Abstract
Since 2008, Mainland China has undergone widespread outbreaks of hand, foot, and mouth disease (HFMD). In order to determine the characteristics of epidemics and enteroviruses (EV) associated with HFMD in Tianjin, in northern China, epidemiological and virological data from routine surveillance were collected and analyzed. In Tianjin, a persistent epidemic of HFMD was demonstrated during 2008–2013, involving 102,705 mild, 179 severe, and 16 fatal cases. Overall, 8234 specimens were collected from 7829 HFMD patients for EV detection during 2008–2013. Enterovirus 71 (EV-A71) and coxsackievirus A16 (CV-A16) were the dominant serotypes during 2008–2012, and they were replaced by CV-A6 as the major causative agent in 2013. Phylogenetic analysis based on complete VP1 nucleotide sequences revealed that multiple CV-A6 lineages co-circulated in Tianjin, which grouped together with strains from China and other countries and split into two distinct clusters (clusters 1 and 2). Most Tianjin strains grouped in cluster 1 and were closely related to strains from several eastern and southern provinces of China during 2012 and 2013. Estimates from Bayesian MCMC analysis suggested that multiple lineages had been transmitted silently before the outbreaks at an estimated evolutionary rate of 4.10 × 10−3 substitutions per site per year without a specific distribution of rate variances among lineages. The sudden outbreak of CV-A6 in Tianjin during 2013 is attributed to indigenous CV-A6 lineages, which were linked to the wide spread of endemic strains around eastern and southern China.
Introduction
Hand, foot, and mouth disease (HFMD) is a common infectious disease caused by human enteroviruses (EVs) that usually attacks children. Enterovirus 71 (EV-A71) and coxsackievirus A16 (CV-A16) are the major pathogens causing HFMD [1, 3, 11, 12, 22, 29]. Other EVs, such as CV-A4, CV-A5, CV-A6, CV-A10, and CV-A12, are also often associated with HFMD [7, 8, 10, 16, 26, 28, 30]. Notably, circulation of CV-A6 and CV-A10 has become more active recently, causing several HFMD outbreaks around the world since 2008 [7, 8, 13, 15, 16, 21, 27].
In Mainland China, HFMD was classified as a notifiable disease in 2008, and nationwide surveillance has been performed since then. Tianjin is one of the four directly controlled municipalities, located in northern China. As part of national surveillance, both epidemiological and virological surveillance for HFMD have been carried out in Tianjin since 2008 and have indicated a persistent HFMD epidemic.
In this study, epidemiological and virological investigations were performed to characterize the epidemics of HFMD in Tianjin during 2008–2013. Analyses based on complete VP1 nucleotide sequences were performed to determine the evolutionary trajectory of emerging CV-A6.
Materials and methods
Collection of epidemiological data
As a notifiable disease in China, demographic and epidemiologic data from HFMD cases are collected using a standard case investigation form and reported online to the China Information System for Disease Control and Prevention [24, 29]. Epidemiological data in this study were retrieved from this national database. In this study, an epidemic season of HFMD was defined as a period of ≥2 consecutive weeks when the weekly number of reported cases accounted for ≥2.0 % of the cases reported in that year. The epidemic peak is the week when the number of weekly reported cases is the highest.
Mild cases of HFMD have good prognosis, without serious complications. In a few patients, the central nervous system (CNS) is involved, and these cases are considered severe. For clinical classification, we followed Guidelines for Diagnosis and Management of Hand, Foot and Mouth Disease (2010) (http://www.moh.gov.cn/publicfiles/business/htmlfiles/mohyzs/s3586/201004/46884.htm), released by China Ministry of Health.
Specimen collection and processing
Because HFMD is notifiable, as part of routine virological surveillance, specimens were routinely collected from clinically diagnosed HFMD cases in Tianjin city within 7 days of the onset of illness. From 8 April 2008 to 3 December 2013, a total of 8234 specimens (7631 stools, 1 CSF, 80 sera, and 522 throat swabs) were collected from 7829 patients for EV detection (Fig. 1a). Stool specimens were processed as described previously [17] for subsequent viral RNA extraction. Specimens of other types were used directly for viral RNA extraction. As a public-health surveillance activity, ethical review was not required.
Fig. 1.
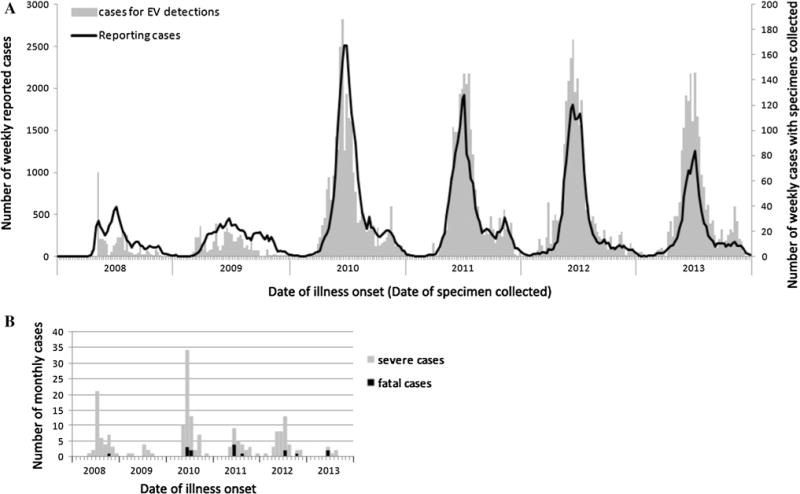
Temporal distribution of reported cases of HFMD and cases of EV detected in Tianjin, 2008–2013. (A) Weekly distribution of reported cases and cases with specimens collected for EV detection, indicated on the left and right axis, respectively. (B) Monthly distribution of severe and fatal cases
Viral RNA extraction and detection
Viral RNA was extracted using a QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA, USA). EV-A71, CV-A16, and pan-EV RNA detection were routinely performed. Previously described conventional RT-PCR methods [32] were used during 2008 to 2009, and commercial serotype-specific real-time RT-PCR (rRT-PCR) kits were employed beginning in 2010 (from 2010 to 2012, Taitaigen, Shenzhen, China; in 2013, Mole, Taizhou, China).
In 2013, additional detection was performed using commercial CV-A6- and CV-A10-specific rRT-PCR kits (Shuoshi, Taizhou, China) because of the increased number of other EV serotypes. rRT-PCR was performed according to the manufacturer’s instructions.
VP1 sequencing of CV-A6
Degenerate primer pairs (486/488 and 040/012/011) were used for amplification of a partial VP1 sequence of CV-A6 [18, 19]. Also, the complete VP1-encoding region of CV-A6 was amplified from some of the specimens, using the forward primer 5′-CTTCGTAGTGCCACCAGATA-3′ (nucleotides 2317–2336; all of the nucleotide positions in this study correspond to those of CV-A6/Gdula: AY421764) and the reverse primer 5′-GTGGCGAGATGTCGGTTTA-3′ (nucleotides 3408–3426). PCR products were purified and sequenced as described previously [32]. Sequences were edited and assembled by using Sequencher 5.0 software (Gene Codes, Ann Arbor, MI, USA). All of the sequences determined (n=73) were deposited in Gen-Bank, with accession numbers KJ774060-KJ774103 and KJ848296-KJ848324.
Evolutionary analysis of CV-A6
Multiple sequence alignments, estimation of genetic distance, and construction of maximum-likelihood (ML) trees were performed using MEGA 5.1 software [6, 23]. Branch lengths were estimated using the general time-reversible (GTR) nucleotide substitution model [31] and a gamma distribution of rates among sites (Γ4) as estimated by Modeltest [20]. A majority-rule consensus tree was obtained after 1,000 pseudo-replicates.
Bayesian Markov chain Monte Carlo (MCMC) methods were used to estimate evolutionary characteristics using BEAST v1.8 (http://beast.bio.ed.ac.uk/) with the relaxed clock model [5]. Two independent MCMC chains were run for each BEAST analysis (80 million generations each) and sampling efficiency was measured using the effective sampling size (ESS) function in TRACER v1.5 (http://tree.bio.ed.ac.uk/software/tracer/). Confidence intervals for evolutionary estimates were obtained as 95 % high posterior density intervals (95 % HPD). Results from the two independent chains were combined into a maximum clade credibility (MCC) tree. Phylogenetic trees were displayed and annotated by using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Epidemics of HFMD
From 1 January 2008 to 31 December 2013, a total of 102,705 HFMD cases were reported in Tianjin, including 179 (1.74‰) severe cases with CNS complications and 16 (0.16‰) fatal cases (Table 1). Incidence rates ranged from 70.3 to 229.4 cases per 100,000 individuals. Based on the data from 2010 to 2013, the epidemic season in Tianjin extended from as early as week 19 to as late as week 33 (May to August), with an epidemic peak around weeks 24 to 27, when the proportion of weekly reported cases relative to annually reported cases ranged from 8.6 % to 9.2 % (Fig. 1a). Overall, 73.8 % (75,848/102,705) of all cases, 78.8 % (141/179) of severe cases, and 87.5 % (14/16) of fatal cases occurred in the epidemic season during 2008 to 2013 (Fig. 1).
Table 1.
Annual number and incidence of HFMD cases reported in Tianjin, 2008–2013
| Year | No. of cases reported | Incidence (per 100,000) | No. of severe cases | No. of fatal cases |
|---|---|---|---|---|
| 2008 | 7839 | 70.3047 | 44 | 1 |
| 2009 | 10125 | 86.0969 | 9 | 0 |
| 2010 | 28178 | 229.4325 | 62 | 5 |
| 2011 | 22280 | 172.2006 | 22 | 5 |
| 2012 | 20705 | 152.8517 | 38 | 3 |
| 2013 | 13578 | 96.0832 | 4 | 2 |
| Total | 102705 | – | 179 | 16 |
Children <6 years old constituted the majority of subjects affected by HFMD, accounting for 88.2 % (90,558/102,705) of reported cases and all of the fatal cases. Children aged 1–4 years showed higher age-specific incidence rates, ranging from 882.9 to 4583.4 cases per 100,000 per year (Fig. 2). Children aged <1 year showed the highest fatality rate (39 deaths per 100,000 reported cases).
Fig. 2.
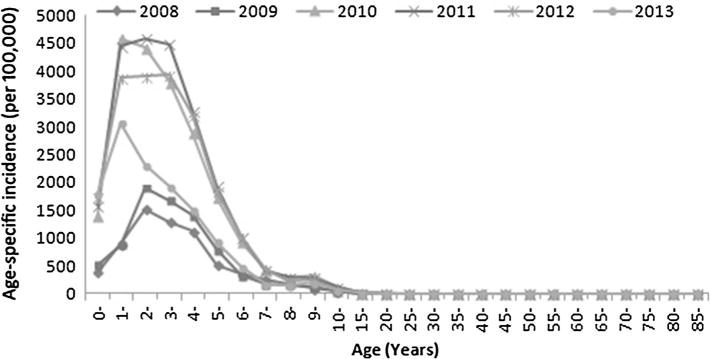
Incidence of HFMD in Tianjin in 2008–2013 according to age
EV detection
During 2008–2013, assays for EV were performed on samples from 7,829 patients (7.6 % of reported cases), and 72.8 % were EV positive (Table 2). Virus surveillance in Tianjin revealed a serotype change during 2008–2013. During 2009–2012, EV-A71 was the most commonly detected serotype, and CV-A16 was the second most commonly detected, except in 2011 (Table 2). In 2013, detection of other EVs increased sharply, accounting for 64.7 % of all EVs detected (Table 2). CV-A6 was found to have displaced both EV-A71 and CV-A16 as the dominant serotype in 2013, accounting for 54.3 % (741/1364) of total EVs detected (Table 2).
Table 2.
Results of EV detection from HFMD cases in Tianjin, 2008–2013
| Year | Number of cases detected
|
||||||
|---|---|---|---|---|---|---|---|
| EV positive (%)
|
EV negative | Total | |||||
| EV-A71a | CV-A16a | EV-A71 + CV-A16a | Other EVa | Totalb | |||
| 2008 | 142 (85.5) | 16 (9.6) | 0 | 8 (4.8) | 166 (60.6) | 108 | 274 |
| 2009 | 128 (46.5) | 88 (32.0) | 7 (2.5) | 52 (18.9) | 275 (73.9) | 97 | 372 |
| 2010 | 564 (43.0) | 535 (40.8) | 22 (1.7) | 191 (14.6) | 1312 (72.9) | 487 | 1799 |
| 2011 | 646 (49.1) | 285 (21.7) | 11 (0.8) | 373 (28.4) | 1315 (72.8) | 492 | 1807 |
| 2012 | 623 (40.5) | 565 (36.7) | 0 | 351 (22.8) | 1539 (81.6) | 348 | 1887 |
| 2013 | 277 (20.3) | 200 (14.7) | 4 (0.3) | 883 (64.7)c | 1364 (80.7) | 326 | 1690 |
| Total | 2380 (39.9) | 1689 (28.3) | 44 (0.7) | 1858 (31.1) | 5971 (76.3) | 1858 | 7829 |
Values in parentheses are the percentage of the total that were EV positive
Values in parentheses are the EV positive rate (total number of EV positive/total number of cases detected)
Out of 883 other EVs in 2013, 741 were identified as CV-A6 and 28 were identified as CV-A10 by serotype-specific rRT-PCR, and 114 were untyped
Evolutionary analysis of CV-A6
Analysis of partial VP1 nucleotide sequences (nucleotides 2980–3355) indicated that CV-A6 strains in this study (n = 73) had 82.5 %–87.6 % nucleotide sequence identity to the CV-A6 prototype (CV-A6/Gdula, AY421764). In the ML tree (Figure S1), out of 73 CV-A6 strains in Tianjin, 66 grouped in one cluster, and the other seven grouped in another cluster, with estimated pairwise nucleotide sequence identities of 92.1–100 % and 94.2–100 % within the respective clusters. Nucleotide sequence identity between clusters was 85.9–92.1 % (mean, 89.7 %).
To determine phylogenetic relationships among CV-A6 strains, we constructed an ML tree based on complete VP1 nucleotide sequences (915 nucleotides) of Tianjin strains (n = 29) and strains from elsewhere in China (n = 21) and other countries (n = 14) during 2008–2013 (Fig. 3). To facilitate molecular epidemiological descriptions, genogroup designations were used to indicate distinct monophyletic clades.
Fig. 3.
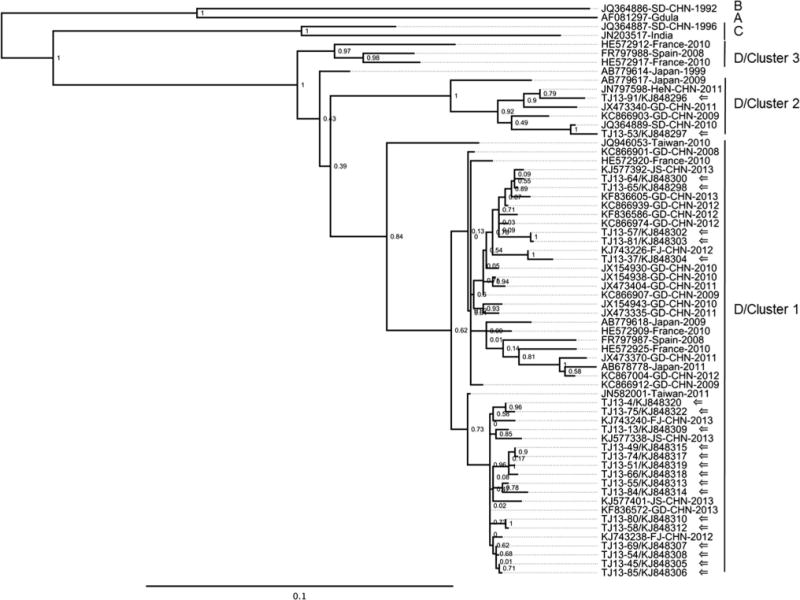
Maximum-likelihood tree based on complete VP1 nucleotide sequences (915 nucleotides) of CV-A6 strains from this study and references from GenBank. Tianjin sequences are indicated by arrows. The other sequences are indicated by GenBank accession number, country and year (where available). Chinese provinces: FJ, Fujian; GD, Guangdong; HeN, Henan; JS, Jiangsu; SD, Shandong
In the ML tree, the prototype strain CV-A6/Gdula and three other strains from China and India were distantly related to the Tianjin sequences and separated into distinct clades (genogroups A, B, and C). The Tianjin strains and the rest of the reference strains (31/35) grouped in another monophyletic clade (genogroup D, 100 % bootstrap support). The topology of the tree revealed at least three different clusters of CV-A6 strains within genogroup D (indicated as clusters 1, 2, and 3), which were supported by bootstrap values of ≥84 %. Most Tianjin strains (n = 27) grouped in cluster 1, and the other two Tianjin strains grouped in cluster 2 (Fig. 3). All previous CV-A6 strains from China were also grouped in clusters 1 (n = 16) and 2 (n = 4), except JQ364886. The Tianjin strains were closely related to strains from Fujian, Guangdong, Jiangsu, Henan, and Shandong provinces during 2008–2013. The prevalent lineages in Guangdong province described previously [10, 13] were also grouped in cluster 1, indicating the prevalence of cluster 1 in China. In clusters 1 and 2, there were also strains from Japan, Spain, France and Taiwan during 1999–2013, which were associated with HFMD, herpangina, or onychomadesis after HFMD [2, 4, 7, 16]. CV-A6 strains from Spain in 2008 and France in 2010 constituted cluster 3. Genogroups A, B, and C contained very few members, which might be due both to a lack of complete VP1 nucleotide sequence data and limited virus circulation.
We estimated pairwise distances (p-distance) among sequences shown in the phylogenetic tree (Fig. 3). Inter-genogroup distances ranged from 12.4 % to 18.4 %. Inter-cluster distances within genogroup D ranged from 8 % to 11 %, whereas pairwise distances within a cluster were ≤7 %.
To investigate the evolutionary details, an MCC tree was constructed based on complete VP1 nucleotide sequences of Tianjin strains (n = 29) and strains related closely to the Tianjin strains in the ML tree (n = 12) (Fig. 4). In the topology of the MCC tree (Fig. 4), the strains (n = 41) split into two main clusters (clusters 1 and 2), in agreement with the topologies of the ML (Fig. 3) and neighbor-joining trees (not shown). Each cluster further split into several individual lineages. Prevalent Tianjin lineages were grouped in cluster 1 (Fig. 3 and 4). In cluster 1, the lineages from Tianjin showed a close relationship to strains isolated during 2012–2013 from Fujian, Jiangsu, and Guangdong provinces, located in eastern or southern China (Fig. 4), indicating that the CV-A6 lineages causing the outbreak in this study were co-circulating widely around eastern and southern China rather than being localized to Tianjin. CV-A6 strains from other provinces were interspersed on the coalescent path of Tianjin strains in the MCC tree, and infections with the most recent common ancestor of strains in other provinces and strains in Tianjin were estimated to have occurred between 2011 and 2012 (Fig. 4). The MCMC results suggested that one to two years before the outbreak in 2013, CV-A6 of cluster 1 had been transmitted actively and widely in these areas. In general, the confidence intervals for time estimates were reasonable (as shown in node bars in Figure 4), except in nodes with posterior probabilities <0.75, for which confidence intervals were not determined. Estimates of the time to the most recent common ancestor of members of cluster 1 suggested that these viruses circulated endemically within the region for at least 6 years (95 % HPD: 3–9 years). Clusters 1 and 2 share a deep node with an estimated time to the most recent common ancestor of 19 years (95 % HPD: 13–26 years). The estimated mean evolutionary rate from the MCC tree was 4.10 × 10−3 substitutions per site per year (95 % HPD: 2.46 × 10−3 – 5.73 × 10−3), which is consistent with the evolutionary rate estimated from expanded CV-A6 surveys (>300 VP1 sequences) and rates estimated previously [10].
Fig. 4.
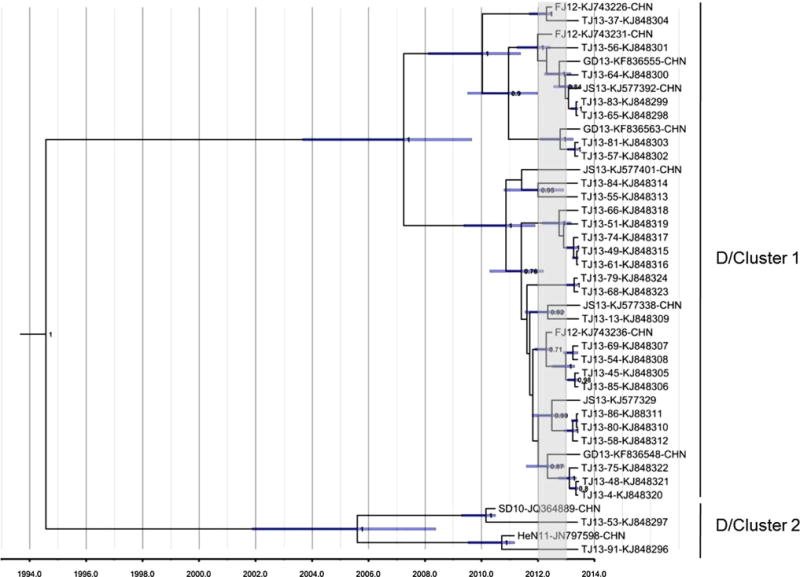
MCC tree of complete VP1 nucleotide sequences (915 nucleotides) of CV-A6 strains representing D1 and D2 clusters. The scale is in units of evolutionary time in years, using the relaxed clock model [5], with an estimated mean rate of 4.49 × 10−3 substitutions/site/year. The time period from year 2012 to 2013 is indicated by a gray rectangle
Discussion
HFMD is a common infectious childhood disease that is associated with EV infections. Numerous outbreaks have been reported in the Western Pacific Region [1, 11, 12, 22]. In Mainland China, HFMD was classified as a class “C” notifiable disease in 2008. Enhanced surveillance demonstrated persistent HFMD outbreaks throughout China during the last 6 years, with nine million cases reported. Based on nationwide data, EV-A71 and CV-A16 were the predominant pathogens during 2009–2012 [29]. However, a remarkable increase in the proportion of CV-A6 was detected in Shenzhen city since September, 2012 [10], followed by emergence of CV-A6 outbreaks reported in both southern and northeastern China in 2013 [9, 13]. In this study, surveillance for HFMD in Tianjin revealed an emerging outbreak of CV-A6 in 2013 in northern China. Although CV-A6 has been associated with several outbreaks of HFMD in Finland, Spain, Japan, Thailand, the United States, and Taiwan since 2008 [7, 8, 13, 15, 16, 21, 27], it was detected only sporadically in Mainland China before 2013 [14, 25, 30].
In previous studies on CV-A6, molecular epidemiological analyses were all based on partial VP1 nucleotide sequences [10, 16, 27]. In this study, we analyzed complete VP1 nucleotide sequences of CV-A6 strains from Tianjin as well as those obtained in other surveys in Asia and Europe. The dataset used for analysis was limited due to low accessibility of complete VP1 sequences and incomplete surveillance of CV-A6. Nevertheless, this study revealed that the genetic group encompassing Tianjin sequences was the most prevalent worldwide, was associated with several global outbreaks since 2008 [2, 4, 7, 16], and included most of the sequences from Mainland China. CV-A6 was associated with onychomadesis, herpangina, typical and atypical HFMD during previous outbreaks [7, 8, 15, 16, 27]. Surveillance programs for HFMD and acute flaccid paralysis (AFP; to detect poliovirus) are the only nationwide surveillance programs for EV in China. It is uncertain whether CV-A6 also causes other diseases in China.
In our study, multiple lineages of CV-A6 from two distinct genetic clusters (clusters 1 and 2) were found to be co-circulating independently in Tianjin (Fig. 3 and 4). Cluster 1 encompassed not only the major lineages from the outbreak in Tianjin but also major lineages from an outbreak in Guangdong [10], indicating the prevalence of cluster 1 in China. Because serotyping for other EV was not required for the surveillance system in China, national data on CV-A6 were not available. However, the analyses revealed the widespread circulation of CV-A6 in several regions of China. Moreover, a remarkable increase in the prevalence of other EVs (≥50 %) was identified in 15 provinces in 2013 according to unpublished surveillance data on HFMD. The activity of other EVs in China requires further attention.
HFMD primarily affects young children, for whom the age-specific incidence can be quite high [29]. In Tianjin, the age group with the highest incidence of HFMD varied from year to year, but this disease was commonly found in children aged between 1 and 3 years (Fig. 2). However, in 2013, when CV-A6 emerged, the highest incidence was seen in 1-year-old children, as was also observed in another outbreak of CV-A6 [15]. It is possible that CV-A6 circulated for several years in Tianjin before 2013 and that the older age group might have been immunized due to natural infection with CV-A6 during previous years. Additional studies of CV-A6 seroprevalence might be necessary for further clarification. In 2013, the rate of severe cases was much lower (295 per 100,000) than that during the previous three years (987 to 2200 per 100,000), when the proportion of EV-A71 was much higher (Table 1), suggesting that the virulence of CV-A6 is lower than that of EV-A71. The reasons for this are unknown.
Supplementary Material
Acknowledgments
We appreciate the pediatricians reporting cases to the surveillance system for HFMD in the Tianjin metropolitan area. We thank all the staff members who were responsible for specimen collection and shipment at the CDCs of all districts of Tianjin for their excellent assistance. We also thank Dr. Steve Oberste (CDC, Atlanta) for the revision of this manuscript. This work was supported by grants from the Key Technologies Research and Development Program from the Ministry of Science and Technology (grant numbers: 2013ZX10004-202, 2012ZX10004201-003) and Tianjin Municipal Center for Disease Control and Prevention (Grant number: CDCKY1303).
Footnotes
Conflict of interest None reported.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of CDC and other contributing agencies.
Electronic supplementary material The online version of this article (doi:10.1007/s00705-015-2340-3) contains supplementary material, which is available to authorized users.
Contributor Information
Xiaojuan Tan, Key Laboratory of Medical Virology of Ministry of Health, Chinese Center for Disease Control and Prevention, Institute for Viral Disease Control and Prevention, 155 Changbai Road, Beijing 102206, China.
Li Li, Tianjin municipal Center for Disease Control and Prevention, 6 Huayue Road, Tianjin 300171, China.
Baomin Zhang, Key Laboratory of Medical Virology of Ministry of Health, Chinese Center for Disease Control and Prevention, Institute for Viral Disease Control and Prevention, 155 Changbai Road, Beijing 102206, China.
Jaume Jorba, Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, 1600 Clifton Road NE, Atlanta, GA 30333, USA.
Xu Su, Email: suxu@cdctj.gov.cn, Tianjin municipal Center for Disease Control and Prevention, 6 Huayue Road, Tianjin 300171, China.
Tianjiao Ji, Key Laboratory of Medical Virology of Ministry of Health, Chinese Center for Disease Control and Prevention, Institute for Viral Disease Control and Prevention, 155 Changbai Road, Beijing 102206, China.
Dongjing Yang, Tianjin municipal Center for Disease Control and Prevention, 6 Huayue Road, Tianjin 300171, China.
Likun Lv, Tianjin municipal Center for Disease Control and Prevention, 6 Huayue Road, Tianjin 300171, China.
Jiameng Li, Tianjin municipal Center for Disease Control and Prevention, 6 Huayue Road, Tianjin 300171, China.
Wenbo Xu, Email: wenbo_xu1@aliyun.com, Key Laboratory of Medical Virology of Ministry of Health, Chinese Center for Disease Control and Prevention, Institute for Viral Disease Control and Prevention, 155 Changbai Road, Beijing 102206, China.
References
- 1.AbuBakar S, Chee HY, Al-Kobaisi MF, Xiaoshan J, Chua KB, Lam SK. Identification of enterovirus 71 isolates from an outbreak of hand, foot and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res. 1999;61:1–9. doi: 10.1016/s0168-1702(99)00019-2. [DOI] [PubMed] [Google Scholar]
- 2.Bracho MA, Gonzalez-Candelas F, Valero A, Cordoba J, Salazar A. Enterovirus co-infections and onychomadesis after hand, foot, and mouth disease, Spain, 2008. Emerg Infect Dis. 2011;17:2223–2231. doi: 10.3201/eid1712.110395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan LG, Parashar UD, Lye MS, Ong FG, Zaki SR, Alexander JP, Ho KK, Han LL, Pallansch MA, Suleiman AB, Jegathesan M, Anderson LJ. Deaths of children during an outbreak of hand, foot, and mouth disease in Sarawak, Malaysia: clinical and pathological characteristics of the disease. For the Outbreak Study Group. Clin Infect Dis. 2000;31:678–683. doi: 10.1086/314032. [DOI] [PubMed] [Google Scholar]
- 4.Chen YJ, Chang SC, Tsao KC, Shih SR, Yang SL, Lin TY, Huang YC. Comparative genomic analysis of coxsack-ievirus A6 strains of different clinical disease entities. PLoS One. 2012;7:e52432. doi: 10.1371/journal.pone.0052432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 7.Fujimoto T, Iizuka S, Enomoto M, Abe K, Yamashita K, Hanaoka N, Okabe N, Yoshida H, Yasui Y, Kobayashi M, Fujii Y, Tanaka H, Yamamoto M, Shimizu H. Hand, foot, and mouth disease caused by coxsackievirus A6, Japan, 2011. Emerg Infect Dis. 2012;18:337–339. doi: 10.3201/eid1802.111147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopalkrishna V, Patil PR, Patil GP, Chitambar SD. Circulation of multiple enterovirus serotypes causing hand, foot and mouth disease in India. J Med Microbiol. 2012;61:420–425. doi: 10.1099/jmm.0.036400-0. [DOI] [PubMed] [Google Scholar]
- 9.Han JF, Xu S, Zhang Y, Zhu SY, Wu DL, Yang XD, Liu H, Sun BX, Wu XY, Qin CF. Hand, foot, and mouth disease outbreak caused by coxsackievirus A6, China, 2013. J Infect. 2014;69:303–305. doi: 10.1016/j.jinf.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 10.He YQ, Chen L, Xu WB, Yang H, Wang HZ, Zong WP, Xian HX, Chen HL, Yao XJ, Hu ZL, Luo M, Zhang HL, Ma HW, Cheng JQ, Feng QJ, Zhao DJ. Emergence, circulation, and spatiotemporal phylogenetic analysis of coxsackievirus a6- and coxsackievirus a10-associated hand, foot, and mouth disease infections from 2008 to 2012 in Shenzhen, China. J Clin Microbiol. 2013;51:3560–3566. doi: 10.1128/JCM.01231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho M, Chen ER, Hsu KH, Twu SJ, Chen KT, Tsai SF, Wang JR, Shih SR. An epidemic of enterovirus 71 infection in Tai-wan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med. 1999;341:929–935. doi: 10.1056/NEJM199909233411301. [DOI] [PubMed] [Google Scholar]
- 12.Khanh TH, Sabanathan S, Thanh TT, le Thoa PK, Thuong TC, Hang V, Farrar J, Hien TT, Chau N, van Doorn HR. En-terovirus 71-associated hand, foot, and mouth disease, Southern Vietnam, 2011. Emerg Infect Dis. 2012;18:2002–2005. doi: 10.3201/eid1812.120929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Zeng H, Zheng H, Yi L, Guo X, Liu L, Sun L, Tan X, Li H, Ke C, Lin J. Hand, foot and mouth disease in Guangdong, China, in 2013: new trends in the continuing epidemic. Clin Microbiol Infect. 2014;20:O442–O445. doi: 10.1111/1469-0691.12468. [DOI] [PubMed] [Google Scholar]
- 14.Lu QB, Zhang XA, Wo Y, Xu HM, Li XJ, Wang XJ, Ding SJ, Chen XD, He C, Liu LJ, Li H, Yang H, Li TY, Liu W, Cao WC. Circulation of Coxsackievirus A10 and A6 in hand-foot-mouth disease in China, 2009–2011. PLoS One. 2012;7:e52073. doi: 10.1371/journal.pone.0052073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathes EF, Oza V, Frieden IJ, Cordoro KM, Yagi S, Howard R, Kristal L, Ginocchio CC, Schaffer J, Maguiness S, Bayliss S, Lara-Corrales I, Garcia-Romero MT, Kelly D, Salas M, Oberste MS, Nix WA, Glaser C, Antaya R. “Eczema coxsackium” and unusual cutaneous findings in an enterovirus outbreak. Pediatrics. 2013;132:e149–e157. doi: 10.1542/peds.2012-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirand A, Henquell C, Archimbaud C, Ughetto S, Antona D, Bailly JL, Peigue-Lafeuille H. Outbreak of hand, foot and mouth disease/herpangina associated with coxsackievirus A6 and A10 infections in 2010, France: a large citywide, prospective observational study. Clin Microbiol Infect. 2012;18:E110–E118. doi: 10.1111/j.1469-0691.2012.03789.x. [DOI] [PubMed] [Google Scholar]
- 17.Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberste MS, Maher K, Williams AJ, Dybdahl-Sissoko N, Brown BA, Gookin MS, Penaranda S, Mishrik N, Uddin M, Pallansch MA. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J Gen Virol. 2006;87:119–128. doi: 10.1099/vir.0.81179-0. [DOI] [PubMed] [Google Scholar]
- 20.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 21.Puenpa J, Chieochansin T, Linsuwanon P, Korkong S, Thongkomplew S, Vichaiwattana P, Theamboonlers A, Poovorawan Y. Hand, foot, and mouth disease caused by cox-sackievirus A6, Thailand, 2012. Emerg Infect Dis. 2013;19:641–643. doi: 10.3201/eid1904.121666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seiff A. Cambodia unravels cause of mystery illness. Lancet. 2012;380:206. doi: 10.1016/s0140-6736(12)61200-8. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan X, Huang X, Zhu S, Chen H, Yu Q, Wang H, Huo X, Zhou J, Wu Y, Yan D, Zhang Y, Wang D, Cui A, An H, Xu W. The persistent circulation of enterovirus 71 in People’s Republic of China: causing emerging nationwide epidemics since 2008. PLoS One. 2011;6:e25662. doi: 10.1371/journal.pone.0025662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian H, Zhang Y, Sun Q, Zhu S, Li X, Pan Z, Xu W, Xu B. Prevalence of multiple enteroviruses associated with hand, foot, and mouth disease in Shijiazhuang City, Hebei province, China: outbreaks of coxsackieviruses a10 and b3. PLoS One. 2014;9:e84233. doi: 10.1371/journal.pone.0084233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tseng FC, Huang HC, Chi CY, Lin TL, Liu CC, Jian JW, Hsu LC, Wu HS, Yang JY, Chang YW, Wang HC, Hsu YW, Su IJ, Wang JR. Epidemiological survey of enterovirus infections occurring in Taiwan between 2000 and 2005: analysis of sentinel physician surveillance data. J Med Virol. 2007;79:1850–1860. doi: 10.1002/jmv.21006. [DOI] [PubMed] [Google Scholar]
- 27.Wei SH, Huang YP, Liu MC, Tsou TP, Lin HC, Lin TL, Tsai CY, Chao YN, Chang LY, Hsu CM. An outbreak of coxsackievirus A6 hand, foot, and mouth disease associated with ony-chomadesis in Taiwan, 2010. BMC Infect Dis. 2011;11:346. doi: 10.1186/1471-2334-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Yeo A, Phoon MC, Tan EL, Poh CL, Quak SH, Chow VT. The largest outbreak of hand; foot and mouth disease in Singapore in 2008: the role of enterovirus 71 and coxsackievirus A strains. Int J Infect Dis. 2010;14:e1076–e1081. doi: 10.1016/j.ijid.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Xing W, Liao Q, Viboud C, Zhang J, Sun J, Wu JT, Chang Z, Liu F, Fang VJ, Zheng Y, Cowling BJ, Varma JK, Farrar JJ, Leung GM, Yu H. Hand, foot, and mouth disease in China, 2008–12: an epidemiological study. Lancet Infect Dis. 2014;14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F, Zhang T, Hu Y, Wang X, Du J, Li Y, Sun S, Sun X, Li Z, Jin Q. Survey of enterovirus infections from hand, foot and mouth disease outbreak in China, 2009. Virol J. 2011;8:508. doi: 10.1186/1743-422X-8-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z. Estimating the pattern of nucleotide substitution. J Mol Evol. 1994;39:105–111. doi: 10.1007/BF00178256. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Tan XJ, Wang HY, Yan DM, Zhu SL, Wang DY, Ji F, Wang XJ, Gao YJ, Chen L, An HQ, Li DX, Wang SW, Xu AQ, Wang ZJ, Xu WB. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J Clin Virol. 2009;44:262–267. doi: 10.1016/j.jcv.2009.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


