Abstract
K-Ras is a monomeric GTPase that controls cellular and tissue homeostasis. Prior studies demonstrated that mutationally activated K-Ras (K-RasG12D) signals through MEK to promote expansion and hyperproliferation of the highly mitotically active transit-amplifying cells (TACs) in the intestinal crypt. Its effect on normally quiescent stem cells was unknown, however. Here, we have used an H2B-Egfp transgenic system to demonstrate that K-RasG12D accelerates the proliferative kinetics of quiescent intestinal stem cells. As in the TAC compartment, the effect of mutant K-Ras on the quiescent stem cell is dependent upon activation of MEK. Mutant K-Ras is also able to increase self-renewal potential of intestinal stem cells following damage. These results demonstrate that mutant K-Ras can influence intestinal homeostasis on multiple levels.
Keywords: K-Ras, MEK, quiescent stem cells, label retention, proliferation
1.1 Introduction
Homeostasis in the intestinal epithelium requires the concerted action of multiple stem and progenitor populations. Some of these populations are highly proliferative, while others are typically quiescent [1]. Highly-proliferative, or fast-cycling, stem cells are present throughout the intestine and are responsible for the daily maintenance of the epithelium, and therefore represent the “work-horse” for intestinal homeostasis [2]. Slowly-cycling, or quiescent, stem cells are less prevalent and have been shown to be able to replace fast-cycling stem cells after injury, and therefore are considered clonogenic reserve cells that can maintain intestinal homeostasis [3, 4]. While the precise relationships between the different types of stem and progenitor cells remain to be determined, it is clear that altering their growth properties can lead to pathology, for example cancer [5]. We demonstrated previously that activation of the K-Ras oncoprotein promotes expansion and hyper-proliferation within the intestinal transit amplifying cells (TACs), a population that is normally highly proliferative [6]. Similarly, oncogenic K-Ras has been shown to accelerate the rate of cell division in Lgr5+ stem cells in the small intestine [7]. Here, we have studied if/how mutant K-Ras can affect the proliferative and self-renewal properties of intestinal stem cells that are normally quiescent.
1.2 Material and Methods
Animals and Tissues
All mice were cared for according to the guidelines of the Hospital Subcommittee on Animal Research (SRAC). Fabpl-Cre ; K-RasLSL-G12D/+ animals have been described previously (3). In these animals, Cre is expressed only in the distal small intestinal and colonic epithelia, so mutant K-Ras is expressed only in those regions. Rosa26-M2-rtTA; ColA1-H2B-Egfp transgenic mice have been described previously (6). In this system, the M2 reverse tetracycline transactivator (M2-rtTA) is knocked into the Rosa26 locus, which is expressed in all cells in the adult mouse. Expression of the H2B-Egfp chimeric cDNA from the collagen 1a1 locus is regulated by a doxycycline (DOX) response element. In the absence of DOX, the H2B-Egfp fusion protein is not expressed. When animals are administered DOX in the drinking water, M2-rtTA binds to the response element and induces expression of H2B-Egfp. Both control (Fabpl-Cre ; Rosa26-M2-rtTA ; ColA1-H2B-Egfp) and K-Ras mutant (Fabpl-Cre ; K-RasLSL-G12D ; Rosa26-M2-rtTA ; ColA1-H2B-Egfp) animals (4-6 week old mice at the beginning of treatment) were given a pulse of DOX (Sigma-Aldrich, 2mg/ml in water) in the drinking water for 2 weeks. DOX was then removed from the drinking water and animals were sacrificed at defined time points: 1, 3, 5,7, 9,10, 11, 13, 15, 20, and 25 days. At least 3 animals were analyzed at each time point following DOX removal.
Control and K-Ras mutant animals that were treated with PD0325901were also given a 2 week pulse of DOX in the drinking water. Following removal of DOX, animals were given a daily intraperitoneal injection of PD0325901 (ChemieTek, 12.5 mg/kg in 10% DMSO) or 10% Dimethyl Sulfoxide (DMSO) and then sacrificed at defined time points: 5, 7, and 13 days. Five animals were analyzed at each time point.
For experiments in which dextran sodium sulfate (DSS) was used to induce epithelial damage, percent damage was determined by dividing the summed length of all damaged regions by the total length of the distal colon. Damaged tissue was defined as any region that was not healthy. Healthy tissue was taken to be any contiguous region that had at least 5 undamaged crypts. For all animals, distal colon was taken to be 5 cm of colon from the distal end.
For tissue collection, animals were sacrificed and the entire intestinal tract was removed and washed with cold PBS. For immunohistochemical analysis, tissues were fixed in formalin overnight. For western blot analysis tissue was lysed in RIPA buffer and stored at −80°C. The proximal small intestine (duodenum) was obtained from tissue immediately adjacent to the stomach. The distal small intestine (ileum) was obtained from tissue immediately adjacent to the cecum. The colon was obtained from tissue immediately adjacent to the rectum.
Flow cytometry, RNA preparation, and qRT-PCR
Rosa26-M2-rtTA ; ColA1-H2B-Egfp transgenic mice were given a pulse of doxycycline in the drinking water for 2 weeks. DOX was then removed from the drinking water and animals were sacrificed at 25 days post induction. Lgr5-Egfp-IRES-CreERT2 mice were 8-10 weeks of age. Following sacrifice, colons were removed, opened longitudinally and cut into 3-5mm segments. Tissue was placed in cold PBS (Ca and Mg-free) and vortexed 6 times for 5 seconds to remove debris and fecal matter before incubation in PBS with 20mM EDTA at 37°C for 30min. The tissue was then transferred to cold PBS and vortexed vigorously to release the crypts. Crypt-containing supernatants were supplemented with 10% FBS, pelleted and resuspended in DMEM for one additional wash. Following spin, crypt pellet was resuspended in 10ml TrypLE Express (Invitrogen #12605-010) with DNase (1000u/10ml, Sigma #D5025), and incubated at 37°C for 45 min with intermittent mixing to disassociate the crypts into single cells. Cells were filtered through a 40 μm cell strainer (BD Bioscience #352340) into DMEM (Corning #10-013-CV) media containing 5mM EDTA. Cells were washed twice with DMEM and resuspended in DMEM with 4mM MgCl2, 200u/ml DNase, and Propidium Iodide. Because H2B-Egfp animals also show expression of Egfp in immune cells, cells from those animals were stained with CD45-APC-Cy7 (Biolegend #103116) for 10 minutes at room temperature before resuspension in sorting media. Using the BD Aria flow cytometer, cells were gated on PI negative, CD45 negative populations and then the high Egfp expressing populations (~2% total epithelial cells) and Egfp negative populations were sorted directly into trizol. RNA was isolated using TRIzol reagent according to manufacturer’s instructions. Subsequently, cDNA was made using High Capacity cDNA Reverse Transcriptase kit (Applied Biosystems) and quantitative real time PCR was performed using TaqMan PCR assays (Applied Biosystems) to detect the expression of Egfp (Assay ID: Mr04097229-mr), Leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5, Mm00438890_m1), Olfactomedin 4 (Olfm4, Mm01320260_m1), Achaete-scute complex homolog 2 (Ascl2, Mm01268891_g1), Axin 2 (Mm00443610_m1), Mucin 2 (Muc2, Mm01276696_m1), Alkaline phosphatase (Alph, Mm01285814_g1), and Chromogranin A (Chga, Mm00514341_m1). All samples were run in duplicate and normalization was carried out using the 2ΔΔCT method relative to 18S rRNA (Mm03928990).
Immunohistochemistry
Tissues were processed for histology via standard protocols and tissue sections were cut to 5 μm. Tissue sections were stained with mouse anti-GFP antibody (Clontech, 632380), rabbit anti-phospho-Histone H3 antibody (Ser10, #9701), and the Vectastain ABC Kit (Vector Laboratories) according to manufacturer’s instructions. Reactions were visualized with 3,3′-diaminobenzidine (DAB) and counterstained with hematoxylin.
Immunoblotting
Polyacrylamide gels (10%) were loaded with 40μg of mouse epithelial intestinal protein. Immunoblotting was performed using rabbit anti-Erk1/2 (Cell Signaling Technology #4696), mouse anti-phospho-Erk1/2 (Cell Signaling Technology #9101), and mouse anti-α-Tubulin (Sigma-Aldrich #T6074). Membranes were incubated with primary antibodies overnight at 4°C and incubated for 1 hour at room temperature with secondary antibodies (Rockland) diluted at 1:10,000. Membranes were scanned using a LI-COR Odyssey infrared imaging system.
Statistical Analysis
The ratio qRT-PCR values depicted in Fig. 1C were computed for the ith animal as θi/ αi, where θi was the Egfp+ qRT-PCR signal and αi was the Egfp- qRT-PCR signal. Ratios were averaged over i=[1,2,3] (N=3 animals), then log2 transformed. Error bars represent mean ± SEM of the log2 transformed values. A signal was considered to be enriched if its log2 transformed values were significantly greater than 0 (corresponding to ratio values greater than 1). Statistical significance was computed on the log2 transformed values for each quantified gene using a one-sample t-test (GraphPad Prism 6). Egfp half-life in Fig. 2 was calculated using a sigmoidal dose-response curve with variable slope (Prism). Statistical significance between half-life for each experimental group (control/K-Ras) was assessed from the mean ± SE values for the logEC50 (half-life). The logEC50s were compared using the F-test (GraphPad Prism 6) to assess significance. Statistical significance for the DSS-induced damage datat presented in Fig. 3D was assessed from the mean ± SE values for each group (Graphpad Prism 6). A two-tailed t-test was used to determine the P-value.
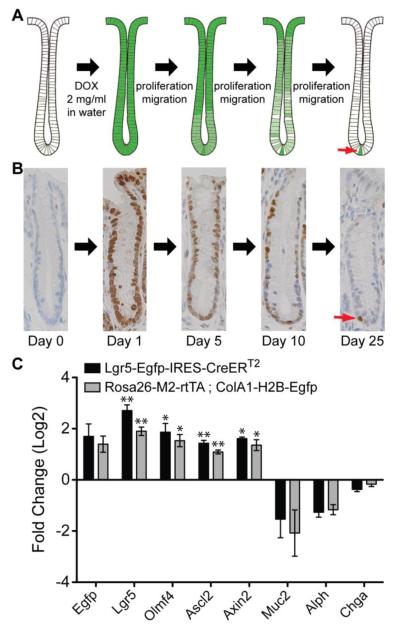
Figure 1.
H2B-Egfp label retention in the intestinal epithelium. (A) Schematic representation of the label retention time course. Rosa26-M2-rtTA; ColA1-H2B-Egfp transgenic mice have been described previously [10]. In this system, the M2 reverse tetracycline transactivator (M2-rtTA) is knocked into the Rosa26 locus, which is expressed in all cells in the adult mouse. Expression of the H2B-Egfp chimeric protein from the collagen 1a1 locus is regulated by a doxycycline response element. In the absence of DOX, H2B-Egfp is not expressed. When animals are administered DOX in the drinking water, M2-rtTA binds to the response element and induces expression of H2B-Egfp. (B) Immunohistochemistry for Egfp over the time course of the labeling experiment. Over time, only a single Egfp-positive cell remains at the bottom of the crypt, which is the slowly cycling stem cell (red arrow). Egfp immunohistochemistry is shown for crypts in the distal colon. (C) qRT-PCR data comparing expression patterns of stem cell markers in putative Egfp+ stem cells. In Egfp+ colonic epithelial cells from both Rosa26-M2-rtTA-ColA1-H2B-Egfp and Lgr5-Egfp-IRES-CreERT2 mice, there is a 3-4 fold enrichment of stem cell markers and a negative enrichment of differentiation markers. The qRT-PCR ratio values were computed as the normalized qRT-PCR signal from Egfp+ cells divided by the normalized qRT-PCR signal Egfp- cells from the same animal. Ratios were averaged (N=3 animals), then log2 transformed. Error bars represent +/− SEM values for each group. Statistical significance of each gene was computed using a one sample t-test of the log2 transformed ratio values relative to 0 (see Materials and Methods section). * represents P = 0.01, ** represents P < 0.01.
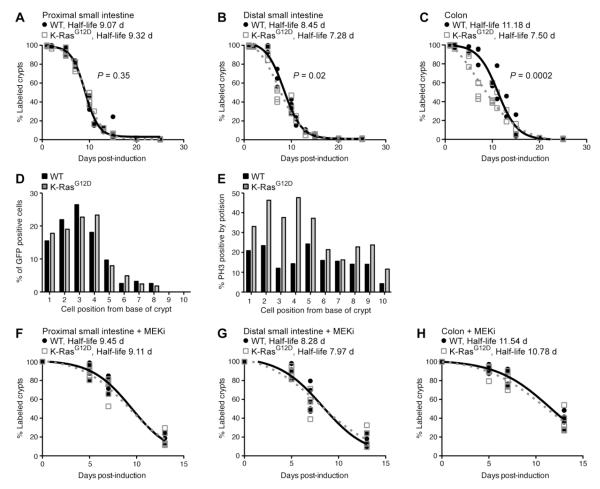
Figure 2.
K-RasG12D accelerates the loss of Egfp label in the intestinal epithelium. (A-C) Determination of Egfp half-life in animals expressing WT or mutant K-Ras. Animals were exposed to DOX (2 mg/ml) in the drinking water for 2 weeks and then sacrificed at defined time points. Immunohistochemistry for Egfp was used to identify crypts that included at least a single Egfp+ cell. Using these data, the Egfp half-life for control (black line) and K-Ras mutant animals (gray dotted line) was determined for the proximal small intestine (A), distal small intestine (B) and colon (C). A minimum of 3 animals were used for each experimental group at each time point, with approximately 50 crypts analyzed per animal. The Egfp half-life was significantly reduced by K-RasG12D in the distal small intestine (P = 0.0212) and colon (P = 0.0002), but not in the proximal small intestine (P=0.3510). (D) Position of Egfp label-retaining cells in WT and mutant animals. At time points near the half-life for each condition, immunohistochemistry for Egfp was used to determine the relative positions of Egfp+ cells within crypts that contain a single labeled cell. A minimum of 200 crypts was counted for each experimental group. Both experimental groups illustrated a similar distribution of Egfp positive cells within the crypt. (E) Distribution of mitotic cells in WT and mutant animals. The relative positions of phospho-Histone H3 (PH3) positive cells within crypts was determined by immunohistochemistry for PH3. A minimum of 200 cells at each position were counted for each experimental group. K-Ras mutant animals (gray line) displayed a higher percentage of PH3-positive mitotic cells than control animals (black line), particularly at cell position 1-5, were stem cells reside. (F-H) Determination of Egfp half-life after MEK inhibition in animals expressing WT or mutant K-Ras. Animals were treated daily with PD0325901 (MEKi) for either 5, 7, or 13 days following DOX pulse. The half-life for both control (black line) and K-Ras mutant animals (gray dotted line) was determined for the proximal small intestine (F), distal small intestine (G) and colon (H). 5 animals per time point were used for each experimental group. In the presence of MEKi, K-RasG12D does not affect label retention.
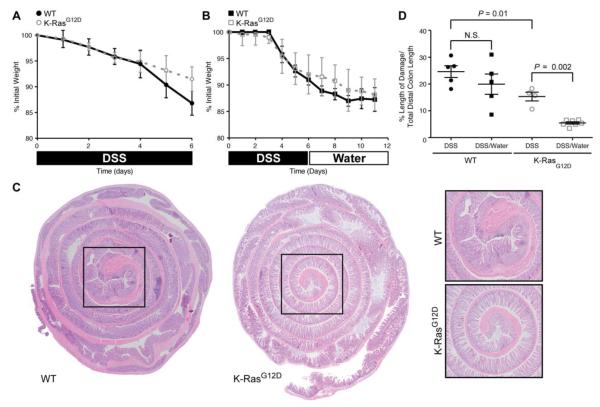
Figure 3.
K-RasG12D animals recover more efficiently from DSS-induced damage than control animals. (A) Mouse body weight changes following treatment with 3% DSS (6 days). Both experimental groups (WT, K-RasG12D) lose weight over the course of treatment, with animals expressing K-RasG12D losing slightly less weight. (B) Mouse body weight changes following treatment with 3% DSS (6 days) followed by water (6 days). Both experimental groups (WT/K-Ras) lose weight over the course of treatment. (C) Hematoxylin and eosin staining of the colonic epithelium from control (WT) and K-Ras mutant animals following treatment with 3% DSS (6 days) followed by water (6 days). The epithelial damage is restricted to the distal colon (black box), which is magnified in the panels on the right. (D) Quantification of histological damage following DSS treatment (DSS). The damage was significantly reduced in K-RasG12D mutant animals both immediately after DSS exposure (P = 0.01) and following recovery from injury (P = 0.002).
1.3 RESULTS
Quiescent cells can be identified in vivo by virtue of their ability to retain label. Historically, this was achieved by treating animals with nucleotide analogues, for example 5-bromo-2′-deoxyuridine (BrdU) or tritiated thymidine, that are incorporated into DNA during replication [8]. More recently, transgenic systems have been developed in which enhanced green fluorescent protein (Egfp)-labeled histones are inducibly expressed and then incorporated into chromatin [9]. We utilized a mouse strain that allows for ubiquitous, doxycycline (DOX)-inducible expression of an H2B-Egfp fusion protein [10] (Figure 1A). Immediately following a pulse of DOX, the entire intestinal epithelium is Egfp-positive. As the cells at the base of the crypt divide and migrate up toward the lumen, Egfp signal is progressively diluted. Within a short period of time, around 25 days, a single Egfp-positive cell typically remains near the bottom of the crypt (Figure 1B).
To validate that the Egfp-positive quiescent cells are really stem cells, we used qRT-PCR to measure the expression of both stem cell (Lgr5, Olmf4, Ascl2, and Axin2) and differentiation markers (Muc2, Alph, and Chga) in epithelial cells isolated from Rosa26-M2-rtTA ; ColA1-H2B-Egfp animals 25 days post DOX induction. Egfp+ epithelial cells from these animals were enriched for stem cell markers relative to Egfp− epithelial cells, while the Egfp− cells were enriched for markers of differentiation (Figure 1C). As a control, we performed the same experiment on Lgr5-Egfp-IRES-CreERT2 mice, where the Egfp+ cells represent bona fide intestinal epithelial stem cells. In these animals, as in the Rosa26-M2-rtTA ; ColA1-H2B-Egfp animals, Egfp+ epithelial cells were enriched for stem cell markers (Figure 1C). These data indicate that the isolated label retaining cells present at the bases of the crypts of Rosa26-M2-rtTA ; ColA1-H2B-Egfp animals 25 days after DOX stimulation are indeed intestinal stem cells.
In order to determine whether mutant K-Ras (K-RasG12D) affects label retention, we crossed mice carrying the inducible histone H2B-Egfp transgene to animals expressing KRasG12D in the distal small intestinal and colonic epithelia [6]. Both control and K-Ras mutant animals were exposed to DOX in the drinking water for 14 days to ensure that all cells of the colonic epithelium were labeled. After this initial pulse, DOX was removed from the drinking water and intestinal epithelia were analyzed at different time points in order to detect cells that retained the H2B-Egfp label. Egfp-positive epithelial cells were quantified and tabulated as the number of positive cells per crypt. These experiments were used to determine the “half-life” of the H2B-Egfp label in both control and K-RasG12D epithelium, which we defined as the time-point at which 50% of the colonic crypts retain at least a single labeled cell. As expected, control and mutant animals illustrated little difference in Egfp half-life at the proximal small intestine, since K-RasG12D is expressed only in the distal small intestine and colon (Figure 2A). By contrast, the retention of label was significantly different between control and mutant animals in the distal small intestinal (a half-life difference of 1.2 days) and colonic epithelia (a half-life difference of 3.7 days), with K-RasG12D tissues exhibiting a significantly reduced Egfp half-life (Figure 2B,C). While mutational activation of K-Ras altered the kinetics of label retention in the colonic epithelium, it did not alter the distribution of Egfp+ cells, the majority of which were found in positions 1-5 at the bottom of the crypt (Figure 2D). Consistent with the idea that mutant K-Ras alters the kinetics of label retention through its effect on proliferation, animals expressing K-RasG12D exhibited an increased frequency of mitotic stem cells relative to controls (Figure 2E). Taken together, these data indicate that K-RasG12D promotes hyperproliferation of normally quiescent intestinal stem cells.
In our prior studies, we showed that K-RasG12D promotes proliferation in the intestinal epithelium by activating MEK, but that K-RasG12D promotes proliferation of colorectal cancer cells through a MEK-independent mechanism [6]. To determine whether K-RasG12D promotes quiescent stem cell proliferation through a MEK-dependent or -independent mechanism, we treated control and mutant animals with PD0325901, an orally active MEK inhibitor (MEKi), [11] and then repeated label retention studies. Our pilot experiments indicated that PD0325901 was highly effective in suppressing MEK activity in vivo in the colonic epithelium (Supplemental Figure 1). Similar to what we observed in our initial experiment, control and mutant animals treated with MEKi did not differ in label retention in the proximal small intestine (Figure 2F). By contrast, while MEK inhibition did not alter label retention in the small intestine and colon of control animals, MEKi reverted the K-RasG12D label retention phenotype to that of control animals (Figure 2G,H). These data strongly suggest that mutant K-Ras promotes hyperproliferation of quiescent intestinal stem cells via a MEK-dependent pathway.
Quiescent intestinal stem cells are also known to play a central role in mucosal regeneration upon tissue injury [12]. Following injury induced by cytotoxic or genotoxic agents, the quiescent stem cells become mitotically active, leading to restoration of normal epithelial architecture and barrier function [13]. In order to determine whether mutant K-Ras can affect quiescent stem cells following injury, animals expressing WT or mutant K-Ras were exposed to 3% dextran sodium sulfate (DSS) in their drinking water for 6 days and then sacrificed, or else allowed to recover for 6 days and then sacrificed. Exposure to DSS induces mucosal injury and subsequent inflammation in the distal colon of rodents [14]. Significant weight loss was evident in both experimental groups following acute DSS treatment, although animals expressing K-RasG12D tended to lose less weight (Figure 3A,B). To assess whether the weight loss phenotypes correlated with epithelial damage, we performed a histologic analysis of the colonic epithelium following DSS exposure. Both groups demonstrated alterations in the epithelial structure, neutrophil and lymphocyte infiltration into the mucosal and submucosal areas, and crypt loss (Figure 3C), with control animals displaying a 1.6 fold increase in the amount of initial damage in the distal colon relative to animals expressing K-RasG12D (Figure 3D). Following a 5-day recovery from DSS treatment, both experimental groups exhibited features of tissue repair, with K-Ras mutant animals exhibiting a more dramatic recovery in epithelial morphology than animals expressing WT K-Ras (Figure 3D). These results suggest that animals expressing mutant K-Ras are able to recover from epithelial damage more efficiently than are control animals.
1.4 Discussion
Proper intestinal homeostasis requires tight regulation of proliferation in epithelial stem and progenitor cells. Nevertheless, the mechanisms controlling cell division in distinct populations of epithelial cells are not completely deciphered. We previously demonstrated that mutant K-Ras, a monomeric GTPase that functions as binary switch to control proliferation, differentiation, apoptosis, cell adhesion, and cell migration, increases the proliferative index in TACs [6]. Activating missense mutations are common in cancer and they impair GTPase activity, locking K-Ras into its GTP-bound (active) state and allowing it to constitutively activate downstream signaling [15]. Expression of mutant K-Ras also accelerates the division rates of Lgr5+ stem cells, leading to clonal expansion of crypts expressing the oncoprotein [7], suggesting that isolated mutational activation of K-Ras could lead to field effects prior to neoplastic initiation. Here, we have found that K-RasG12D also enhances proliferation of intestinal stem cells that are normally quiescent. These quiescent cells have been shown to differentiate into Paneth cells and enteroendocrine cells under homeostatic conditions, and then to give rise to all epithelial lineages following damage [3]. Interestingly, small intestinal crypts expressing mutant K-Ras do not develop Paneth cells [16], suggesting that the quiescent state may be important for maturation into this differentiated cell type.
Paradoxically, we found that mutant K-Ras increases proliferation of quiescent intestinal stem cells (Figure 2), but also increases repair following damage (Figure 3). This phenomenon is similar to the role that oncogenic N-Ras plays in hematopoietic stem cells (HSCs) [17]. In this context, N-RasG12D promotes proliferation and self-renewal of HSCs. Going forward, it will be interesting to determine if this “bimodal” response of stem cells to Ras activation is specific to the Ras family of oncogenes, or whether it is a common feature of other oncogenic mutations.
In our prior work, we found that mutant K-Ras reprograms TACs to be entirely dependent upon MEK signaling for proliferation; inhibition of MEK did not affect proliferation of wild-type TACs, but K-Ras mutant TACs exhibited almost no proliferative capacity when exposed to MEKi [6]. Similar to our prior work, we now find that mutant K-Ras signals through MEK to promote hyperproliferation of the normally quiescent intestinal stem cell (Figure 2). In contrast to our previous studies, however, mutant K-Ras did not reprogram the quiescent cell to be entirely dependent upon MEK. Instead, inhibition of MEK in the context of mutant K-Ras simply restored label retention to that of control animals. Together, these observations suggest that mutant K-Ras differentially influences the molecular program controlling proliferation in distinct epithelial cell types in the intestine. These observations may be highly relevant to the use of MEK inhibitors for colorectal cancers expressing mutant K-Ras.
Supplementary Material
Highlights.
Mutationally activated K-Ras promotes proliferation of intestinal stem cells
Mutationally activated K-Ras decreases label retention in quiescent intestinal stem cells
MEK activity is required for K-Ras to affect the proliferative kinetics of stem cells
Mutationally activated K-Ras promotes mucosal repair after injury
Acknowledgements
The authors thank Konrad Hochedlinger for sharing the Rosa26-rtTA ; ColA1-H2B-Egfp mice.
Funding: This work was supported by a Research Scholar Grant from the American Cancer Society (MGO-114877 to K.M.H.) and by grants from the National Institutes of Health (5R01CA151566 to K.M.H., 5R01CA084056 to D.T.B., 5P30HD18655 to the Children’s Hospital Intellectual and Developmental Disabilities Research Center, and 5P30DK034854 to the Harvard Digestive Disease Center). J.J.G. is supported by a postdoctoral fellowship, PF-13-081-01-TBG, from the American Cancer Society.
Abbreviations used in this paper
- DOX
doxycycline
- Egfp
enhanced green fluorescent protein
- TACs
transit amplifying cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors disclose no conflicts.
Author contributions: K.M.H., J.J.G., and D.T.B. designed the study; J.J.G., J.L., M.S.S., and C.G. acquired data; K.M.H., J.J.G., J.L., and C.G. analyzed and interpreted data; K.M.H. and J.J.G. wrote the manuscript.
References
- 1.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15(1):19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 2.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 3.Buczacki SJ, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495(7439):65–9. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 4.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478(7368):255–9. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 6.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–8. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snippert HJ, et al. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 2014;15(1):62–9. doi: 10.1002/embr.201337799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris RJ, Potten CS. Slowly cycling (label-retaining) epidermal cells behave like clonogenic stem cells in vitro. Cell Prolif. 1994;27(5):279–89. doi: 10.1111/j.1365-2184.1994.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 9.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–63. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foudi A, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27(1):84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bain J, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potten CS, Grant HK. The relationship between ionizing radiation-induced apoptosis and stem cells in the small and large intestine. Br J Cancer. 1998;78(8):993–1003. doi: 10.1038/bjc.1998.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth S, et al. Paneth cells in intestinal homeostasis and tissue injury. PLoS One. 2012;7(6):e38965. doi: 10.1371/journal.pone.0038965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vowinkel T, et al. Impact of dextran sulfate sodium load on the severity of inflammation in experimental colitis. Dig Dis Sci. 2004;49(4):556–64. doi: 10.1023/b:ddas.0000026298.72088.f7. [DOI] [PubMed] [Google Scholar]
- 15.Ahearn IM, et al. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13(1):39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Y, et al. Mutant KRAS promotes hyperplasia and alters differentiation in the colon epithelium but does not expand the presumptive stem cell pool. Gastroenterology. 2011;141(3):1003–1013. e1–10. doi: 10.1053/j.gastro.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature. 2013;504(7478):143–7. doi: 10.1038/nature12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.