Abstract
Background and aims
Alcohol is a primary cause of liver disease and an important co-morbidity factor in other causes of liver disease. A common feature of progressive liver disease is fibrosis, which results from the net deposition of fibril-forming extracellular matrix (ECM). The hepatic stellate cell (HSC) is widely considered to be the major cellular source of fibrotic ECM. We determined if HSC are responsive to direct stimulation by alcohol.
Methods
HSC undergoing transdifferentiation were incubated with ethanol and expression of fibrogenic genes and epigenetic regulators measured. Mechanisms responsible for recorded changes were investigated using ChIP-Seq and bioinformatics analysis. Ethanol induced changes were confirmed using HSCs isolated from mouse alcohol model, ALD patient liver and precision cut liver slices.
Results
HSCs responded to ethanol exposure by increasing profibrogenic and ECM gene expression including elastin. Ethanol induced altered expression of multiple epigenetic regulators indicative of a potential to modulate chromatin structure during HSC transdifferentiation. MLL1, a histone 3 lysine 4 (H3K4) methyltransferase, was induced by ethanol and recruited to the elastin gene promoter where it was associated with enriched H3K4me3, mark of active chromatin. Chromatin immunoprecipitation sequencing (ChIPseq) revealed that ethanol has broad effects on the HSC epigenome and identified 41 gene loci at which both MML1 and its H3K4me3 mark were enriched in response to ethanol.
Conclusions
Ethanol directly influences HSC transdifferentiation by stimulating global changes in chromatin structure resulting in increased expression of ECM proteins. The ability of alcohol to remodel epigenome during HSC transdifferentiation provides mechanisms for it to act as a comorbidity factor in liver disease.
Keywords: ALD, HSC, elastin, MLL1, H3K4me3, epigenetics
Introduction
Chronic alcohol consumption is both a direct cause of liver disease as well as a major comorbidity factor in the progression of liver disease resulting from other primary causes such as viral hepatitis [1, 2]. Mechanisms explaining the hepatotoxicity of alcohol are beginning to be understood and help to explain its impact on liver disease. Alcohol and its metabolites, in particular reactive oxygen species (ROS), hydroxyethyl radical and nitric oxide are a major cause of hepatocellular damage [3]. Furthermore these metabolites can induce hepatic inflammation, which via cytokines such as TNFα are an important indirect cause of alcohol-induced hepatocyte damage and death. In addition, ROS-mediated lipid peroxidation has been proposed to cause damage to hepatocyte proteins, which can drive antigenic responses that perpetuate inflammation and liver damage [4].
Hepatocellular damage and inflammation stimulate the transdifferentiation of resident perisinusoidal HSC into α-smooth muscle actin (αSMA)-positive myofibroblasts. These so-called “activated” HSC (aHSC) are the major hepatocellular source of fibrotic ECM proteins and promote the net deposition of fibrotic ECM in chronic liver disease [5]. The molecular processes that promote the progression of liver disease to severe fibrosis remain poorly defined, however the continued production of ECM proteins by aHSC is a major contributory factor. In addition, aHSC secrete proteins that promote cross-linking, maturation and insolubility of the fibrotic ECM such as elastin. Elastin is a target for cross-linking catalysed by lysyl-oxidase or tissue transglutaminase [6] and its accumulation in the fibrotic ECM limits its potential for degradation and impacts on the degree of reversibility of the fibrotic tissue. Hence, factors that stimulate the expression of ECM maturation proteins such as elastin are likely to determine fibrosis progression in liver disease.
Recent studies from our laboratory and other investigators have revealed the importance of epigenetic signalling events in HSC transdifferentiation and fibrogenesis. Experimental manipulation of epigenetic signatures such as DNA methylation, histone acetylation/methylation and the activities of proteins that either annotate or interpret these epigenetic marks can have profound effects on the HSC phenotype [7]. The concept that alcohol can stimulate epigenetic changes in liver tissue is already well established. Ethanol impairs the normal metabolism of methionine and in turn affect the availability of methyl groups in the form of S-adenosylmethioine (SAMe) for DNA and histone methylation [8]. Kendrick and colleagues showed that ethanol or acetate reduced histone deacetylase activity and up-regulated expression of acetyl-CoA synthetases resulting in increases in histone acetylation and transcriptional activity at inflammatory genes [9]. Less is understood concerning the potential for alcohol to influence the HSC epigenome and expression of ECM proteins.
Here we investigate the impact of alcohol on the expression of epigenetic regulators during HSC transdifferentiation and report that histone-modifying enzymes such as the H3K4 methyltransferase MLL1 are up-regulated in HSC exposed to ethanol and leads to altered expression of profibrogenic genes including elastin. We provide a combinatorial chromatin immunoprecipitation sequencing (ChIPseq) analysis on the H3K4me3 signature and MLL1 in ethanol-stimulated HSC that reveals widespread changes in chromatin structure confirming a profound genome-wide effect of alcohol on HSC transdifferentiation. We suggest that our findings provide new insights into the mechanisms by which alcohol acts as a co-morbidity factor and a stimulator of fibrosis in chronic liver disease.
Materials and methods
Ethics
Authors hold appropriate licences for animal experiments which were issued/approved by local ethical committee and UK Home Office.
Human subjects
Use of human tissue was approved by Newcastle and North Tyneside Local Research Ethics (approval number H10/H0906/41). All samples were collected and used subject to patient’s written consent.
Cell isolation and culture
All rats were purchased from Charles River, UK. Rat hepatic stellate cells (rHSC) were isolated from normal livers of 350g Sprague-Dawley rats by sequential perfusion with collagenase and pronase, followed by discontinuous density centrifugation in 11.5% Optiprep (Sigma-Aldrich, Gillingham, Dorset, UK). Cells were cultured on plastic in Dulbecco’s modified Eagle’s medium, supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, and 16% fetal calf serum and maintained in an incubator at 37°C at an atmosphere of 5% CO2. HSC were treated at day 1, 2 and 10 of culture for 24h and 48h with 86mM ethanol (dose equivalent to heavy drinking in humans) [10]. In order to create atmosphere saturated with ethanol and prevent evaporation from culture media, 0.5% ethanol in water was added to the incubator 24h before the treatment. Acetate treatment- day 2 cultures of HSC were treated with 1mM sodium acetate, which is an acetate concentration found in a person that is metabolizing ethanol at the concentration of 86mM [9].
Animal model
Male Col1a1-GFP mice (12 wk old) were fed ad libitum for 2 wk with a liquid "Western diet" (WD) high in cholesterol (1% or 0.5%) and saturated fat (27%Cal or 22%Cal as lard). Then the diet was switched to an ethanol containing Western diet for 8 weeks. Ethanol intake was gradually increased from 1% (w/v) on day 1 to 4.5% (w/v) on day 12 until the end of the end of 8 wk feeding. From the second week of ethanol feeding, a weekly binge dose of alcohol was given via a stomach tube and repeated 7 times. The binge dose was gradually increased from 3.5g/kg to 4.5g/kg. For control mice, age and gender matched Col1a1-GFP mice were fed regular chow.
HSC isolation from the model
A non-parenchymal liver cell fraction was collected by pronase-collagenase perfusion and gradient ultracentrifugation. GFP fluorescence was excited at 488 nm by an argon laser and measured through a 530 nm filter and the vitamin A UV fluorescence excited at 350 nm by a krypton laser and measured through a 350 nm filter. UV-/GFP+ cells (aHSCs) were collected in tubes containing the medium with DNAse.
Crosslinked chromatin immunoprecipitation (XChIP) assay
Chromatin immunoprecipitation (ChIP) assays for MLL1 and H3K4me3 binding were carried out by using 50μg cross-linked chromatin prepared by fixing cells in 1% formaldehyde, then sonicating lysed cells for 5 minutes in Diagenode Bioruptor. Precleared chromatin was incubated with 5μg anti MLL1 or H3K4me3 antibodies, the complexes were precipitated, washed and eluted. Crosslinks were reversed and genomic DNA purified. Each PCR reaction was performed in duplicate and the analysis was repeated at least three times from independent ChIP experiments. A signal intensity value for each sample was calculated from the average of the experiments. Average values of eluates were normalized to average values of control antibody sample and expressed as fold enrichment above background (i.e. control antibody). Quantitative PCR amplification was carried out using primers listed in Table 2 and 3.
ChIP-seq and bioinformatic analysis
ChIP assays were carried out as above. 500ng of genomic DNA was then used to construct a library according to TruSeq DNA (LS) protocol by Illumina. The DNA fragments were selected for 550bp insert size. In order to increase the final concentration of the library, ten cycles of PCR reaction were performed. ChIP-seq results were validated by standard ChIP assays and quantitative PCR performed using primers in Table 3. ChIPseq data were analyzed using Model-based Analysis of ChIP-Seq (MACS) software, with bowtie2 alignments against the annotated rat genome from Ensembl (Rnor_5.0), and compared against the control for each immunoprecipitation. Peaks were called as significant with a p-value of 1e-07. The resulting peak list was visualised using IGV browser (http://www.broadinstitute.org/igv/home).
Immunohistochemistry, SDS-PAGE and Immunoblotting, Quantitative PCR (RTqPCR analysis), and Micrococcal nuclease digestion are in Supplementary Material and Methods
Results
Ethanol promotes HSC transdifferentiation and stimulates expression of histone modifying enzymes in transdifferentiating HSC
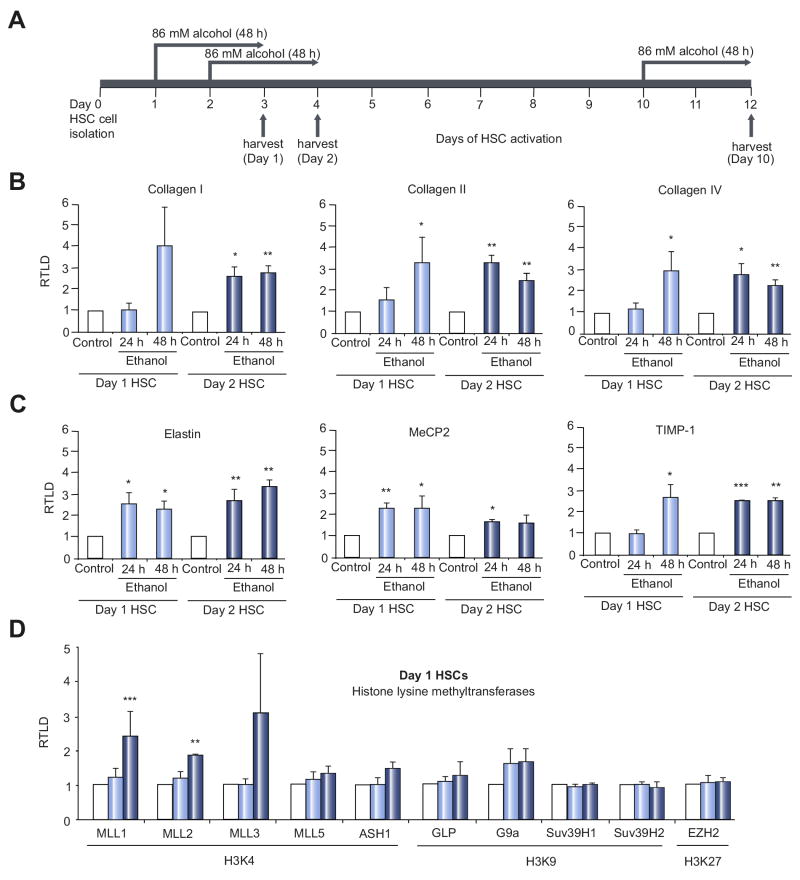
Culture-induced activation of primary rat HSC is associated with time-dependent increases in the transcriptional activity and expression of ECM genes including Type I and III collagen, tropoelastin (Elastin) and tissue inhibitor of metalloproteinase-1 (TIMP-1) (Suppl Fig 1). Freshly isolated (day 1) HSC expressed low or undetectable levels of transcript for these genes, which were subsequently induced to detectable levels by day 3 of culture and continued to rise in expression with further culture as the cells acquired their myofibroblastic phenotype. To determine the influence of alcohol on HSC transdifferentiation, we exposed early transitioning HSCs (culture days 1 and 2) to ethanol (Fig 1A). Ethanol treatment of transitioning HSC enhanced the expression of collagen genes (I, III and IV), elastin and TIMP-1 (Fig 1B/C). Ethanol also enhanced expression of the methyl-DNA binding protein MeCP2, which functions as a master epigenetic regulator of HSC transdifferentiation [11]. In contrast to these stimulatory effects ethanol had no impact on gene expression in myofibroblastic aHSC (Suppl Fig 2A). The lack of effect may be explained by observation that level of metabolising enzymes alcohol dehydrogenase (ADH1) and CYP2E1 are both markedly reduced in aHSCs, although aldehyde dehydrogenase (ALDH2) remains upregulated (Suppl Fig 2B).
Figure 1. Ethanol accelerates HSC transdifferentiation and induces expression of ECM, profibrogenic genes and epigenetic regulators.
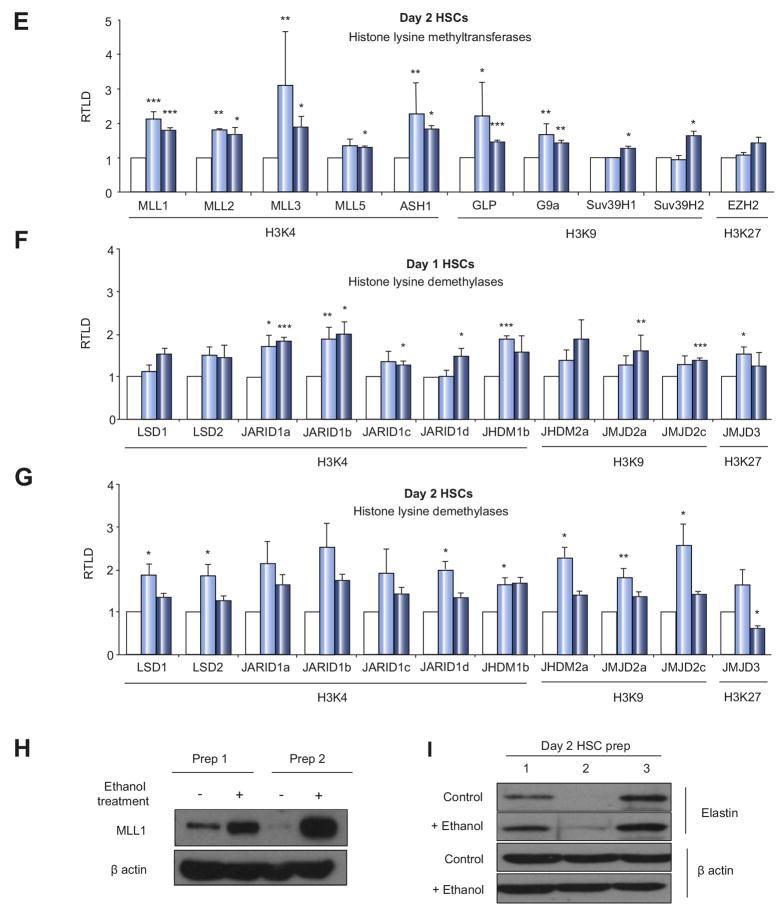
(A) Schematic presentation of ethanol treatment during HSC activation time course. Freshly isolated rat HSCs were treated for 48 hours with 86mM ethanol starting either at day 1, day 2 or day 10 of cell culture induced activation. (B-G) mRNA levels of collagen I, III, IV (B), elastin, MeCP2 and TIMP-1 (C) histone lysine methyltransferases (D, E) and histone lysine demethylases (F, G) were quantified by RT-PCR in at least four separate preparations of primary rat HSCs treated at day 1 and 2 of culture for 24h or 48h with 86mM ethanol. Error bars represent mean values ± standard error of the mean (SEM). *p<0.05; **p<0.005; ***p<0.001. (H, I) Thirty μg whole cell protein extract from at least 2 preparations of rat HSCs treated at day 2 of culture for 48h with 86mM ethanol were separated by SDS-PAGE and immunoblotted for MLL1 (H) or elastin (I). β-actin was used as loading control.
HSC transdifferention is under tight epigenetic control, therefore we were interested to discover effects of ethanol on epigenetic regulators that may underlie its ability to enhance HSC transdifferentiation. Quantitative RT-PCR analysis indicated a broad stimulatory effect of ethanol on the expression of transcripts for histone lysine methyltransferases and demethylases in transitioning HSC (Fig 1D-G). Of particular interest was the enhanced expression of histone 3 lysine 4 (H3K4) methyltransferases (MLL1, MLL2, MML3, MML5 and ASH1) since this chromatin modification is often associated with transcriptionally active genes [12]. MLL1 was the H3K4 methyltransferase that underwent the most significant and reproducible induction in response to ethanol and moreover this effect was reproduced at the protein level (Fig 1H). We also confirmed upregulation of elastin under same conditions (Fig 1I).
MLL1 and H3K4 methylation are enriched at the elastin gene in response to ethanol
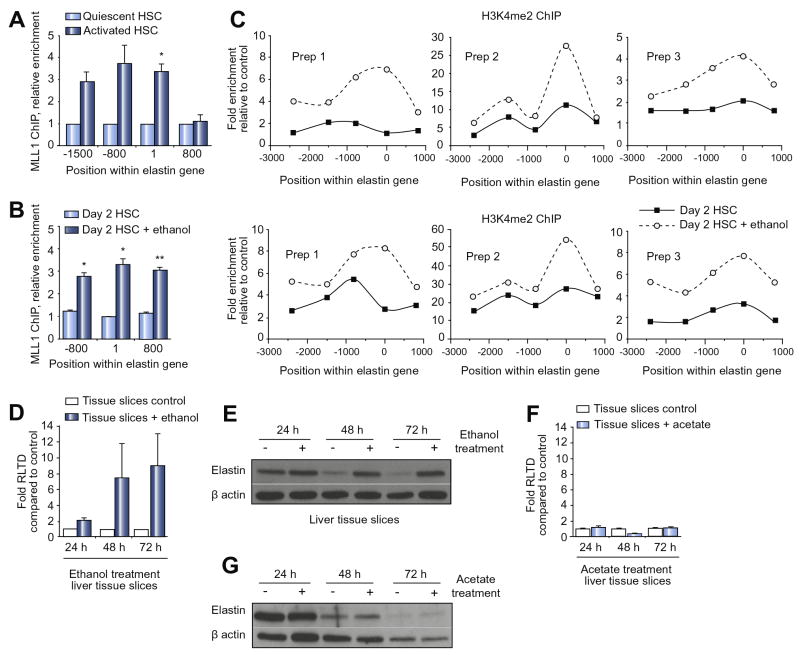
As both elastin and MLL1 underwent similar increases in expression in response to ethanol we investigated a functional relationship. ChIP assays for MLL1 at the elastin gene demonstrated enriched binding in aHSC compared with qHSC, which was localised to upstream regulatory regions (Fig 2A). A modest trend towards enriched ASH1 binding was also observed in aHSC, whereas MLL5 association with the elastin gene was unchanged between qHSC and aHSC (Suppl Fig 3). These data suggest selective and dynamic changes in the occupancy of H3K4 methyltransferases with the elastin gene during HSC transdifferentiation with MLL1 likely to play a prominent regulatory role. Treatment of transitioning HSC with ethanol brought about 3-fold enhanced binding of MLL1 with regulatory regions of the elastin gene (Fig 2B). To determine the functional consequence of ethanol-induced MLL1 association we examined the H3K4me2 and H3K4me3 landscapes of the elastin gene in control and ethanol-treated transitioning HSC (Fig 2C). Both H3K4me2 and H3K4me3 signatures were enhanced by ethanol treatment (dotted lines) and in particular at regions surrounding the transcriptional start site (-500 to +500) of the elastin gene where they would be expected to facilitate a transcriptionally active state of chromatin. It was possible that these changes in H3K4 methylation were simply a consequence of gross alterations in local chromatin structure. However, we did not observe any obvious ethanol-induced changes in nucleosomal spacing at the elastin gene, as determined by micrococcal nuclease digestion profiling (Suppl Fig 4), this arguing against non-specific effects of ethanol on chromatin.
Figure 2. Ethanol induces binding of MLL1 to elastin promoter and increases trimethylation of H3K4 as well as elastin transcript and protein expression.
(A) Fiftyμg of crosslinked chromatin from quiescent rat HSC or fully activated myofibroblasts was incubated with 5μg of anti-MLL1 antibody and ChIP assay carried out. (B-C) ChIP assay for MLL1 (in B) or H3K4me2 and H3K4me3 (in C) using crosslinked chromatin from day 2 HSCs treated for 48h with 86mM of ethanol was carried out as in (A). Following ChIP, immunoprecipitated genomic DNA was used as template in qPCR reactions using primers specific for different regions of elastin gene (A-C). (D) mRNA levels of elastin were quantified by qPCR in three separate preparations of PCLS and treated with 86mM ethanol for 24h, 48h or 72h. (E) Thirty μg whole cell protein from slices as in (E) were immunoblotted for elastin and β-actin. (F) mRNA levels of elastin were quantified by qPCR in three separate preparations of precision cut normal rat liver tissue slices were treated with 1mM sodium acetate for 24h, 48h or 72h. (G) Thirty μg whole cell protein from slices as in (H) immunoblotted for elastin and β-actin. Error bars in relevant panels represent mean values ± standard error of the mean (SEM). *p<0.05; **p<0.005; ***p<0.001.
Elastin expression is directly induced by ethanol in liver tissue
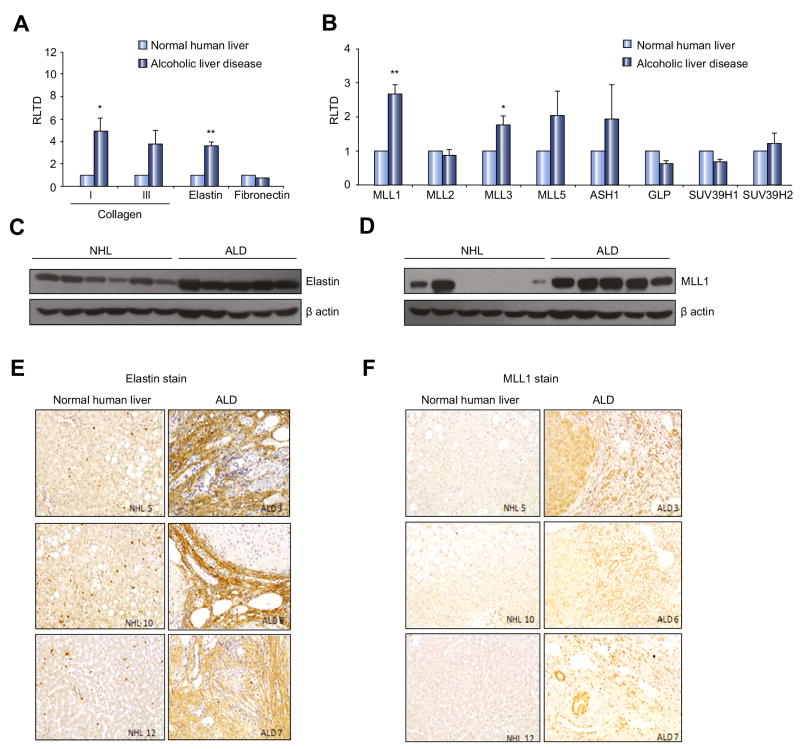
To validate the physiological relevance of our discovery that ethanol induces elastin expression we asked if this effect could be reproduced in a more complex tissue culture system. To this end we generated precision cut liver slices (PCLS) from normal rat liver which were cultured in the absence or presence of ethanol for 24, 48 or 72hrs. At the transcript level we observed induction of elastin expression in ethanol-treated PCLS after 48 and 72 hrs (Fig 2D). This effect was associated with maintenance of high level elastin protein expression which in control cultures was diminished at 48 and 72hrs of culture (Fig 2E). It is widely thought that most of the toxic effects of ethanol including its pro-fibrogenic properties are mediated by its metabolites acetaldehyde and acetate [13, 14]. Hence in PCLS it would be difficult to distinguish between the direct effects of ethanol on elastin expression versus the indirect effects of its main metabolites. We therefore repeated the PCLS experiment but replaced ethanol with its stable metabolite acetate. Acetate treatment was without effect on elastin expression at either the mRNA or protein level indicating that in this complex culture system ethanol has direct effects on elastin gene transcription (Fig 2F and G). We next examined the expression of elastin and its regulator MLL1 in human ALD. As anticipated elastin mRNA expression along with collagen I and III was elevated in ALD explant liver tissue compared with normal human liver (Fig 3A). Expression profiling of histone lysine methyltransferases revealed that ALD is associated with significantly increased expression of MLL1 and MLL3 transcripts and a trend towards higher expression of MLL5 and ASH1 (Fig 3B). Increased expression of elastin and MLL in ALD tissue was confirmed at the protein level (Fig 3C and D). IHC staining for elastin (Fig 3E) and MLL1 (Fig 3F) revealed the former to be mainly confined to fibrotic tissue in ALD, whereas the histone methyltransferase was expressed within hepatocytes, bile ducts and fibrotic scars.
Figure 3. ALD is associated with an increase in MLL1 and elastin expression.
(A) mRNA levels of extracellular components collagen I, III, elastin and fibronectin were quantified by qPCR in six separate preparations of normal human liver or alcoholic liver disease explant. (B) Histone lysine methylatransferases were quantified by qPCR as in (A). Error bars in relevant panels represent mean values ± standard error of the mean (SEM). *p<0.05; **p<0.005. (C-D) Thirty μg whole cell protein from six normal human liver (NHL) or five alcoholic liver disease explanted tissue samples were immunoblotted for MLL1, elastin and β-actin. (E) Representative sections showing MLL1 and elastin immunochistochemical staining in either normal human liver or ALD explanted liver.
To test whether the increase in MLL1 expression was specifically related to ALD rather than presence of cirrhosis, we tested for MLL1 and elastin expression in explanted, cirrhotic primary sclerosing cholingitis (PSC) livers (Suppl Fig 5A and B). There was no detectable MLL1 expression in cirrhotic PSC livers, while elastin levels were similar to those found in normal human livers (Suppl Fig 5A and B). These data suggest that increase in MLL1 expression, as well as downstream events, are related to etiology of ALD rather than the presence of advanced cirrhosis.
Ethanol modifies the global chromatin landscape of the transitioning HSC
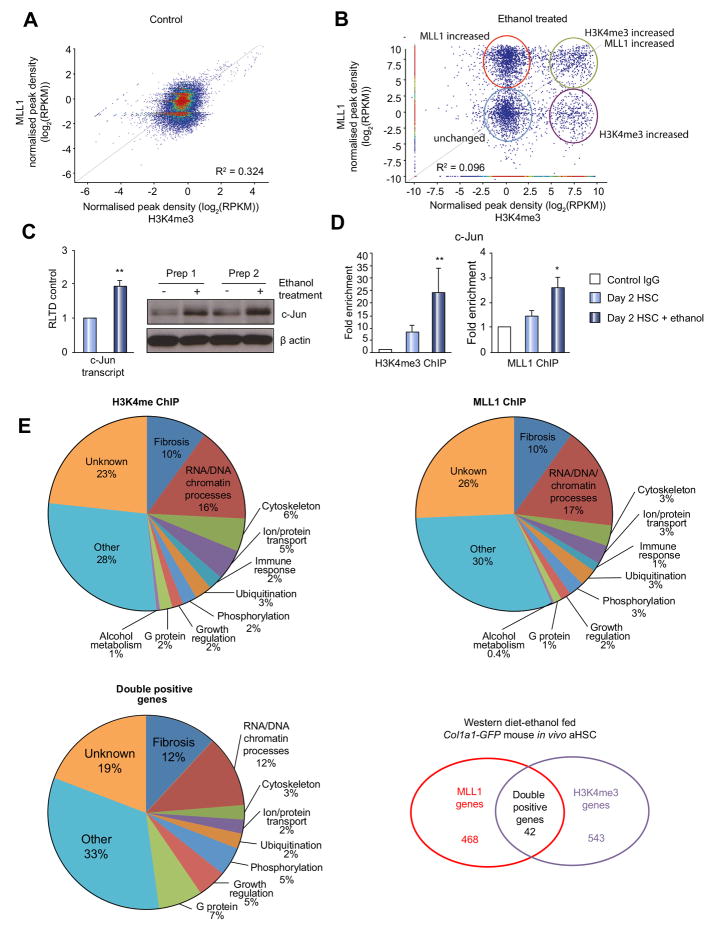
It is expected that HSC transdifferention is underpinned by global remodelling of the HSC epigenome and in particular histone modifications associated with gene activation and silencing. However, the data presented above are restricted to a single gene, elastin, and as such do not provide sufficient support for ethanol having a broader influence on HSC chromatin remodelling. To address this deficiency we carried out ChIPseq for both MLL1 and H3K4me3 comparing control transitioning HSC with ethanol-treated transitioning HSC. From the raw ChIPseq data obtained (Suppl Fig 6), we carried out a combinatorial bioinformatics analysis of MLL1 and H3K4me3 peaks across the genome. In control HSC cultures there appeared to be a close correlation between MLL1 and H3K4me3 peaks (Fig 4A). Ethanol treatment dramatically remodelled this tight MLL1/H3K4Me3 association with the result that subsets of peaks displayed induced increases in either MLL1 alone, H3K4me3 alone or MLL1/H3K4me3 as a tight combination (Fig 4B). Interrogation of these plots revealed 41 gene loci at which ethanol induced a combined enrichment of MLL1 and H3K4me3 (Table 4). Selecting two of these MLL1/H3K4me3 targets, namely proto-oncogene c-Jun and fibroblast growth factor binding protein 3 (FGFBP3) we confirmed that their expression is stimulated by ethanol treatment (Fig 4C and Suppl Fig 7A). In addition, loci-specific ChIP assays validated that both genes underwent ethanol-induced combinatorial enrichments in MLL1 binding and the H3K4me3 modification (Fig 4D and Suppl Fig 7B). Having identified the epigenetic modifications of HSCs by ethanol in vitro, we wondered whether similar changes also occur in vivo after ethanol consumption. To this end, we compared the epigenetic modifications of HSCs isolated by FACS sorting from Col1a1-GFP mice subjected to WD+alcohol feeding plus weekly binge or regular chow feeding using ChIPseq for both MLL1 and H3K4me3 modification. Rather than obtaining control HSCs from either the WD or alcohol fed mice (which would constitute most appropriate experimental controls), we deliberately used HSCs isolated from Col1a1-GFP mice on regular chow as this presents the most normal state that emphasizes the changes resulting from complex ALD pathogenesis in vivo. Following bioinformatics analysis of the data obtained, we were able to confirm that in vivo that in vivo treatment with the combination of WD+ethanol had similar effect on HSC epigenome as observed in rat HSC in vitro cultures (Table 5 and Fig 4E). Analysis revealed 543 genes associated with higher H3K4me3 enrichment; 468 genes were associated with MLL1 in WD+ethanol fed mice and 42 of those showed combined enrichment of H3K4me3 and MLL1 (Table 5 and Figure 4E). Importantly, around a quarter of genes identified in all groups are functionally involved in fibrosis and chromatin modifying processes (Fig 4E, all panels).
Figure 4. ChIP-Seq reveals complexities of MLL1 and H3K4 trimethylation in early transitioning HSC treated with ethanol.
(A-B) ChIP assay for MLL1 or H3K4me3 using crosslinked chromatin from control (A) or day 2 HSCs treated for 48h with 86mM of ethanol (B) was carried out and gDNA sequenced on Illumina sequencer MiSeq. Graph showing correlation analysis of MLL1 and H3K4me3 binding to sequences within the rat HSC genome, red clusters represent regions of high sequence similarity. Correlation analysis of MLL1 and H3K4me3 binding to sequences within the rat genome of control (A) and ethanol treated HSC (B). Red circle (upper left) denotes regions of MLL1 only enrichment after ethanol exposure, light blue circle (lower left) shows regions where binding is unchanged to that found in control HSCs; green circle (upper right) highlights areas where both MLL1 and H3K4me3 binding is increased with ethanol treatment and purple circle (lower right) shows regions where H3K4me3 binding only is increased following ethanol exposure. (C) mRNA levels of c-Jun were quantified by qPCR in four separate preparations of control or rat HSCs treated at day 2 of culture for 48h with 86mM ethanol (left panel). Thirty μg whole cell protein from same samples were separated by SDS-PAGE and immunoblotted for c-Jun and β-actin. (D) ChIP assay validation for MLL1 and H3K4me3 using crosslinked chromatin from control or day 2 HSCs treated for 48h with 86mM of ethanol was carried out and binding to c-Jun promoter tested. Error bars represent mean values ± standard error of the mean (SEM). *p<0.05; **p<0.005. E) ChIP assay as in A-B was carried out on chromatin from HSC isolated from WD+ethanol-fed or control Col1a1-GFP mice. Pie charts show numbers and percentage functional clustering of genes found in MLL1, H3K4me3 and double positive ChIP-seq group.
A number of genes listed were found to overlap with the in vitro ChIPseq results; these are highlighted in red in Table 5. Furthermore, in vivo analysis identified elastin as one of the targets for epigenetic changes induced by in vivo WD+ethanol feeding (Table 5, highlighted in blue).
Discussion
Progression of liver disease to cirrhosis is highly dynamic and variable between patients. This variability is partially explained by genetic factors, age-of-onset of disease, sex and a wide variety of environmental co-morbidity factors including diet, smoking and alcohol consumption. Understanding how these genetic, age/sex and environmental influences combine to determine the course of disease is an important challenge that may improve disease prognosis and patient stratification. Alcohol is both an important primary cause of liver disease and a secondary co-morbidity factor that can influence the progression of chronic liver disease of distinct primary causes such as viral, autoimmune and metabolic injuries [1, 2]. Understanding how alcohol influences core mechanisms such as inflammation and fibrosis that are common to liver disease progression irrespective of aetiology is therefore important. HSC transdifferentiation is widely considered to be a pivotal event in liver fibrosis irrespective of the cause of liver injury and inflammation [15]. Hence, cues in the microenvironment that modulate HSC transdifferentiation have the potential to alter the fibrogenic reaction and in turn the rate of progression of liver disease. Here we have shown that during the early phase of HSC transdifferentiation, remodelling of the HSC epigenome can be influenced by ethanol to bring about genome-wide alterations in gene expression including the enhanced expression of ECM. Therefore HSC triggered to undergo activation in response to other primary insults such as viruses, immune attack or metabolic factors would effectively experience a “second hit” by ethanol that operates at the epigenetic level to modify gene expression and fibrogenesis.
Previous studies have shown that ethanol exerts its gene inducing effects in liver via a number of potential mechanisms that converge on increase in lysine acetylation within histone H3. These mechanisms include oxidative stress (via ROS), which in addition to inducing changes in H3K9 acetylation also affects ALD1 transcription. Further studies have delineated H3K9 and H3K23 as direct targets of ethanol related epigenetic modulation, which at least in hepatocytes may be downstream of ERK signalling [16-18].
In order to understand the nature of the epigenetic changes induced by ethanol in our study, we initially focussed on ECM genes as readouts for ethanol-induced modifications in transitioning HSC with the aim of determining if the fibrogenic phenotype of HSC is susceptible to influence by alcohol. We were particularly intrigued by robust augmentation of elastin expression in response to ethanol exposure. Previous studies have reported that elastin is expressed by HSC from the onset of liver injury but only accumulates in the tissue in the later phases of injury [19]. Deposition of elastin into scar tissue is mainly controlled by the elastase-degrading activity of macrophage-derived MMP12, which in the late phases of injury is suppressed by TIMP-1, this enabling net accumulation of elastin. We observed that ethanol not only stimulates increased expression of elastin but also augments expression of TIMP-1 by HSC, as such both the synthesis and stability of elastin can be enhanced by ethanol exposure. Of note, in our PCLS model we reproducibly observed time-dependent loss of elastin expression occurring between 24 and 48 hrs of culture (Fig 2E and G). However, elastin protein expression was stable for up to 72hrs in ethanol-treated PCSL and by contrast was not stabilised by the ethanol metabolite acetate. Our interpretation of these findings is that the elevated expression of elastin and TIMP-1 in ethanol-exposed PCLS would contribute at least in-part to continued expression and deposition of elastin, which in control and acetate-treated PCLS is presumably subject to rapid degredation by elastases. But taken together with our findings in HSC monocultures we can conclude that ethanol has a direct stimulatory effect on tropoelastin gene transcription, elastin protein expression and TIMP-1 gene transcription that has the potential to promote elastin accumulation. The pathophysiological relevance of this would be the augmented incorporation of elastin into the fibrotic ECM, this promoting maturation and insolubility of the scar tissue through elastin-mediated cross-links [20]. In the context of iterative injury characteristic of chronic liver disease it would be expected that there is on-going de novo activation and transdifferention of qHSC as well as the maintenance of mature HSC-derived myofibroblasts. Newly activating HSC would therefore have the potential to contribute to maturation of existing fibrotic ECM under the influence of ethanol.
Discovering an epigenetic basis to ethanol-stimulation of ECM gene expression in HSC indicated the potential for a more global impact on the HSC epigenome and phenotype. To interrogate this concept we adopted an unbiased ChIPseq approach to identify genes that are associated with alterations in MLL1 and its histone modification H3K4me3, which we had initially identified as being induced by ethanol at the tropoelastin gene. While ChIPseq is potentially a powerful technology, when it is applied to epigenetic regulators or histone modifications the acquired datasets require informed bioinformatic analysis and validation if biologically meaningful discoveries are to be reported [21, 22]. In particular, it is important to recognise that ChIPseq analysis of individual histone signatures or modifying enzymes can be misleading due to inherent complexity in the epigenetic code as well as redundancy in the specificities of the regulatory enzymes [23, 24]. We therefore carried out combinatorial analysis of MLL1 and H3K4me3 ChIPseq data obtained from control and ethanol-exposed HSC. From this we identified genes that displayed changes in MLL1 and H3K4me3 associations either independently or in combination. We assume that those genes displaying changes in H3K4me3 alone are likely to be annotated at this residue by other ethanol-regulated H3K4 methylases such as MLL2, MLL3, MLL5 or ASH1 all of which were upregulated in ethanol-treated HSC (Fig 1D/E). Genes with alterations in MLL1 alone may be subject to additional regulatory mechanisms such as opposing H3K4 demethylase activities, and again we observed ethanol-induced expression of several including multiple members of the JARID1 family (Fig 1F/G). A total of 41 genes were found to acquire combinatorial enrichment of MLL1/H3K4me3 in response to ethanol. Selecting 2 of these genes, c-Jun and FGFBP3, we confirmed the MLL1/H3K4me3 signature by loci-specific ChIP and importantly showed that this epigenetic remodelling was associated with increased gene expression.
Although in vitro studies produce robust data, we were particularly interested to know whether prolonged exposure to ethanol in vivo would yield similar epigenetic changes in HSCs. To address this question, we have used the two-hit model of liver fibrosis based on consumption of alcohol and Western diet. Consumption of the Western control liquid diet or a low fat/alcohol diet does not induce steatohepatitis or pericellular/perisinosoidal liver fibrosis. However, combination of Western diet and alcohol induces alcoholic steatohepatitis with liver fibrosis, confining the prevailing notion of the two-hit or multiple-hit pathogenesis of alcoholic liver disease. It should be noted that for this study, which focuses on epigenetic regulation of activated hepatic stellate cells from the model, we used HSCs isolated from chow-fed animals as the cells presenting the most normal state to highlight changes detected in activated HSCs from the two-hit diet/alcohol model. Combinatorial ChIPseq analysis of MLL1 and H3K4me3 binding in HSCs that were FACS sorted from ethanol-fed or control Col1a1-GFP mice identified a number of overlapping genes between the in vitro and in vivo studies. Furthermore, it also identified numerous genes that were not found in the in vitro study, presumably because in vivo effects are likely to be much more complex as both ethanol and its metabolites (acetaldehyde and acetate) contribute to the in vivo effect. Importantly, in vivo approach clearly shows that a quarter of alcohol induced genes have a fibrosis or chromatin remodelling related function (Fig 4E). Furthermore, in vivo study showed mouse elastin gene to be associated with increased H3K4me3. Despite a plethora of data showing ethanol-induced enrichment of MLL1/H3K4me3 at rat elastin promoter in our candidate gene approach study, we did not identify rat elastin as a target gene in our in vitro MLL1/H3K4me3 ChIP-seq. This discrepancy highlights an important, but not unexpected limitation of the ChIPseq analysis that arises due to differences in the sequencing and PCR technologies and the downstream data analysis platforms for ChIPseq versus loci-specific ChIP protocols that were used in this study.
In summary, we have discovered that ethanol can directly influence the process of epigenetic reprogramming that underpins the transdifferention of qHSC to aHSC with consequences on the HSC phenotype. This finding sheds new light onto the molecular mechanisms, by which ethanol contributes to ALD but also as a co-morbidity factor in other chronic liver diseases.
Supplementary Material
Acknowledgments
Financial Support: The present study was supported by grants from Medical Research Council (MRC) (to D.A.M and J.M), National Institute on Alcohol Abuse and Alcoholism (NIAAA) grant U01AA018663 (H.T, D.A.M and J.M), Welcome Trust and Newcastle Biomedical Research Centre (to J.M), NIAAA grant P50011199 (Animal Core) and R24AA012885 (Non-Parenchymal Liver Cell Core) (to H.T), and 1I01BX001991 (to HT)
List of Abbreviations
- ALD
alcoholic liver disease
- ASH1
absent, small, or homeotic disc 1
- ChIP
chromatin immunoprecipitation
- ECM
extra cellular matrix
- H3K4
lysine 4 histone 3
- HSC
hepatic stellate cells
- MeCP2
methyl-CpG binding protein 2
- MLL1
myeloid/lymphoid, or mixed-lineage, leukemia
- MNase
mononuclease
- NHL
normal human liver
- PPARγ
peroxisome proliferator-activated receptor-γ
- qHSC
quiescent hepatic stellate cells
- rHSC
rat hepatic stellate cells
- qPCR
quantitative polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- αSMA
alpha smooth muscle actin
- TGF-β1
transforming growth factor beta1
- TIMP-1
tissue inhibitor of metalloproteinase-1
Footnotes
Agata Page – generation and analysing of majority of results
Pier P. Paoli – construction of libraries for ChIP-seq, performing ChIP-seq and bioinformatic analysis
Stephen J. Hill – preparation and ethanol treatment of precision cut liver slices (PCLS)
Rachel Howarth – assisting with animal experiments
Hidekazu Tsukamoto – discussions and contribution to ideas; establishing alcoholic liver fibrosis model and FACS-based HSC isolation method; reading and revising the manuscript.
Raymond Wu – assisting with the animal model experiment and FACS isolation of HSC
Soo-Mi Kweon – FACS isolation and characterization of HSC from Col1a1-GFP mice
Jeremy French – providing human liver samples
Steve White – providing human liver samples
Derek A. Mann – help with experimental design, interpretation of the data and drafting of the article
Jelena Mann – main contribution to the conception and design of the project and interpretation of the data; writing of the paper
Conflict of interest: Authors report no conflict of interest
References
- 1.Osna NA. Alcohol and liver, 2010. World journal of gastroenterology : WJG. 2010;16:1303. doi: 10.3748/wjg.v16.i11.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szabo G, Wands JR, Eken A, Osna NA, Weinman SA, Machida K, et al. Alcohol and hepatitis C virus--interactions in immune dysfunctions and liver damage. Alcoholism, clinical and experimental research. 2010;34:1675–1686. doi: 10.1111/j.1530-0277.2010.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieber CS. Ethanol metabolism, cirrhosis and alcoholism. Clinica chimica acta; international journal of clinical chemistry. 1997;257:59–84. doi: 10.1016/s0009-8981(96)06434-0. [DOI] [PubMed] [Google Scholar]
- 4.McClain CJ, Song Z, Barve SS, Hill DB, Deaciuc I. Recent advances in alcoholic liver disease. IV. Dysregulated cytokine metabolism in alcoholic liver disease. American journal of physiology Gastrointestinal and liver physiology. 2004;287:G497–502. doi: 10.1152/ajpgi.00171.2004. [DOI] [PubMed] [Google Scholar]
- 5.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nature communications. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato F, Wachi H, Ishida M, Nonaka R, Onoue S, Urban Z, et al. Distinct steps of cross-linking, self-association, and maturation of tropoelastin are necessary for elastic fiber formation. Journal of molecular biology. 2007;369:841–851. doi: 10.1016/j.jmb.2007.03.060. [DOI] [PubMed] [Google Scholar]
- 7.Mann DA. Hepatology. Baltimore, Md: 2014. Epigenetics in liver disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato JM, Lu SC. Alcohol, DNA methylation, and cancer. Alcohol research : current reviews. 2013;35:25–35. [PMC free article] [PubMed] [Google Scholar]
- 9.Kendrick SF, O’Boyle G, Mann J, Zeybel M, Palmer J, Jones DE, et al. Hepatology. Vol. 51. Baltimore, Md: 2010. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis; pp. 1988–1997. [DOI] [PubMed] [Google Scholar]
- 10.Jones AW. The drunkest drinking driver in Sweden: blood alcohol concentration 0.545% w/v. Journal of studies on alcohol. 1999;60:400–406. doi: 10.15288/jsa.1999.60.400. [DOI] [PubMed] [Google Scholar]
- 11.Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, et al. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705–714. 714 e701–704. doi: 10.1053/j.gastro.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelato KA, Fischle W. Role of histone modifications in defining chromatin structure and function. Biological chemistry. 2008;389:353–363. doi: 10.1515/BC.2008.048. [DOI] [PubMed] [Google Scholar]
- 13.Nordback IH, MacGowan S, Potter JJ, Cameron JL. The role of acetaldehyde in the pathogenesis of acute alcoholic pancreatitis. Annals of surgery. 1991;214:671–678. doi: 10.1097/00000658-199112000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita H, Kaneyuki T, Tagawa K. Production of acetate in the liver and its utilization in peripheral tissues. Biochimica et biophysica acta. 2001;1532:79–87. doi: 10.1016/s1388-1981(01)00117-2. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annual review of pathology. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 16.Choudhury M, Park P-H, Jackson D, Shukla SD. Evidence for the role of oxidative stress in the acetylation of histone H3 by ethanol in rat hepatocytes. Alcohol. 2010;44:531–540. doi: 10.1016/j.alcohol.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park PH, Miller R, Shukla SD. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochemical and biophysical research communications. 2003;306:501–504. doi: 10.1016/s0006-291x(03)01040-4. [DOI] [PubMed] [Google Scholar]
- 18.Kim JS, Shukla SD. Alcohol and alcoholism. Vol. 40. Oxford, Oxfordshire: 2005. Histone h3 modifications in rat hepatic stellate cells by ethanol; pp. 367–372. [DOI] [PubMed] [Google Scholar]
- 19.Pellicoro A, Aucott RL, Ramachandran P, Robson AJ, Fallowfield JA, Snowdon VK, et al. Hepatology. Vol. 55. Baltimore, Md: 2012. Elastin accumulation is regulated at the level of degradation by macrophage metalloelastase (MMP-12) during experimental liver fibrosis; pp. 1965–1975. [DOI] [PubMed] [Google Scholar]
- 20.Sato S, Adachi A, Wakamatsu K, Sasaki Y, Satomura K, Asano G. Abnormal elastic system fibers in fibrotic human liver. Medical electron microscopy : official journal of the Clinical Electron Microscopy Society of Japan. 2000;33:135–142. doi: 10.1007/s007950000013. [DOI] [PubMed] [Google Scholar]
- 21.Schulz S, Haussler S. Methods in molecular biology. Clifton, NJ: 2014. 1149. Chromatin Immunoprecipitation for ChIP-chip and ChIP-seq; pp. 591–605. [DOI] [PubMed] [Google Scholar]
- 22.Nair NU, Kumar S, Moret BM, Bucher P. Bioinformatics. Oxford, England: 2014. Probabilistic partitioning methods to find significant patterns in ChIP-Seq data. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein T, Allis CD. Science. Vol. 293. New York, NY: 2001. Translating the histone code; pp. 1074–1080. [DOI] [PubMed] [Google Scholar]
- 24.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.