamamoto 1998; Pérez-Ortín et al. 2002; Yuasa et al. 2004). Recently, another translocation between chromosome XV and XVI was identified in several wine strains through quantitative trait loci (QTL) mapping for lag phase duration in the alcoholic fermentation of grape juice, and this translocation increased the expression of this gene (Zimmer et al. 2014). The VIII-t-XVI and XV-t-XVI translocations (Fig. 3) have only been observed in wine yeasts, and 88% of 36 wine strains analyzed possess at least one of these translocations. The VIII-t-XVI translocation is the more frequent and the XV-t-XVI form was only found in commercial starter wine strains, suggesting a recent event. Both translocations conferred a selective advantage by shortening the growth lag phase in medium containing SO2. Thus, the wide use of sulfites since the Middles Ages (Pérez-Ortín et al. 2002) likely caused an evolutionary bottleneck, favoring convergent evolutionary rearrangements that confer a growth advantage to strains carrying the SSU1 recombinant forms.
Figure 3.
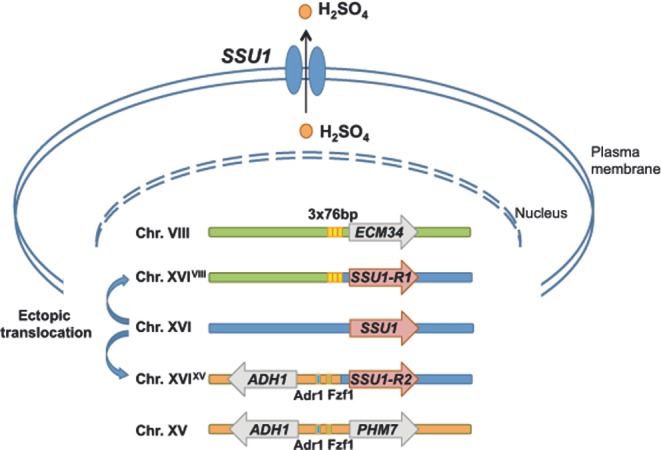
Mechanisms of sulfite resistance through reciprocal translocations. Two ectopic translocations resulting in increased expression of SSU1, encoding a plasma membrane sulfite pump enabling yeast cells to resist to sulfite have been described. A first translocation (VIII-t-XVI, the most frequent form) was mediated through crossing-over mediated by microhomology within the promoters of ECM34 and SSU1. Several 76-bp (in yellow) repeats (3–6 tandem repeats) were found in the promoters of non-recombinant ECM34 and recombinant SSU1-R1. A direct relationship between the number of 76-bp repeats and sulfite tolerance has been described (Pérez-Ortín et al. 2002). A second reciprocal translocation (XV-t-XVI) involves Adr1 (blue rectangle) and Fzf1 (green rectangle) binding regions of the promoter of ADH1 and SSU1, respectively, resulting in the SSU1-R2 allele having increased expression during the first hours of alcoholic fermentation (Zimmer et al. 2014).
Another potential domestication-related trait is the acquisition of resistance to copper sulfate. Elevated copper tolerance in the European and Sake lineages has been associated with a CNV of CUP1 encoding the copper-binding metallothionein (Warringer et al. 2011). The emergence of the CUP1 CNV in these lineages, but not in other S. cerevisiae populations or in S. paradoxus, strongly suggests that the CUP1 CNV reflects convergent evolution due to human selection for industrial production (Warringer et al. 2011). Consistent with a previous study using a restricted number of strains (Fay et al. 2004), recent studies based on a higher number of strains revealed increased resistance to CuSO4 associated with a higher number of copies of CUP1 among wine strains compared with oak or other isolates (Almeida et al. 2015). These data suggest that the acquisition of this trait could be associated with the use of copper sulfate in vineyards, which has been used as a fungicide against powdery mildew since the 1880s (Fay et al. 2004).
Recently, a promoter variant of CUP1 with increased expression variability was identified in the wine yeast strain EC1118. This promoter provides a benefit under environmental stress conditions, suggesting that modulation of gene expression is another potential adaptation mechanism in yeast (Liu et al. 2015).
In addition to these examples, genome-wide studies have provided more complete insights into structural variation, revealing the existence of many CNV in wine yeasts, corresponding to genes encoding transporters or dehydrogenases or genes involved in drug response (Dunn, Levine and Sherlock 2005; Carreto et al. 2008; Borneman et al. 2011; Warringer et al. 2011).
Adaptive loss of aquaporins in wine yeasts
The water transporters aquaporins AQY are critical for surviving freeze–thaw stress. It was suggested that rapid export of water increases freeze–thaw survival by preventing intracellular shearing due to water crystallization (Tanghe et al. 2002). On the other hand, Wills et al. (2010) showed that loss of AQYs function provides a major fitness advantage on high-sugar substrates to overcome the effect of high osmolarity. Laboratory and industrial strains as well as several vineyard isolates harbor non-functional alleles of AQY2 while several strains also harbor a non-functional version of AQY1 (Bonhivers et al. 1998; Laizé et al. 2000). These paralogs have been lost at least six independent times through missense and frame-shift mutations (Will et al. 2010). However, Malysian strains which are not associated with domestication events show unique non-functional AQY alleles, indicating that loss of aquaporins is not strictly driven by domestication. The antagonistic effect of AQYs contributes to the maintenance of both functional and nonfunctional alleles in S. cerevisiae.
Introgressions from Saccharomyces sp. in wine yeasts
Several S. paradoxus and S. mikatae introgressions were identified in S. cerevisiae wine strains (Dunn et al. 2012). A large S. paradoxus introgressed region, identified in commercial wine yeast strains, spans a region corresponding to the SUC2 region of the S. cerevisiae genome and includes not only the S. paradoxus SUC2 gene, which encodes sucrose-hydrolyzing invertase, but also a gene similar to S. cerevisiae HPF1, encoding a glucan alpha-1,4-glucosidase that, when overexpressed, reduces protein haze formation in white wines (Brown et al. 2007). Furthermore, this introgressed region also contains AWA1, a gene present in S. cerevisiae sake strains but absent from S288C, encoding a putative GPI-anchored protein localized to the cell wall and conferring hydrophobicity to the cell surface for foam formation in sake mash (Miyashita et al. 2004). This evidence suggests that some adaptive or industrially desirable qualities might be conferred by S. paradoxus genes to these wine strains (Dunn et al. 2012).
In addition to S. cerevisiae, the cryotolerant species, S. uvarum, is also used for wine and cider fermentation. A recent study of the population structure and diversity of this species revealed multiple introgressions from other Saccharomyces species, and those from S. eubayanus were prevalent in European strains associated with human-driven fermentations (Almeida et al. 2014). This study suggests that the anthropic habitats colonized by S. uvarum in Europe might have favored the hybridization of S. uvarum with S. eubayanus, followed by subsequent introgression through backcrossing to S. uvarum. These introgressed regions are enriched in functions involving nitrogen metabolism, suggesting that these regions might confer an advantage under nitrogen-limiting wine fermentation conditions.
Horizontal transfer and evolutionary advantage of FOT genes
In the last decade, comparative genomics revealed the previously unsuspected contribution of horizontal gene transfer (HGT) to the adaptation of wine yeasts. The genome of the commercial S. cerevisiae wine yeast EC1118 unexpectedly contained three large chromosomal segments, A, B and C (120 kb in total), acquired through independent HGT events from distant yeast species (Novo et al. 2009). These regions have primarily been identified in wine yeasts and mosaic genomes (Borneman et al. 2008, 2011; Novo et al. 2009). Zygosaccharomyces bailii, a major contaminant of wine fermentations, was identified as the donor of region B (Novo et al. 2009; Galeote et al. 2011). Multiple copy insertions and different arrangements of region B have been identified in various wine strains, suggesting that a circular intermediate is involved in the amplification and expansion of this region (Borneman et al. 2011; Galeote et al. 2011). Recently, Marsit et al. (2015) showed that region C also results from a recent transfer, dated approximately 2000 years ago, from Torulaspora microellipsoides, a distant yeast species identified in the wine environment. Thus, recurring transfer from distant yeasts have shaped the genome of wine yeasts, indicating the evolutionary advantage and biological relevance of HGT genes.
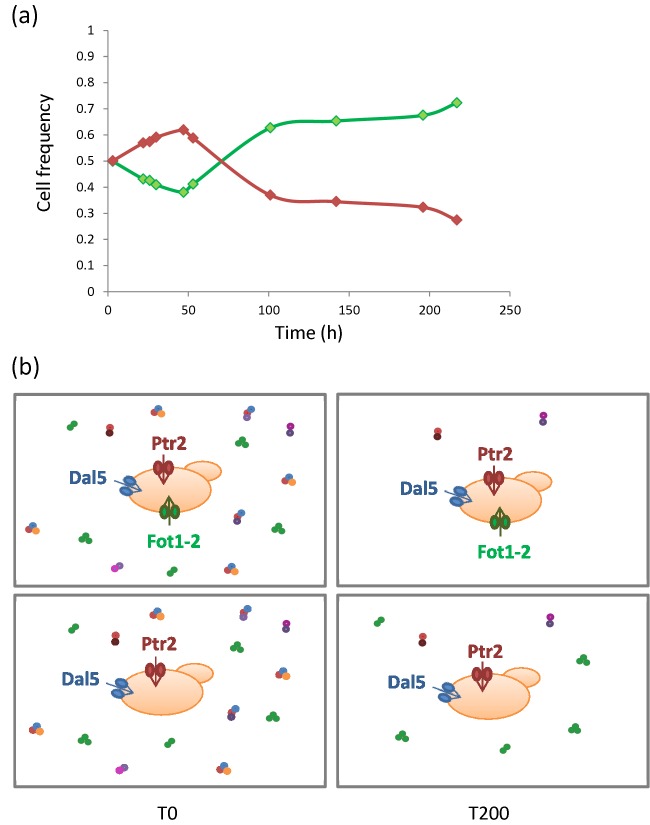
The three initially identified large genomic regions comprise 39 genes (including 5 pseudogenes) encoding functions potentially important for winemaking, such as sugar and nitrogen metabolism (Novo et al. 2009). The functions of several genes of the T. microellipsoides region were characterized in detail. For example, FSY1 encodes a high-affinity fructose/H+ symporter that might be advantageous at the end of wine fermentation, when fructose is the most abundant sugar (Galeote et al. 2010). Another gene, XDH1, encodes a putative xylitol dehydrogenase involved in xylose metabolism (Wenger, Schwartz and Sherlock 2010). Two other tandem duplicated genes FOT1–2 encode oligopeptide transporters, which considerably increase the range of oligopeptides typically transported by the carrier proteins Ptr2p and Dal5p in S. cerevisiae (Damon et al. 2011).
Comparative genomics has provided new insights into the evolutionary history of region C. This region is widespread among wine strains and underwent several rearrangements, including gene losses and gene conversion through FOT genes, resulting in a patchy distribution among various strains (Marsit et al. 2015). FOT1–2 genes are strongly conserved among region C genes, which suggest that they might have an evolutionary advantage. Using competition experiments, Marsit et al. (2015) demonstrated that the presence of FOT genes provides a strong competitive advantage on a natural grape must (Fig. 4). These genes facilitate the transport of a broader range of oligopeptides present in grape juice, particularly those rich in glutamate, which are the most abundant, resulting in improved biomass formation, fermentation efficiency and cell viability during winemaking (Marsit et al. 2015). Furthermore, Fot-mediated peptide uptake substantially affects the central pathways of carbon and nitrogen metabolism, amino acid and protein biosynthesis and the oxidative stress response. In particular, the glutamate node and the NADPH/NADP+ balance are markedly modified, resulting in decreased acetic acid production and increased ester formation, which might improve the organoleptic balance of wines (Marsit S, Galeote V and Dequin S, unpublished data). In addition, several FOT alleles, generated through gene conversion from the FOT genes of T. microellipsoides, were identified in wine yeasts. These variants might have acquired potentially specialized functions, which remain uncharacterized.
Figure 4.
Competitive advantage of FOT genes acquired through HGT in wine yeasts during grape must fermentation. (a) Frequency of cocultured wine strains with (green) or without (red) FOT genes labeled with different fluorochromes and monitored through flow cytometry during fermentation. (b) Wine strains with FOT genes use a broader range of the oligopeptides present in grape juice, particularly those rich in glutamate (colored in green), compared with strains without FOT genes (Marsit et al. 2015).
Interestingly, the ability to use different types of ditripeptides as nitrogen sources considerably varied from one strain to another (Homann et al. 2005). The presence of FOT genes likely contributes to this phenotypic variation in S. cerevisiae.
In addition to FOT genes, several genes present in new regions acquired by wine yeasts have putative functions associated with nitrogen metabolism, including asparaginase, oxoprolinase, an ammonium transporter, an allantoate transporter and two transcription factors associated with the biosynthesis enzymes involved in lysine and proline utilization (Novo et al. 2009). Several introgressions from S. eubayanus in S. uvarum wine strains also contain genes involved in nitrogen metabolism (exopeptidase, L-asparaginase) (Almeida et al. 2014). These genes might facilitate the utilization of nitrogen resources, which is limiting in grape must, providing a competitive advantage to wine yeasts for nutrients during winemaking. These data suggest the concerted evolution of the genome of wine yeasts associated with nitrogen metabolism. Consistently, it was recently suggested that life stage performances have evolved in concert with nitrogen use (Ibstedt et al. 2015).
Little is known about the mechanisms at the origin of an introgression between two different yeast species. In plants, introgressions frequently result from hybridization, followed by successive backcrossing. In nature, the succession of backcrosses seems unlikely, considering the limited frequency of the yeast sexual cycle compared with clonal division, according to Ruderfer et al. (2006). In addition, the sequence divergence between the different species increases the number of necessary backcrosses because this phenomenon reduces the frequency of meiotic recombination. Other mechanisms have been suggested, such as the unidirectional transfer of a single chromosome, chromosome fragment or an episome from one nucleus to another in a newly formed hybrid prior to karyogamy (Morales and Dujon 2012). An alternative explanation is hybridization followed by the loss of most chromosomal material of one of the parents. Many artificial hybrids have been constructed so far, including from distantly related yeasts (Morales and Dujon 2012). These studies showed that hybrid lines generally undergo progressive genome stabilization, during which large genomic rearrangements occur, including aneuploidization, chromosomal translocation and partial or total chromosome loss. Marinoni et al. (1999) tried to obtained interspecific hybrids by crossing yeasts belonging to the genus Saccharomyces, including species of the former sensu stricto and sensu lato groups. They observed that in the case of more distantly related parents, the frequency of interspecific zygotes was lower and that only one parental set, and perhaps some fragments of the other one, could be found in genetically stabilized hybrid lines. Anticipating later findings, they concluded that if Saccharomyces isolates could mate freely in nature, horizontal transfer of genetic material could have occurred during the evolution of modern yeast species.
Wine yeast hybrids
Interspecific hybridization provides new combinations of genes and might confer selective advantages over the parental species (Masneuf et al. 1998; Dujon 2010; Morales and Dujon 2012). In recent decades, a growing number of natural hybrids between two or more Saccharomyces species have been identified in yeast. The best known example is the brewing yeast S. pastorianus, a hybrid between S. cerevisiae and S. eubayanus (Libkind et al. 2011). The molecular characterization of wine and cider yeasts also revealed many hybrids formed independently between S. cerevisiae/S. kudriawzevii (Bradbury et al. 2006; González et al. 2006; Lopandic et al. 2007; Sipiczki 2008; Arroyo-López et al. 2009; Gangl et al. 2009; Borneman et al. 2012; Erny et al. 2012), S. cerevisiae/S. uvarum (Masneuf et al. 1998; Masneuf et al. 2002; Le Jeune et al. 2007; Sipiczki 2008) or between S. cerevisiae/S. kudriawzevii/S. uvarum (Naumova et al. 2005; González et al. 2006).
Hybrids might present several advantages over non-hybrids in wine fermentation (González et al. 2007; Arroyo-López et al. 2009; Gangl et al. 2009; Tronchoni et al. 2009). These hybrids often show more robust features than the parents, such as tolerance to various stresses during fermentation (Belloch et al. 2008; Morales and Dujon 2012). For example, S. kudriavzevii and S. bayanus are better adapted to growth at low temperatures compared with S. cerevisiae wine strains, whereas S. cerevisiae is more alcohol tolerant. The natural hybrids between these species have adapted to growth under ethanol and temperature stress through the inheritance of competitive traits from one or another parental species (Belloch et al. 2009). In recent years, winemakers have preferred the fermentation of white wines at low temperatures, ranging from 10 to 15°C, to minimize the loss of aromatic volatile compounds. Therefore, these hybrids have potential value under these conditions. These inherited traits might also influence the aromatic complexity of wine. Saccharomyces cerevisiae × Saccharomyces kudriavzevii hybrids have been described as greater producers of esters and higher alcohols depending on grape variety (González et al. 2007; Lopandic et al. 2007; Gangl et al. 2009). These hybrids also release much higher amounts of the fruity thiol 4-mercapto-4-methylpentan-2-one from grape-derived non-aromatic precursors than other commercial wine yeast strains (Dubourdieu et al. 2006; Swiegers et al. 2009). The abundance of these hybrids could reflect an adaptive advantage, but it is also possible that stressful conditions trigger hybridization events (Replansky et al. 2008). Hybrid lines generally undergo progressive genome stabilization, during which chromosomal rearrangements and modifications of the genetic contribution of relative parents, aneuploidy or partial chromosome losses were observed (Antunovics et al. 2005; Querol and Bond 2009; Kunicka-Styczyńska and Rajkowska 2011; Borneman et al. 2012; Morales and Dujon 2012).
The molecular characterization of 24 S. cerevisiae–S. kudriavzevii hybrids from Northern European winemaking environments (including commercial strains) revealed multiple ploidy levels (from 2n to 4n) and various amounts of S. kudriavzevii genetic content (Erny et al. 2012). These strains result from multiple hybridization events between several S. cerevisiae wine yeast isolates and various S. kudriavzevii strains (Erny et al. 2012). Another commercial wine strain, Vin7, is an almost complete allotriploid interspecific hybrid containing a heterozygous diploid S. cerevisiae genome and a haploid S. kudriavzevii genome with several homologous recombination and genomic substitution between the two genomes (Borneman et al. 2012). Both parental strains were of European origin, and the S. cerevisiae parent was closely related to, but distinct from, the commercial wine yeasts QA23 and EC1118 (Borneman et al. 2012). Strikingly, S. kudriavzevii yeast strains have never been isolated from wine fermentation, but have been initially isolated from decaying leaves in Japan. Thus, it is unclear how this species formed the hybrids identified in Europe (Naumov et al. 2000). However, recent environmental sampling identified S. kudriavzevii in Portugal and France (Ardèche), but associated with oak bark (Sampaio and Gonçalves 2008; Erny et al. 2012). Despite a common European origin, it remains unknown where and when S. cerevisiae and S. kudriavzevii hybridization occurs.
A recent study provided experimental evidence of evolutionary innovations resulting from hybrid formation. Interspecific hybrids between S. cerevisiae and S. uvarum were de novo generated and subjected to experimental evolution under ammonium limitation conditions. A rearranged interspecific fusion of MEP2, encoding a high-affinity ammonium permease, was shown to confer enhanced fitness under these conditions (Dunn et al. 2013). This rearrangement resulted from the introgression of several bases from S. uvarum in chromosome XIV of S. cerevisiae. The architecture of MEP2 rearrangements suggests a rapid introgression model, which does not require repeated backcrossing with the parental species (Dunn et al. 2013).
PERSPECTIVES
Recent advances in genome-wide analyses and next-generation sequencing have provided unprecedented insights into the population structure and evolutionary history of Saccharomyces, revealing the impact of yeast domestication. Compelling evidence of adaptation in wine yeast strains has been provided, showing that wine yeasts use a variety of mechanisms, including nucleotide and structural variations, introgressions and HGT, to adapt to the winemaking environment. Expanding the whole-genome sequence dataset of strains from the wine environment and other anthropic niches will provide a better understanding of the evolutionary history of domesticated strains and the frequency of these mechanisms, particularly HGT. In addition, the availability of a higher number of genome sequences might facilitate the identification of allelic variants and other divergent regions involved in the adaptation to wine making, which first genomic population approaches have not been able to detect. For example, flor and wine strains belonging to closely related groups with contrasting lifestyles, such as aerobic respiration versus sugar fermentation, might constitute a relevant model to identify divergent regions that might explain the adaptation to these niches. Although QTL mapping strategies have been successfully used in recent decades to decipher the genotype–phenotype associations among well-defined sets of parental strains, sequence information will also increase the number of variants, enabling genome-wide association strategies.
Despite the clear evidence that wine yeast strains have been selected and domesticated from wild strains and subsequently dispersed, little is known about the ecological life cycle and natural history of S. cerevisiae. Indeed, how yeast cells survive in the absence of rich sugar sources in natural environments, particularly during the winter, remains puzzling. A role for birds and insects (Drosophila, bees) as vectors for S. cerevisiae has been suggested (Goddard et al. 2010; Francesca et al. 2012; Buser et al. 2014). A recent study demonstrated a role for social wasps as vectors and natural reservoirs for S. cerevisiae during all seasons, and these authors suggested a multidirectional flow of S. cerevisiae between wineries and vineyards (Stefanini et al. 2012). A major challenge in the future will be to understand how these processes influence the observed population patterns. Metapopulation genomics studies quantifying ecological-scale population processes might provide information to increase the current understanding of gene flow between populations (Knight and Goddard 2015).
Acknowledgments
We thank Jean-Luc Legras for helpful comments on the manuscript.
FUNDING
SM is supported by a fellowship from the Agence Nationale de la Recherche (Grant ANR-13-BSV6-0006 AcrossTrait).
Conflict of interest. None declared.
REFERENCES
- Alexander MA, Jeffries TW. Respiratory efficiency and metabolite partitioning as regulatory phenomena in yeasts. Enzyme Microb Tech. 1990;12:2–19. [Google Scholar]
- Alexandre H. Flor yeasts of Saccharomyces cerevisiae–their ecology, genetics and metabolism. Int J Food Microbiol. 2013;167:269–75. doi: 10.1016/j.ijfoodmicro.2013.08.021. [DOI] [PubMed] [Google Scholar]
- Alexandre H, Rousseaux I, Charpentier C. Ethanol adaptation mechanisms in Saccharomyces cerevisiae. Biotechnol Appl Bioc. 1994;20:173–83. [PubMed] [Google Scholar]
- Almeida P, Barbosa R, Zalar P, et al. A Population Genomics Insight into the Mediterranean Origins of Wine Yeast Domestication. Mol Ecol. 2015;6 doi: 10.1111/mec.13341. [DOI] [PubMed] [Google Scholar]
- Almeida P, Gonçalves C, Teixeira S, et al. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat Commun. 2014;5:4044. doi: 10.1038/ncomms5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunovics Z, Nguyen H-V, Gaillardin C, et al. Gradual genome stabilisation by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5:1141–50. doi: 10.1016/j.femsyr.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Arroyo-López FN, Orlić S, Querol A, et al. Effects of temperature, pH and sugar concentration on the growth parameters of Saccharomyces cerevisiae, S. kudriavzevii and their interspecific hybrid. Int J Food Microbiol. 2009;131:120–7. doi: 10.1016/j.ijfoodmicro.2009.01.035. [DOI] [PubMed] [Google Scholar]
- Barrio E, González S, Arias A, et al. Molecular mechanisms involved in the adaptive evolution of industrial yeasts. In: Querol A, Fleet GH, editors. The Yeast Handbook Yeasts. Berlin, Germany: Springer; 2006. pp. 153–74. [Google Scholar]
- Bataillon M, Rico A, Sablayrolles J, et al. Early thiamine assimilation by yeasts under enological conditions: impact on fermentation kinetics. J Ferment Bioeng. 1996;82:145–50. [Google Scholar]
- Bauer EF, Pretorius LS. Yeast stress response and fermentation efficiency: how to survive the making of wine—a review. S Afr J Enol Vitic. 2000;21:27–51. [Google Scholar]
- Bell S, Henschke PA. Implications of nitrogen nutrition for grapes, fermentation and wine. Aust J Grape Wine Res. 2005;11:242–95. [Google Scholar]
- Belloch C, Orlic S, Barrio E, et al. Fermentative stress adaptation of hybrids within the Saccharomyces sensu stricto complex. Int J Food Microbiol. 2008;122:188–95. doi: 10.1016/j.ijfoodmicro.2007.11.083. [DOI] [PubMed] [Google Scholar]
- Belloch C, Pérez-Torrado R, González SS, et al. Chimeric genomes of natural hybrids of Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Appl Environ Microb. 2009;75:2534–44. doi: 10.1128/AEM.02282-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidenne C, Blondin B, Dequin S, et al. Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr Genet. 1992;22:1–7. doi: 10.1007/BF00351734. [DOI] [PubMed] [Google Scholar]
- Bisson LF. Stuck and sluggish fermentations. Am J Enol Vitic. 1999;50:107–19. [Google Scholar]
- Blondin B, Dequin S, Querol A, et al. Genome of Saccharomyces cerevisiae and related yeasts. In: Konig H, Unden G, Frohlich J, editors. Biology of Microorganisms on Grapes, in Must and in Wine. Berlin: Springer-Verlag; 2009. pp. 361–78. [Google Scholar]
- Bonhivers M, Carbrey JM, Gould SJ, et al. Aquaporins in Saccharomyces: genetic and functional distinctions between laboratory and wild-type strains. J Biol Chem. 1998;273:27565–72. doi: 10.1074/jbc.273.42.27565. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, et al. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, et al. The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 2012;12:88–96. doi: 10.1111/j.1567-1364.2011.00773.x. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Forgan AH, Pretorius IS, et al. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 2008;8:1185–95. doi: 10.1111/j.1567-1364.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- Bradbury JE, Richards KD, Niederer HA, et al. A homozygous diploid subset of commercial wine yeast strains. Anton Leeuw. 2006;89:27–37. doi: 10.1007/s10482-005-9006-1. [DOI] [PubMed] [Google Scholar]
- Brown SL, Stockdale VJ, Pettolino F, et al. Reducing haziness in white wine by overexpression of Saccharomyces cerevisiae genes YOL155c and YDR055w. Appl Microbiol Biot. 2007;73:1363–76. doi: 10.1007/s00253-006-0606-0. [DOI] [PubMed] [Google Scholar]
- Buser CC, Newcomb RD, Gaskett AC, et al. Niche construction initiates the evolution of mutualistic interactions. Ecol Lett. 2014;10:1257–64. doi: 10.1111/ele.12331. [DOI] [PubMed] [Google Scholar]
- Camarasa C, Sanchez I, Brial P, et al. Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One. 2011;6:e25147. doi: 10.1371/journal.pone.0025147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreto L, Eiriz MF, Gomes AC, et al. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics. 2008;9:524. doi: 10.1186/1471-2164-9-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri D, McGovern PE, Hartl DL, et al. Evidence for S. cerevisiae fermentation in ancient wine. J Mol Evol. 2003;57:226–32. doi: 10.1007/s00239-003-0031-2. [DOI] [PubMed] [Google Scholar]
- Codón AC, Benítez T, Korhola M. Chromosomal polymorphism and adaptation to specific industrial environments of Saccharomyces strains. Appl Microbiol Biot. 1998;49:154–63. doi: 10.1007/s002530051152. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol. 2007;3:129. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie Ga, Hyma KE, Ludlow CL, et al. Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3 (Bethesda) 2013;3:2163–71. doi: 10.1534/g3.113.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, Vásquez C, Faugeron S, et al. Self-fertilization is the main sexual reproduction mechanism in native wine yeast populations. FEMS Microbiol Ecol. 2009;67:162–70. doi: 10.1111/j.1574-6941.2008.00600.x. [DOI] [PubMed] [Google Scholar]
- Damon C, Vallon L, Zimmermann S, et al. A novel fungal family of oligopeptide transporters identified by functional metatranscriptomics of soil eukaryotes. ISME J. 2011;5:1871–80. doi: 10.1038/ismej.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashko S, Zhou N, Compagno C, et al. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res. 2014;14:826–32. doi: 10.1111/1567-1364.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequin S. The potential of genetic engineering for improving brewing, wine-making and baking yeasts. Appl Microbiol Biot. 2001;56:577–88. doi: 10.1007/s002530100700. [DOI] [PubMed] [Google Scholar]
- Dequin S, Casaregola S. The genomes of fermentative Saccharomyces. C R Biol. 2011;334:687–93. doi: 10.1016/j.crvi.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Dubourdieu D, Tominaga T, Masneuf I, et al. The role of yeasts in grape flavor development during fermentation: the example of Sauvignon blanc. Am J Enol Vitic. 2006;57:81–8. [Google Scholar]
- Dujon B. Yeast evolutionary genomics. Nat Rev Genet. 2010;11:512–24. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- Dunn B, Levine RP, Sherlock G. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics. 2005;6:53. doi: 10.1186/1471-2164-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B, Paulish T, Stanbery A, et al. Recurrent rearrangement during adaptive evolution in an interspecific yeast hybrid suggests a model for rapid introgression. PLoS Genet. 2013;9:e1003366. doi: 10.1371/journal.pgen.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B, Richter C, Kvitek DJ, et al. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 2012;22:908–24. doi: 10.1101/gr.130310.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny C, Raoult P, Alais A, et al. Ecological success of a group of Saccharomyces cerevisiae/Saccharomyces kudriavzevii hybrids in the northern european wine-making environment. Appl Environ Microb. 2012;78:3256–65. doi: 10.1128/AEM.06752-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, McCullough HL, Sniegowski PD, et al. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol. 2004;5:R26. doi: 10.1186/gb-2004-5-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Barrales RR, Ibeas JI, et al. Adaptive evolution by mutations in the FLO11 gene. P Natl Acad Sci USA. 2006;103:11228–33. doi: 10.1073/pnas.0601713103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet G. The microorganisms of winemaking—isolation, enumeration and identification. In: Fleet G, editor. Wine Microbiology and Biotechnology. Switzerland: Harwood Academic Publishers; 1993. pp. 1–25. [Google Scholar]
- Fleet GH, Lafon-Lafourcade S, Ribereau-Gayon P. Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl Environ Microb. 1984;48:1034–8. doi: 10.1128/aem.48.5.1034-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesca N, Canale DE, Settanni L, et al. Dissemination of wine-related yeasts by migratory birds. Environ Microbiol Rep. 2012;4:105–12. doi: 10.1111/j.1758-2229.2011.00310.x. [DOI] [PubMed] [Google Scholar]
- Galeote V, Bigey F, Beyne E, et al. Amplification of a Zygosaccharomyces bailii DNA segment in wine yeast genomes by extrachromosomal circular DNA formation. PLoS One. 2011;6:e17872. doi: 10.1371/journal.pone.0017872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote V, Novo M, Salema-Oom M, et al. FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high-affinity fructose/H+ symporter. Microbiology. 2010;156:3754–61. doi: 10.1099/mic.0.041673-0. [DOI] [PubMed] [Google Scholar]
- Gangl H, Batusic M, Tscheik G, et al. Exceptional fermentation characteristics of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. N Biotechnol. 2009;25:244–51. doi: 10.1016/j.nbt.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Gayevskiy V, Goddard MR. Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J. 2012;6:1281–90. doi: 10.1038/ismej.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard MR, Anfang N, Tang R, et al. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ Microbiol. 2010;12:63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, et al. Life with 6000 Genes. Science (80–) 1996;274:546–67. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- González SS, Barrio E, Gafner J, et al. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 2006;6:1221–34. doi: 10.1111/j.1567-1364.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- González SS, Gallo L, Climent MD, et al. Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int J Food Microbiol. 2007;116:11–8. doi: 10.1016/j.ijfoodmicro.2006.10.047. [DOI] [PubMed] [Google Scholar]
- Goto-yamamoto N, Kazuyoshi K, Kunio S, et al. SSUI-R, a sulfite resistance gene of wine yeast, is an allele of SSUl with a different upstream sequence. J Ferment Bioeng. 1998;86:427–33. [Google Scholar]
- Guillaume C, Delobel P, Sablayrolles J, et al. Molecular basis of fructose utilization by the wine yeast Saccharomyces cerevisiae: a mutated HXT3 allele enhances fructose fermentation. Appl Environ Microb. 2007;73:2432–9. doi: 10.1128/AEM.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Styles Ca, Feng Q, et al. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. P Natl Acad Sci. 2000;97:12158–63. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman A, Piškur J. A study on the fundamental mechanism and the evolutionary driving forces behind aerobic fermentation in yeast. PLoS One. 2015;10:e0116942. doi: 10.1371/journal.pone.0116942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschke PA, Jiranek V. Yeasts—metabolism of nitrogen compounds. In: Fleet GH, editor. Wine Microbiology and Biotechnology. Switzerland: Harwood Academic Publishers; 1993. pp. 77–164. [Google Scholar]
- Homann OR, Cai H, Becker JM, et al. Harnessing natural diversity to probe metabolic pathways. PLoS Genet. 2005;1:e80. doi: 10.1371/journal.pgen.0010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyma KE, Fay JC. Mixing of vineyard and oak-tree ecotypes of Saccharomyces cerevisiae in North American vineyards. Mol Ecol. 2013;22:2917–30. doi: 10.1111/mec.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyma KE, Saerens SM, Verstrepen KJ, et al. Divergence in wine characteristics produced by wild and domesticated strains of Saccharomyces cerevisiae. FEMS Yeast Res. 2011;11:540–51. doi: 10.1111/j.1567-1364.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibstedt S, Stenberg S, Bagés S, et al. Concerted evolution of life stage performances signals recent selection on yeast nitrogen use. Mol Biol Evol. 2015;32:153–61. doi: 10.1093/molbev/msu285. [DOI] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-nejad M, et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science (80–) 2005;309:938–41. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Johnston JR, Baccari C, Mortimer RK. Genotypic characterization of strains of commercial wine yeasts by tetrad analysis. Res Microbiol. 2000;151:583–90. doi: 10.1016/s0923-2508(00)00228-x. [DOI] [PubMed] [Google Scholar]
- Jolly NP, Varela C, Pretorius IS. Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014;14:215–37. doi: 10.1111/1567-1364.12111. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–24. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kelly AC, Shewmaker FP, Kryndushkin D, et al. PNAS Plus: sex, prions, and plasmids in yeast. P Natl Acad Sci USA. 2012;109:E2683–90. doi: 10.1073/pnas.1213449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Goddard MR. Quantifying separation and similarity in a Saccharomyces cerevisiae metapopulation. ISME J. 2015;9:361–70. doi: 10.1038/ismej.2014.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunicka-Styczyńska A, Rajkowska K. Physiological and genetic stability of hybrids of industrial wine yeasts Saccharomyces sensu stricto complex. J Appl Microbiol. 2011;110:1538–49. doi: 10.1111/j.1365-2672.2011.05009.x. [DOI] [PubMed] [Google Scholar]
- Laizé V, Tacnet F, Ripoche P, et al. Polymorphism of Saccharomyces cerevisiae aquaporins. Yeast. 2000;16:897–903. doi: 10.1002/1097-0061(200007)16:10<897::AID-YEA583>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Landry CR, Townsend JP, Hartl DL, et al. Ecological and evolutionary genomics of Saccharomyces cerevisiae. Mol Ecol. 2006;15:575–91. doi: 10.1111/j.1365-294X.2006.02778.x. [DOI] [PubMed] [Google Scholar]
- Le Jeune C, Lollier M, Demuyter C, et al. Characterization of natural hybrids of Saccharomyces cerevisiae and Saccharomyces bayanus var. uvarum. FEMS Yeast Res. 2007;7:540–9. doi: 10.1111/j.1567-1364.2007.00207.x. [DOI] [PubMed] [Google Scholar]
- Legras J, Erny C, Charpentier C. Population structure and comparative genome hybridization of European flor yeast reveal a unique group of Saccharomyces cerevisiae strains with few gene duplications in their genome. PLoS One. 2014;9:e108089. doi: 10.1371/journal.pone.0108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legras J-L, Merdinoglu D, Cornuet J-M, et al. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Libkind D, Hittinger CT, Valério E, et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. P Natl Acad Sci USA. 2011;108:14539–44. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–41. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Martin-Yken H, Bigey F, et al. Natural yeast promoter variants reveal epistasis in the generation of transcriptional-mediated noise and its potential benefit in stressful conditions. Genome Biol Evol. 2015;7:969–84. doi: 10.1093/gbe/evv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopandic K, Gangl H, Wallner E, et al. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 2007;7:953–65. doi: 10.1111/j.1567-1364.2007.00240.x. [DOI] [PubMed] [Google Scholar]
- Luparia V, Soubeyrand V, Berges T, et al. Assimilation of grape phytosterols by Saccharomyces cerevisiae and their impact on enological fermentations. Appl Microbiol Biot. 2004;65:25–32. doi: 10.1007/s00253-003-1549-3. [DOI] [PubMed] [Google Scholar]
- McCusker J. Saccharomyces cerevisiae: an emerging and model pathogenic fungus. In: Heitman J, Edwards JE, Filler SG, et al., editors. Molecular Principles of Fungal Pathogenesis. Washington, DC: ASM Press; 2006. p. 1627. [Google Scholar]
- McGovern P. Ancient Wine: The Search for the Origins of Viniculture. Princeton: Princeton University Press; 2003. [Google Scholar]
- Mc-Govern PE, Donald L, Glusker LJE. Neolithic resinated wine. Nature. 1996;381:480–1. [Google Scholar]
- McGovern PE, Zhang J, Tang J, et al. Fermented beverages of pre- and proto-historic China. P Natl Acad Sci USA. 2004;101:17593–8. doi: 10.1073/pnas.0407921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwene PM. Revisiting Mortimer's Genome Renewal Hypothesis: heterozygosity, homothallism, and the potential for adaptation in yeast. Adv Exp Med Biol. 2014;781:37–48. doi: 10.1007/978-94-007-7347-9_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwene PM, Kayıkçı Ö, Granek JA, et al. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. P Natl Acad Sci USA. 2011;108:1987–92. doi: 10.1073/pnas.1012544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinoni G, Manuel M, Petersen RF, et al. Horizontal transfer of genetic material among Saccharomyces yeasts. J Bacteriol. 1999;181:6488–96. doi: 10.1128/jb.181.20.6488-6496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit S, Mena A, Bigey F, et al. Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Mol Biol Evol. 2015;32:1695–707. doi: 10.1093/molbev/msv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell P, Stratford M, Steels H, et al. Physiological characterization of spoilage strains of Zygosaccharomyces bailii and Zygosaccharomyces rouxii isolated from high sugar environments. Int J Food Microbiol. 2007;114:234–42. doi: 10.1016/j.ijfoodmicro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Masneuf I, Hansen J, Groth C, et al. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl Environ Microb. 1998;64:3887–92. doi: 10.1128/aem.64.10.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masneuf I, Murat M, Naumov G, et al. Hybrids Saccharomyces cerevisiae and Saccharomyces bayanus var. uvarum having a high liberating ability of some sulfur varietal aromas of Vitis vinifera sauvignon blanc wines. J Int Sci Vigne Vin. 2002;36:205–12. [Google Scholar]
- Miyashita K, Sakamoto K, Kitagaki H, et al. Cloning and analysis of the AWA1 gene of a nonfoaming mutant of a sake yeast. J Biosci Bioeng. 2004;97:14–8. doi: 10.1016/S1389-1723(04)70158-9. [DOI] [PubMed] [Google Scholar]
- Morales L, Dujon B. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol Mol Biol Rev. 2012;76:721–39. doi: 10.1128/MMBR.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 2000;10:403–9. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Romano P, Suzzi G, et al. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast. 1994;10:1543–52. doi: 10.1002/yea.320101203. [DOI] [PubMed] [Google Scholar]
- Naumov G, James S, Naumova E, et al. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int J Syst Evol Microbiol. 2000;50:1931–42. doi: 10.1099/00207713-50-5-1931. [DOI] [PubMed] [Google Scholar]
- Naumova ES, Naumov GI, Masneuf-Pomarède I, et al. Molecular genetic study of introgression between Saccharomyces bayanus and S. cerevisiae. Yeast. 2005;22:1099–115. doi: 10.1002/yea.1298. [DOI] [PubMed] [Google Scholar]
- Novo M, Bigey F, Beyne E, et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. P Natl Acad Sci USA. 2009;106:16333–8. doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ough CS, Huang ZDA, Stevens D. Amino acid uptake by four commercial yeasts at two different temperatures of growth and fermentation: effects on urea excretion and reabsorption. Am J Enol Vitic. 1991;42:26–40. [Google Scholar]
- Park H, Bakalinsky A. SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast. 2000;16:881–8. doi: 10.1002/1097-0061(200007)16:10<881::AID-YEA576>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Pasteur L. Mémoire sur la fermentation alcoolique. Ann Chim Phys. 1860;58:323–426. [Google Scholar]
- Pérez-Ortín JE, Querol A, Puig S, et al. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002;12:1533–9. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piškur J, Rozpedowska E, Polakova S, et al. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–6. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Postma E, Verduyn C, Scheffers Wa, et al. Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microb. 1989;55:468–77. doi: 10.1128/aem.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretorius I. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;15:675–29. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Pronk JT, Steensma HY, Van Dijken JP. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–33. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Querol A, Bond U. The complex and dynamic genomes of industrial yeasts. FEMS Microbiol Lett. 2009;293:1–10. doi: 10.1111/j.1574-6968.2008.01480.x. [DOI] [PubMed] [Google Scholar]
- Rachidi N, Barre P, Blondin B. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol Gen Genet MGG. 1999;261:841–50. doi: 10.1007/s004380050028. [DOI] [PubMed] [Google Scholar]
- Replansky T, Koufopanou V, Greig D, et al. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 2008;23:494–501. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Reuter M, Bell G, Greig D. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr Biol. 2007;17:81–3. doi: 10.1016/j.cub.2006.11.059. [DOI] [PubMed] [Google Scholar]
- Rozpędowska E, Hellborg L, Ishchuk OP, et al. Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts. Nat Commun. 2011;2:302. doi: 10.1038/ncomms1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Pratt SC, Seidel HS, et al. Population genomic analysis of outcrossing and recombination in yeast. Nat Genet. 2006;38:1077–81. doi: 10.1038/ng1859. [DOI] [PubMed] [Google Scholar]
- Sablayrolles JM. Fermented beverages: the example of winemaking. In: Pandey A, Larroche C, Soccol CR, editors. Advances in Fermentation Technology. New Delhi: Asiatech Publishers; 2008. pp. 322–47. [Google Scholar]
- Sampaio JP, Gonçalves P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl Environ Microb. 2008;74:2144–52. doi: 10.1128/AEM.02396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, et al. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–5. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller D, Cardoso F, Sousa S, et al. Genetic diversity and population structure of Saccharomyces cerevisiae strains isolated from different grape varieties and winemaking regions. PLoS One. 2012;7:e32507. doi: 10.1371/journal.pone.0032507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard D, Legras J. Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 2011;334:229–36. doi: 10.1016/j.crvi.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Sipiczki M. Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Res. 2008;8:996–1007. doi: 10.1111/j.1567-1364.2008.00369.x. [DOI] [PubMed] [Google Scholar]
- Spor A, Nidelet T, Simon J, et al. Niche-driven evolution of metabolic and life-history strategies in natural and domesticated populations of Saccharomyces cerevisiae. BMC Evol Biol. 2009;9:296. doi: 10.1186/1471-2148-9-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini I, Dapporto L, Legras J, et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. P Natl Acad Sci USA. 2012;109:13398–403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiegers JH, Bartowsky EJ, Henschke PA, et al. Yeast and bacterial modulation of wine aroma and flavour. Aust J Grape Wine Res. 2005;11:139–73. [Google Scholar]
- Swiegers JH, Kievit RL, Siebert T, et al. The influence of yeast on the aroma of Sauvignon Blanc wine. Food Microbiol. 2009;26:204–11. doi: 10.1016/j.fm.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Tanghe A, Van Dijck P, Dumortier F, et al. Aquaporin expression correlates with freeze tolerance in baker's yeast, and overexpression improves freeze tolerance in industrial strains. Appl Environ Microb. 2002;68:5981–9. doi: 10.1128/AEM.68.12.5981-5989.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesnière C, Delobel P, Pradal M, et al. Impact of nutrient imbalance on wine alcoholic fermentations: nitrogen excess enhances yeast cell death in lipid-limited must. PLoS One. 2013;8:e61645. doi: 10.1371/journal.pone.0061645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, et al. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–5. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronchoni J, Gamero A, Arroyo-López FN, et al. Differences in the glucose and fructose consumption profiles in diverse Saccharomyces wine species and their hybrids during grape juice fermentation. Int J Food Microbiol. 2009;134:237–43. doi: 10.1016/j.ijfoodmicro.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Vemuri GN, Eiteman MA, McEwen JE, et al. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. P Natl Acad Sci USA. 2007;104:2402–7. doi: 10.1073/pnas.0607469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezinhet F, Blondin B, Hallet J-N. Chromosomal DNA patterns and mitochondrial DNA polymorphism as tools for identification of enological strains of Saccharomyces cerevisiae. Appl Microbiol Biot. 1990;32:568–71. [Google Scholar]
- Wang Q-M, Liu W-Q, Liti G, et al. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21:5404–17. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- Warringer J, Zörgö E, Cubillos FA, et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger JW, Schwartz K, Sherlock G. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 2010;6:e1000942. doi: 10.1371/journal.pgen.1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will JL, Kim HS, Clarke J, et al. Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet. 2010;6:e1000893. doi: 10.1371/journal.pgen.1000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–13. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Amemiya H, Yokomori Y, et al. Electrophoretic karyotypes of wine yeasts. Am J Enol Vitic. 1991;42:358–63. [Google Scholar]
- Yuasa N, Nakagawa Y, Hayakawa M, et al. Distribution of the sulfite resistance gene SSU1-R and the variation in its promoter region in wine yeasts. J Biosci Bioeng. 2004;98:394–7. doi: 10.1016/S1389-1723(04)00303-2. [DOI] [PubMed] [Google Scholar]
- Zimmer A, Durand C, Loira N, et al. QTL dissection of Lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS One. 2014;9:e86298. doi: 10.1371/journal.pone.0086298. [DOI] [PMC free article] [PubMed] [Google Scholar]