Abstract
The current predominant theapeutic paradigm is based on maximizing drug-receptor occupancy to achieve clinical benefit. This strategy, however, generally requires excessive drug concentrations to ensure sufficient occupancy, often leading to adverse side effects. Here, we describe major improvements to the proteolysis targeting chimeras (PROTACs) method, a chemical knockdown strategy in which a heterobifunctional molecule recruits a specific protein target to an E3 ubiquitin ligase, resulting in the target’s ubiquitination and degradation. These compounds behave catalytically in their ability to induce the ubiquitination of super-stoichiometric quantities of proteins, providing efficacy that is not limited by equilibrium occupancy. We present two PROTACs that are capable of specifically reducing protein levels by >90% at nanomolar concentrations. In addition, mouse studies indicate that they provide broad tissue distribution and knockdown of the targeted protein in tumor xenografts. Together, these data demonstrate a protein knockdown system combining many of the favorable properties of small-molecule agents with the potent protein knockdown of RNAi and CRISPR.
Small molecule–mediated inhibition of protein function is the fundamental paradigm underpinning the efficacy of the vast majority of clinically used agents. Pharmacologically relevant inhibition, however, is often only achieved upon >90% target engagement1, necessitating high dosing levels that can lead to off-target effects. Thus, approaches that directly control cellular protein levels have the potential to offer cellular efficacy not easily achievable with small-molecule inhibitors.
The best-investigated methods of reducing cellular protein levels are genetic knockdown approaches based on antisense oligonucleotides, RNA interference (RNAi), CRISPR/Cas9 or related strategies. Despite the clear therapeutic potential2,3, difficulties in achieving sufficient drug concentrations at the targeted site of action, safety challenges due to off-target effects, and poor metabolic stability remain as major obstacles for routine, systemic delivery of nucleic acid–based protein knockdown agents for therapeutic applications4. There has been some success in developing knockdown strategies not based on nucleic acid technologies, so-called ‘chemical knockdown strategies’5. Chemical knockdown typically use a bifunctional small molecule that binds to a protein target while simultaneously engaging the cellular protein quality control machinery, thus ‘hijacking’ the machinery to degrade the protein target. Various methods have been used to engage cellular quality control mechanisms. The first, initially developed in our lab, uses proteolysis targeting chimeras (PROTACs, Fig. 1a) to directly recruit an E3 ubiquitin ligase, reprogramming the enzyme to ubiquitinate a chosen target protein, which leads to its degradation6–9. Previous work used peptides derived from a key recognition motif of HIF1α that possess exquisite binding specificity toward the von Hippel–Lindau (VHL)–cullin–RING-ligase complex10,11 linked to ligands for various targets such as the androgen receptor, estrogen receptor and aryl hydrocarbon receptor12,13 so as to generate peptide-based PROTAC molecules. A similar bifunctional molecular approach was employed to target proteins to the E3 ligase IAP through the ligand bestatin14,15. Unfortunately, bestatin is a nonspecific ligand with the potential to induce degradation of the IAP proteins required for efficacy16, limiting the bio-orthogonality and maximal potency of the approach.
Figure 1.
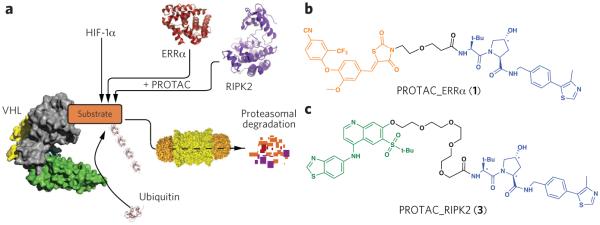
Proteolysis targeting chimeras (PROTACs). (a) Proposed model of PROTAC-induced degradation. Von Hippel–Lindau protein (VHL, gray) is an E3 ubiquitin ligase that, under normoxic conditions, functions with a cullin RING ligase (green and yellow) to degrade HIF1α. PROTACs recruit VHL to target proteins to induce their ubiquitination and subsequent proteasome-mediated downregulation. PROTACs were generated to two target proteins: the orphan nuclear receptor ERRα and the protein kinase RIPK2. (b) Structure of PROTAC_ERRα. The parent ERRα ligand is shown in orange and the modular VHL ligand in blue, with asterisks indicating stereocenter(s) whose inversion (in PROTAC_ERRα_epi) abolishes VHL binding. (c) Structure of PROTAC_RIPK2. The parent RIPK2 ligand is shown in green and the modular VHL ligand in blue, as in b.
Here, we present a significant improvement to the PROTAC technology. This new generation of nonpeptidic PROTAC molecules achieves potent and highly selective downregulation of target proteins in cell culture. Through a series of in vitro and cellular studies, we show that the mechanism is dependent on a ternary complex able to efficiently induce ubiquitination of substrate and allow subsequent proteasomal degradation. We further show a departure from traditional occupancy-limited efficacy whereby each PROTAC molecule is able to induce the degradation of multiple substrate protein molecules. Lastly, in a preliminary mouse study, we show that PROTACs are capable of targeted protein knockdown in various tissues including solid tumors.
RESULTS
PROTAC-mediated protein degradation
To design potent small-molecule PROTACs, we replaced the HIF1α peptide used in previous generations of PROTAC molecules with a recently developed, high-affinity, small-molecule ligand for VHL (Supplementary Results, Supplementary Fig. 1a), which retains the hydroxyproline moiety critical for VHL binding17,18. Crystal structure analyses of VHL bound to the first-generation VHL ligands17,19 suggested that modification of the residue coupled to the N terminus of the hydroxyproline could increase interactions with the HIF1α-binding site on VHL. Introduction of a tert-butyl moiety at this position resulted in a VHL ligand with a Kd of 320 nM, which was incorporated into subsequent PROTACs18. Confident that the VHL ligand would provide an orthogonal means to efficiently recruit an E3 ligase, we first targeted estrogen-related receptor alpha (ERRα), an orphan nuclear hormone receptor that has been implicated as a master regulator of cellular energy homeostasis, regulating expression of genes involved in mitochondrial biogenesis, gluconeogenesis, oxidative phosphorylation and fatty acid metabolism20. Incorporating the compound identified in ref. 17 as compound 29 (Supplementary Fig. 1b), which is a thiazolidinedione-based ligand reported previously to be selective for binding to ERRα over other ERR isoforms21 yet not able to degrade ERRα alone (Supplementary Fig. 2), we generated PROTAC_ERRα (1, Fig. 1b). PROTAC_ERRα was selected as the preferred compound from a small set of five molecules. (For a brief description of PROTAC structure-activity relationships, see the section on RIPK2 below.)
We next developed a PROTAC to target the serine-threonine kinase RIPK2, which functions as an important mediator of innate immune signaling. Once activated, RIPK2 associates with NOD1 and NOD2 to recruit other kinases (TAK1, IKKα, IKKβ, IKKξ) involved in NF-κB and MAPK activation22. Dysregulation of RIPK2-dependent signaling is associated with autoinflammatory diseases including Blau syndrome23 and early-onset sarcoidosis24. We designed PROTAC_RIPK2 (3) by making use of a previously reported cell-active inhibitor (Fig. 1c and Supplementary Fig. 1c). A panel of other RIPK2-targeting PROTACS from a previous generation of RIPK2 PROTACs based on the tyrosine kinase inhibitor vandetanib (Supplementary Fig. 3) explored the structure-activity relationship for different linkers and showed that the 12-atom linker was preferred, so we therefore used this in PROTAC_RIPK2. In a competitive binding fluorescence polarization experiment, PROTAC_RIPK2 was able to compete with a peptide derived from HIF1α for binding to the VHL complex with an IC50 of 660 nM (Supplementary Fig. 4).
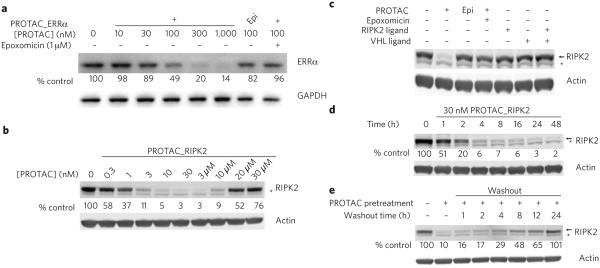
To assess the extent of PROTAC-mediated degradation, we analyzed protein levels by immunoblotting. We found a dose-dependent decrease in ERRα levels in MCF-7 breast cancer cells incubated with PROTAC_ERRα: the maximal level of degradation (Dmax) was 86%, while the concentration at which 50% degradation was observed (DC50) was ~100 nM (Fig. 2a). To confirm that this degradation is VHL dependent, we synthesized PROTAC_ERRa_epi (2), whose stereochemistry is inverted at a key position (indicated by an asterisk in Fig. 1b), thus preventing VHL binding10,11. At concentrations at which the active PROTAC reduced ERRα levels by 50%, PROTAC_ERRα_epi reduced protein levels by only ~20% (Fig. 2a). Finally, to demonstrate that the degradation was mediated by the proteasome, we showed that pretreatment of the cells with epoxomicin25 resulted in only a 5% decrease in normalized ERRα levels (Fig. 2a). We assessed the effect of PROTAC_ERRα on the processing of HIF1α by VHL using luciferase fused with the oxygen-dependent degradation (ODD) domain from HIF1α. PROTAC_ERRα did not stabilize HIF1α at concentrations up to 30 μM (Supplementary Fig. 5).
Figure 2.
PROTACs downregulate the protein levels of their respective targets. (a) PROTAC_ERRα dose-dependently downregulates ERRα protein levels. MCF7 cells were treated with either PROTAC_ERRα or PROTAC_ERRα_epi as indicated for 8 h before harvesting. Where indicated, cells were pretreated with 1 μM of the proteasome inhibitor epoxomicin for 1 h before the treatment. Target protein levels were subsequently detected by western blot analysis. Protein levels were normalized to loading and DMSO controls. Unless otherwise noted, all results in this work are representative of at least three independent experiments. (b) PROTAC_RIPK2 dose-dependently downregulates protein levels and demonstrates an amelioration of efficacy at higher concentrations (‘hook effect’) consistent with a ternary complex–mediated mechanism. THP-1 cells were treated with the indicated amounts of RIPK2_PROTAC for 16 h and then analyzed by western blotting. (c) Degradation by PROTACs is dependent on the proteasome and the presence of the linkage between both targeting ligands. THP-1 cells were treated with the indicated compounds (1 μM for epoxomicin, 30 nM for all others) for 16 h and analyzed by western blotting. (d) RIPK2 is rapidly degraded by PROTAC_RIPK2. THP-1 cells were treated with 30 nM PROTAC_RIPK2 for the indicated times and then analyzed by western blotting. (e) Downregulation by PROTACs is reversible. After a 4-h pretreatment with 30 nM PROTAC_RIPK2, the medium was replaced on THP-1 cells with fresh medium lacking PROTAC and the cells washed thoroughly to remove residual PROTAC. After the indicated times, the cells were analyzed by western blotting. In b–d, * indicates a nonspecific band observed on western blots. For all panels, uncropped blots are shown in the corresponding panels of Supplementary Figure 6 (Supplementary Fig. 6a–e).
Human THP-1 monocytes treated with increasing concentrations of PROTAC_RIPK2 showed a robust, dose-dependent degradation of RIPK2, giving a Dmax of >95% at concentrations of 10 nM and higher, and DC50 of 1.4 nM (mean of four experiments) (Fig. 2b). At concentrations above 3 μM, the protein knockdown was dose-dependently ameliorated, with protein levels returning to basal levels at 30 μM (Fig. 2b). This biphasic response is indicative of a mechanism in which degradation is dependent on a PROTAC-mediated ternary complex (see below). The inactive PROTAC_RIPK2_epi (4) shows no degradation of RIPK2 in THP-1 cells (Supplementary Fig. 6f) and no binding to VHL at concentrations up to 10 μM (Supplementary Fig. 4). We found no effect on ODD-luciferase levels at PROTAC_RIPK2 concentrations up to 3 μM, although modest stabilization of HIF1α occurred at 30 μM (Supplementary Fig. 5). This may suggest a window between RIPK2 degradation and modulation of endogenous VHL function of >1,000 fold. Importantly, this observation suggests that VHL can be efficiently hijacked to allow induced degradation with only a low potential to induce undesired effects through this mechanism. By pretreating the cells with the proteasome inhibitor epoxomicin, we confirmed the proteasomal dependence of the degradation (Fig. 2c). Furthermore, treatment of THP-1 cells with the parent RIPK2 ligand either alone or in combination with the VHL ligand also had no effect on RIPK2 protein levels, suggesting that the ability of the PROTAC to bring RIPK2 and VHL into close proximity is critical for the observed efficacy (Fig. 2c). The lack of effect on RIPK2 protein levels by this inhibitor alone is in contrast to earlier reports that RIPK2 inhibitors may cause nonproteasomal destabilization of RIPK2 (ref. 26).
This efficient degradation of RIPK2 was observed as early as 1 h after treatment and was nearly complete after 4 h (Fig. 2d): a very fast effect given the protein’s half-life of ~60 h27. Furthermore, protein levels recover to pre-treatment levels within 24 h after removal of PROTAC_RIPK2, indicating that the degradation observed is reversible, unlike other nucleic acid–based technologies (Fig. 2e). No cellular toxicity was observed at any concentration (Supplementary Fig. 7).
PROTACs mediate catalytic ubiquitination
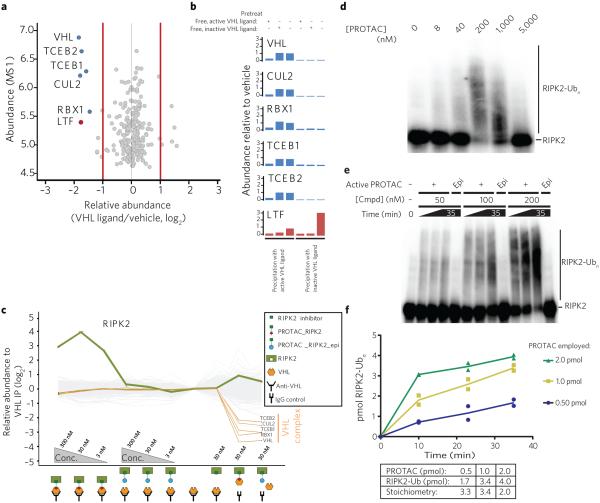
The PROTAC model for induced protein turnover requires the existence of a ternary complex between the target protein, the PROTAC, and the cullin-based VHL E3 ligase. Chemoproteomic pulldown experiments using either the active or inactive VHL ligand tethered to Sepharose as an affinity matrix identified members of the VHL complex—VHL, CUL2, TCEB1, TCEB2 and RBX1—as the only major targets of the active VHL ligand (Fig. 3a, Supplementary Table 1). The enrichment of the VHL complex was abrogated by pretreatment of the lysates with free, active VHL ligand (Fig. 3b). We next sought to characterize the proposed ternary complex by immunoprecipitating VHL from THP-1 cell lysates and subsequently probing for RIPK2 (Fig. 3c, Supplementary Fig. 8 and Supplementary Table 2). The multiplexed quantitative MS analysis using isobaric mass tags (TMT10 labels) revealed selective enrichment of all VHL complex members by the anti-VHL antibody (Fig. 3c). Although addition of PROTAC_RIPK2_epi and the RIPK2 inhibitor alone did not influence the immune-captured proteins or enrich RIPK2 levels, increasing concentrations of PROTAC_RIPK2 lead to co-recruitment of RIPK2 up to 16-fold over background at 30 nM, as shown by the selective enrichment of RIPK2. A large number of nonspecific IgG-binding proteins were not affected under any conditions (Fig. 3c). In broad agreement with cellular observations (discussed above), a dose-dependent increase in RIPK2 capture was observed on increasing the PROTAC_RIPK2 concentration from 3 nM to 30 nM, although at 300 nM, enrichment was reduced by two-fold, suggestive of the previously described diminution of ternary complex formation efficiency28 as increasing concentrations of binary complexes begin to predominate and thus prevent ternary complex formation.
Figure 3.
PROTACs induce the catalytic ubiquitination of their target protein in a reconstituted E1-E2-VHL assay. (a) VHL ligand is highly selective for VHL and associated proteins. The VHL ligand was tethered to Sepharose beads and used to precipitate associated proteins. This was followed by washing of beads, elution of bound proteins and proteomic analysis of over 7,000 proteins. Detailed statistics are available in Online Methods. (b) VHL ligand binds selectively to VHL complex. Active (left three graphs) or inactive (right three graphs) VHL ligands were immobilized onto Sepharose beads and added to THP-1 cell lysates pretreated with active (experiments 1 and 4) or inactive (experiments 2 and 5) free VHL ligand or vehicle (experiments 3 and 6). Immobilized active VHL ligand selectively precipitated members of the VHL E3–dependent ligase complex (compare experiments 3 and 6). This effect was abrogated by prior treatment with free active VHL ligand (compare experiments 1 and 3) but not with inactive, epimeric VHL ligand (compare experiments 2 and 3). The only other protein significantly precipitated was lactotransferrin, which was associated and competed with free ligand when using both active and inactive VHL ligands. (c) PROTAC_RIPK2 mediates the co-immunoprecipitation of VHL and RIPK2. Cell lysates were immunoprecipitated with either IgG control (lanes 9, 10) or an anti-VHL antibody (lanes 1–8) in the presence of PROTAC_RIPK2 (lanes 1–3) or PROTAC_RIPK2_epi (lanes 4–6). The expression levels of over 7,000 proteins were quantified and normalized to the VHL precipitate without PROTAC present (lane 7). (d) PROTAC_RIPK2 mediates direct RIPK2 ubiquitination in vitro. RIPK2 was labeled by autophosphorylation and then incubated with the indicated concentrations of PROTAC and the reconstituted ubiquitination cascade (see Online Methods for more details). Samples were quenched 15 min after initiation of the reaction, and imaged by PAGE and autoradiography. RIPK2-Ubn is indicated. (e) Increasing the PROTAC concentration increases the rate of ubiquitination. Reactions were performed as in d with 50, 100 or 200 nM PROTAC_RIPK2 or PROTAC_RIPK2_epi, quenched at the indicated times and then analyzed by PAGE. (f) PROTACs are able to induce super-stoichiometric ubiquitination of RIPK2. Bands corresponding to ‘Modified RIPK2’ (RIPK2 that had received any number of ubiquitins) were excised, and the number of moles of RIPK2 from two parallel experiments was determined (see Online Methods). Abundance of modified RIPK2 (in pmol) is plotted against time for the three different reactions employing the indicated amounts of PROTAC_RIPK2.
To demonstrate that the PROTAC-mediated ternary complex leads to productive ubiquitination of RIPK2, we next reconstituted the E1-E2-E3 enzymatic cascade in vitro29,30. We then sought to characterize the PROTAC-dependent ubiquitination of the reconstituted kinase domain of RIPK2, radiolabeled by autophosphorylation. Again, in the absence of PROTAC, no ubiquitination of RIPK2 was observed. However, higher-molecular-weight species of RIPK2 were observed upon incubation with increasing concentrations of PROTAC (Fig. 3d). Consistent with biphasic degradation (Fig. 2b) and immunoprecipitation results (Fig. 3c), less RIPK2 ubiquitination was observed at the highest PROTAC concentrations tested.
With direct evidence for the existence of a reversible ternary complex, we then addressed one of the most exciting and fundamental facets of PROTAC action: sub-stoichiometric catalysis. Because PROTACs function by redefining the catalytic properties of the VHL–E2 ligase enzyme complex for a novel substrate, PROTACs should be capable of exerting their induced-ubiquitination substoichiometrically, whereby one molecule of PROTAC is able to induce the ubiquitination and degradation of multiple molecules of RIPK2. To determine the kinetics and stoichiometry of PROTAC_RIPK2-induced ubiquitination of RIPK2, in vitro ubiquitination was performed using three concentrations of PROTAC (50, 100 and 200 nM). Protein bands corresponding to unmodified RIPK2 and modified RIPK2 (that is, protein that has received any number of ubiquitins) were excised, and the absolute amount of RIPK2 was determined by liquid scintillation analysis. Increasing PROTAC concentrations increased the initial rate of the ubiquitination of the RIPK2 substrate (500 nM in each reaction) (Fig. 3e; quantification in Fig. 3f). Furthermore, PROTAC_RIPK2_epi was unable to induce ubiquitination of the target, at any concentration and even at the longest time point (35 min). The ratio of modified RIPK2 to PROTAC would be expected to be greater than 1 if PROTACs were indeed capable of super-stoichiometric ubiquitination. At the final time point, reactions containing 0.50 pmol, 1.0 pmol and 2.0 pmol PROTAC resulted in 1.7 pmol, 3.4 pmol and 4.0 pmol of modified RIPK2, corresponding to stoichiometries of 3.3, 3.4 and 2.0, respectively (Fig. 3f). Although these numbers are clear and positive evidence for the catalytic nature of PROTACs, they represent a very conservative estimate of the stoichiometry, for two reasons. First, in a cellular environment, sufficiently ubiquitinated proteins would be degraded, allowing the PROTAC to ubiquitinate another substrate molecule rather than hyperubiquitinate the first. Second, our analysis assumed that polyubiquitination of a given substrate was complete upon production of a single ternary complex, whereas in fact the final polyubiquitination of a substrate is likely the result of multiple productive ternary complexes formed by each molecule of PROTAC.
PROTACs are highly specific for their targets
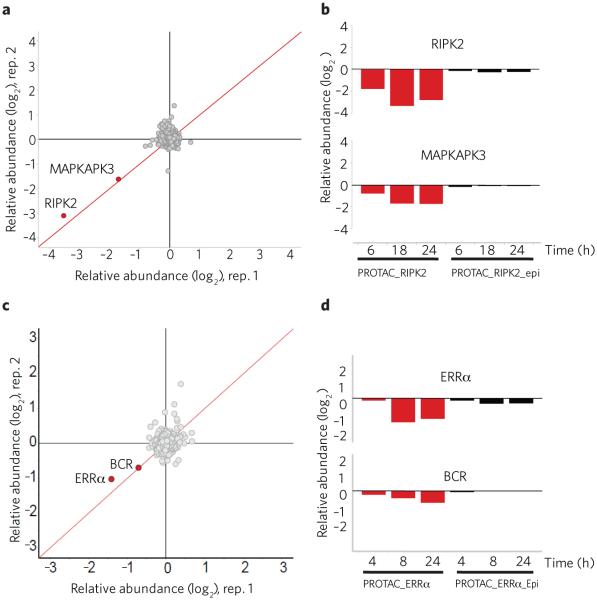
We next sought to determine the specificity for PROTAC-mediated degradation of the target proteins RIPK2 and ERRα. The RIPK2-binding ligand possesses good levels of selectivity over related kinases (Supplementary Fig. 9 and Supplementary Table 4), which would be predicted to result in specific cellular degradation of RIPK2 by PROTAC_RIPK2. To assess the specificity of the induced degradation, we undertook cellular expression proteomic studies to quantify degradation at the proteome level. THP-1 cells were treated for 6, 18 and 24 h with 30 nM of each of PROTAC_RIPK2, PROTAC_RIPK2_epi and RIPK2 ligand alone (Fig. 4a,b, Supplementary Fig. 10a and Supplementary Table 5). PROTAC_RIPK2 induced the degradation of RIPK2 from 6 h onwards, whereas neither the PROTAC_RIPK2_epi nor the RIPK2 ligand affected RIPK2 levels at any time point. Of the ~7,000 proteins quantified in this experiment, only RIPK2 and the unrelated kinase MAPKAPK3 were significantly degraded by PROTAC_RIPK2. Other kinases bound by PROTAC_RIPK2 (such as RIPK3, ABL and TESK; Supplementary Fig. 9b) were not degraded under these conditions. We believe this indicates that PROTACs offer opportunities to modulate functional precision beyond intrinsic binding specificity. MAPKAPK3 binding to PROTAC_RIPK2 was below the limit of detection (Kd > 3 μM), suggesting that this kinase may be particularly susceptible to degradation by this mechanism or that modulation of MAPKAPK3 is via an indirect mechanism dependent on RIPK2 degradation. Further studies are underway to assess the mechanism of MAPKAPK3 degradation.
Figure 4.
PROTACs are highly specific for their respective target. (a) PROTAC_RIPK2 is highly selective for RIPK2 degradation. THP-1 cells were treated for 18 h with 30 nM PROTAC_RIPK2 in biological duplicate and protein levels quantified (see Online Methods). Data is plotted as fold change (log2) of replicate 1 versus replicate 2. The red diagonal line represents proteins whose changes in protein levels were reproducible between the two experiments. A total of 7,640 proteins were quantified. (b) THP-1 cells were treated for the indicated times with 30 nM of either PROTAC_RIPK2 or PROTAC_RIPK2_epi as in a, and the quantified levels of RIPK2 and MAPKAPK3 are shown. (c) MCF-7 cells were treated for 24 h with 500 nM PROTAC_ERRα in biological duplicate and protein levels quantified. Data is plotted as fold change (log2) of replicate 1 versus replicate 2. The red diagonal line represents proteins whose changes in protein levels were reproducible between the two experiments. A total of 7,576 proteins were quantified. (d) MCF-7 cells were treated for the indicated times with 500 nM PROTAC_ERRα or PROTAC_ERRα_epi as in c, and the quantified levels of ERRα and BCR are shown.
Similar expression proteomics experiments were performed for PROTAC_ERRα and the PROTAC_ERRa_epi in MCF-7 cells. After treatment for 4, 8 or 24 h with 100 nM and 500 nM of PROTAC_ERRα, decreased levels of ERRα were observed (Fig. 4c,d, Supplementary Fig. 10b and Supplementary Table 5). Upon prolonged incubation with 500 nM PROTAC_ERRα, the only other protein downregulated was BCR (P < 0.05), potentially indicative of off-target binding of the ERRα ligand (Fig. 4c,d) or of an indirect, ERRα-dependent effect. In contrast, PROTAC_ERRa_epi did not lead to significant degradation of either ERRα or BCR.
Efficient in vivo knockdown in mice
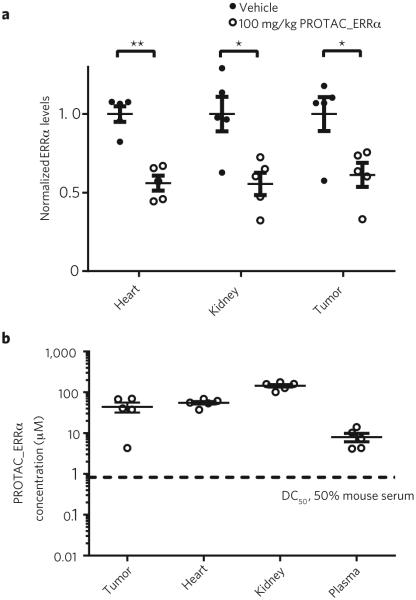
We next determined whether targeted protein degradation of ERRα could be achieved in vivo using PROTAC_ERRα. Mice (n = 5) were injected with either four doses of vehicle (ESD-1) or 100 mg/kg PROTAC_ERRα (3 times per day, intraperitoneally). ERRα levels in mouse heart and kidney and MDA-MB-231 tumors were reduced significantly, by approximately 44%, 44% and 39%, respectively, in mice administered PROTAC_ERRα mice as compared to mice administered an equal volume of ESD-1 vehicle (Fig. 5a). These data demonstrate that the PROTAC_ERRα PROTAC retains its degradation activity in vivo by distributing into tissues and reducing ERRα levels upon target engagement (Fig. 5b).
Figure 5.
PROTAC_ERRα is efficacious in mice. (a) Mice (n = 5) were injected with either vehicle or 100 mg/kg PROTAC_ERRα (3 times per day, intraperitoneally). At ~5 h after the last injection, the mice were killed, and their tissues and tumors were collected and analyzed for ERRα expression by western blotting. Levels of ERRα were normalized to GAPDH levels and plotted, and are shown as mean ± s.e.m. **P <= 0.005, *P < 0.05 by two-tailed, unpaired Student’s t-test. (b) Tissues and plasma from a were analyzed for levels of PROTAC_ERRα by LC/MS. The dashed line represents the DC50 of PROTAC_ERRα when the in vitro degradation experiments were performed in 50% mouse serum. Each data point represents the levels of PROTAC_ERRα from a single mouse and tissue. Data are plotted and shown as mean ± s.e.m.
DISCUSSION
In this study, we sought to improve a system combining the powerful effects of protein knockdown associated with RNAi and CRISPR/Cas9 with the facile cell uptake and in vivo activity of small-molecule agents. The results presented indicate that PROTACs are such a technology, capable of producing potent, selective and reversible cellular protein knockdown as demonstrated in both cellular and in vivo animal studies. Although small-molecule inducers of protein degradation have been previously reported31, the advantages of our PROTAC technology lie in its modular, rationally designed selective engagement of an E3 ligase to induce target protein ubiquitination.
As shown in the in vitro ubiquitination studies, PROTACs are rare among intracellularly acting small molecules in their ability to function catalytically via recruitment of a physiological enzyme cascade rather than via a traditional occupancy-based mechanism. Another ‘catalytic’ approach relies on a ruthenium photocatalyst capable of producing singlet oxygen, which potently inactivates a target protein when conjugated to a targeting ligand32. Under this ‘event-driven’ (in contrast to occupancy-driven) paradigm, the maximal efficacy of a PROTAC is no longer solely determined by the level of equilibrium target occupancy. Rather, the efficiency of protein knockdown depends on a number of kinetic factors: (i) the efficiency of ubiquitin transfer within the ternary complex as determined by the spatial orientation and alignment of E3 ligase and substrate; (ii) the rate at which the ubiquitinated target is trafficked to and processed by the proteasome, along with any competition from deubiquitinase activity; (iii) the basal expression level of the protein to be degraded; and (iv) rates of de novo protein synthesis. This event-driven paradigm offers the possibility of designing small molecules that act with the efficiency of enzymes and may produce cellular and in vivo effects at levels of potency and efficiency hitherto impossible to achieve with traditional, reversible, equilibrium binding–based intervention. Future work will focus on elucidating the contribution of each of these steps in the PROTAC context and optimizing the efficiency of the induced degradation such that the rate of substrate processing approaches that of the rapid HIF1α protein turnover33.
PROTACs function analogously to other strategies broadly defined as ‘substrate switching’. For example, HIV uses a PROTAC-like mechanism to induce the degradation of the host viral restriction protein APOBEC3a via its recruitment to an E3 ubiquitin ligase by the viral protein Vif34. Similarly, PROTAC-induced enzyme reprogramming parallels the mechanism by which cyclosporine and tacrolimus (FK-506) bind to their respective immunophilin targets (cyclophilin and FKBP12) and facilitate subsequent association with calcineurin. This concept of induced functional interaction of two proteins to upregulate a specific biological pathway may find numerous other applications; indeed, it has also been demonstrated, in an extracellular sense, with the recruitment of antibodies35 mediated by PROTAC-like chimeric agents that bind cell surface receptors and display hapten, promoting the binding of antibodies and subsequent immune system mobilization.
Beyond the proteins targeted here, the modular nature of the PROTAC technology may also allow the degradation of a range of other proteins. This, however, requires suitable ligands possessing both sufficient target binding affinity and a solvent-exposed position for introduction of the linker moiety, as exemplified by the RIPK2 and ERRα ligands incorporated into the PROTACs described in this study. Several screening strategies can be used to identify novel protein ligands, including biophysical direct binding approaches (NMR, SPR, thermal shift) commonly used in fragment-based hit identification. Other affinity-based hit identification strategies include DNA-encoded libraries of small molecules36 and small-molecule microarrays37,38. Because these technologies do not rely on enzymatic inhibition, they may facilitate the discovery of ligands for protein targets that are ‘undruggable’ by current small-molecule means. Though these ligands may not be intrinsically biologically active, their incorporation into PROTAC molecules would allow the modulation of proteins for which identification of functional inhibitors has proved challenging or impossible.
PROTAC linker length and composition are additional factors that impact optimal PROTAC-mediated ubiquitin transfer. We have demonstrated that efficient degradation can be observed using both shorter (PROTAC_ERRα) and longer (PROTAC_RIPK2) linkers, but the efficiency of knockdown may be greatly affected by linker identity, and it is likely that optimal linker length and composition will vary for different proteins targeted. The linker may also be able to offer another layer of target specificity: although a PROTAC may be able to bind to multiple targets (most likely determined by the binding specificity of the parent ligand), only a subset of potential targets may be optimally presented for efficient ubiquitination and subsequent degradation. Thus, linker selection and PROTAC architecture may introduce functional selectivity into a nonselective binding ligand upon incorporation into a PROTAC molecule. Finally, to aid the development of suitable therapeutic agents and tools, the linker composition can also be used to modulate properties of the PROTAC such as membrane permeability, aqueous solubility, metabolic stability and biodistribution.
In summary, PROTACs offer a robust method to achieve in vivo protein knockdown with potential therapeutic applications. This approach combines the efficacy typically associated with nucleic acid– based approaches with the flexibility, titratability, temporal control and drug-like properties associated with small-molecule agents. This advance promises to herald the discovery of new classes of transformational medicines using mechanisms not possible today.
ONLINE METHODS
Competition binding assay with VHL ligand
In order to generate a probe matrix of the active and inactive VHL ligand, an amine-functionalized derivative of the VHL ligand was immobilized on NHS-activated Sepharose 4 Fast Flow beads (Amersham Biosciences) at a ligand density of 0.5 μM. Derivatized beads were incubated overnight at room temperature in darkness on an end-over-end shaker and unreacted NHS groups were blocked by incubation with aminoethanol at room temperature on the end-over-end shaker, overnight. Beads were washed with 10 ml of DMSO and were stored in isopropanol at −20 °C. Prior to use, beads were washed three times with 5–10 volumes of DP buffer (50 mM Tris-HCl, 0.8% (v/v) Igepal-CA630, 5% (v/v) glycerol, 150 mM NaCl, 1.5 mM MgCl2, 25 mM NaF, 1 mM sodium vanadate, 1 mM dithiothreitol, Complete EDTA-free protease inhibitor tablet (Roche), pH 7.5), collected by centrifugation for 1 min at 311g in a Heraeus centrifuge and finally re-suspended in DP buffer to prepare a 5% beads slurry.
Affinity profiling assays were carried out as described previously39–40 with minor modifications. MCF-7 lysate was diluted with DP buffer to a protein concentration of 5 mg ml−1 and cleared by centrifugation at 145,000g. Aliquots of cell extracts (1 ml) were incubated with test compounds (5 μM active VHL ligand, inactive VHL ligand or vehicle) for 45 min, then 35 ml derivatized Sepharose beads were added per sample and incubated on an end-over-end shaker for 1 h at 4 °C. Beads were transferred to disposable columns (MoBiTec), washed with DP buffer containing 0.2% Igepal CA-630 and eluted with 50 ml 2× SDS sample buffer. Proteins were alkylated with 200 mg/ml iodoacetamide for 30 min, partially separated on 4–12% NuPAGE (Invitrogen), and stained with colloidal Coomassie before trypsin digestion and mass spectrometric analysis (see below).
Cell culture
Human MDA-MB-231 and MCF7 cells (purchased authenticated ATCC cell lines) were cultured in complete growth medium (Dulbecco’s modified Eagle’s medium (DMEM)/F12 supplemented with 10% FBS; Life Technologies) and grown at 37 °C with 5% CO2. THP-1 cells (ATCC) were cultured in RPMI 1640 + 10% FBS and were grown at 37 °C with 5% CO2. For cellular degradation of ERRα, MCF7 cells were seeded in a 24-well plate at 70% confluency, allowed to attach overnight, and incubated with the indicated compounds. For cellular degradation of RIPK2, 5 × 106 cells were incubated with the indicated compounds. When indicated, a 1-hour pretreatment with 1 μM epoxomicin was performed before the addition of compound. When indicated for washout studies, after PROTAC treatment, cells were washed repeatedly with PBS and incubated with complete medium for the indicated time before harvesting. The THP1 cell line has been authenticated using the Promega GenePrint10 kit to generate a STR Profile for comparison to the expected profile reported by ATCC. Cells were also routinely tested and negative for mycoplasma. Cell lines were routinely tested for mycoplasma contamination at Clongen (Gaithersburg, MD).
Antibodies
Anti-estrogen-related receptor alpha antibodies were purchased from Millipore (EPR46Y, 1:1,000 dilution) or Cell Signaling Technology (E1G1J, 1:1,000 dilution). Anti-RIPK2 (D10B11, 1:500 dilution), anti-GAPDH antibodies (D16H11, 1:2,500 dilution), anti-mouse IgG-HRP, and anti-rabbit IgG-HRP were purchased from Cell Signaling Technology and used at 1:10,000 dilution. Anti-actin antibody was purchased from Abcam (ab6276, 1:2,000). Anti-Tubulin antibody was purchased from Sigma Aldrich (T9026, 1:5,000 dilution).
Immunoblotting
Total protein concentrations of tissue homogenates and cell lysates were determined by Pierce BCA kit (Thermo Fisher). Homogenates or lysates were separated on 4–12% NuPAGE gels (Life Technologies) and transferred onto PVDF membranes (iBlot; Life Technologies). Membranes were blocked with 5% non-fat milk in TBST (0.1% Tween-20 in Tris-Buffered Saline) before overnight incubation with indicated antibodies. After incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies, the bands were visualized by enhanced chemiluminescence (Pierce West Femto ECL). The intensity of the bands was quantified with Bio-Rad Quantity One software. When the IRDye secondary antibodies were used, the infrared signal was detected using an Odyssey scanner (Li-COR Biosciences) and the densitometry was performed using the Odyssey 2.1 Analyser software. The IRDye secondary antibodies anti-mouse (926-32212) and anti-mouse (926-68072) were purchased from Li-COR Biosciences and used at a 1:5,000 dilution. Statistical analysis of tissue densitometry levels were performed with GraphPad Prism software using the indicated statistical test
Animals
Mice were housed in pathogen-free animal facilities at NELS (New Haven, CT). All experiments were conducted under an approved protocol. Female CD-1 mice were obtained from Taconic Laboratories and implanted subcutaneously with 5 × 106 MDA-MB-231 cells in Matrigel (Corning Life Science). After several weeks, mice bearing >100 mm3 tumors were randomized into two unblended groups with five mice in each group. One group served as a control for dosing vehicle, while the other group was given four administrations of PROTAC_ERRα (100 mg/kg, intraperitoneal, every eight hours). Mice were sacrificed five hours after final dose. Blood was collected, processed to plasma, and flash frozen. Tissues were harvested and flash frozen for further analysis. All studies were conducted in accordance with the GSK Policy on the Care, Welfare and Treatment of Laboratory Animals and were reviewed by the Institutional Animal Care and Use Committee either at GSK or by the ethical review process at the institution where the work was performed. In vivo ERRα degradation using PROTAC_ERRα was reproduced in our laboratories two additional times using the same number of animals, dosing scheme and regimen. Sample size was estimated based on the variation in tissue ERRα levels seen from exploratory work before study initiation.
Tissue homogenization
Frozen kidney, heart, liver, and MDA-MB-231 tumors were thawed on ice, chopped into pieces, and placed into microfuge tubes with homogenization buffer (25 mM HEPES, 50 mM NaCl, 1% NP-40, 0.1% SDS, pH 7.4; 10 ml per mg tissue). Tissues were disrupted with a Qiagen TissueLyser bead miller (5 mm stainless steel bead; 2 min, 25 Hz), and homogenates were clarified (15,000g, 10 min, 4 °C) and transferred to new tubes.
LC/MS for PROTAC concentration determination
The concentration of PROTAC_ERRα was determined from snap-frozen plasma or tissue samples. Liquid chromatography-tandem mass spectrometry (LC-MS) analysis was performed by a contractual service (Drumetix Laboratories, Greensboro, NC). The concentration of PROTAC_ERRα was quantified by use of a standard curve.
In vitro ubiquitination
Ubiquitination reactions were performed in three stages. In the first stage, RIPK2 (final 500 nM) was incubated for 10 min at room temperature with [32P]g-ATP in kinase buffer (25 mM Tris pH 7.5, 50 mM KCl, 2 mM Mg(CH3CO2)2, 2 mM MnCl2, 2 mM DTT) to radiolabel RIPK2 through auto-phosphorylation. In the second stage, ubiquitination buffer (25 mM Tris pH 7.5, 5 mM MgCl2, 100 mM NaCl, 2 mM ATP, 0.1 mg/ml BSA, and 2 mM DTT) was added to complete auto-phosphorylation, and then VHL (final 250 nM) and indicated PROTACs (various concentrations) were added to the mixture to allow ternary complex formation. In parallel, UbE1 (final 25 nM), Ubc4 (final 250 nM), and ubiquitin (final 116 μM) were mixed in ubiquitination buffer to allow charging of the E2 enzyme with activated ubiquitin. In stage three of the reaction, the RIPK2-PROTAC-VHL and UbE1-Ubc4~Ub mixtures were combined, and incubated at room temperature for various times before being quenched with Sample Buffer containing 5% β-mercaptoethanol. Ubiquitinated RIPK2 was then separated by 4–15% SDS-PAGE and imaged using a PhosphoImager Screen overnight.
Calculation of RIPK2:PROTAC stoichiometry
After PhosphoImager analysis, the gels were silver-stained, allowing visualization of the unmodified RIPK2 band. This band, along with the upper portion of each lane, was excised and radioactivity was quantified using PerkinElmer TriCarb 2700TR Liquid Scintillation Analyzer with a 32P efficiency of 73%. By using the specific activity on the day of analysis, this radioactivity was converted to moles of 32P. According to a mass spectrometric analysis, autophosphorylated RIPK2 protein has five phosphorylated tryptic peptides observed after the kinase reaction, which are absent in untreated protein (Supplementary Table 3). Using this information, moles of 32P were converted to moles of RIPK2, which can directly be compared to the moles of PROTAC used in each reaction.
Cell treatment for expression proteomics experiment
THP-1 cells were seeded at a concentration of 3′ 106 cells per well in 12 well plates in 1.5 ml growth medium (RPMI1640 + 10% heat-inactivated FBS). 500 μl of a 4′ compound solution prepared in growth medium (DMSO, RIPK2-binding ligand, active or inactive RIPK2-PROTAC) were added and the cells were treated for the indicated periods (6, 18 or 24 h) at 37 °C, 5% CO2. For harvesting, the cells were collected into 2 ml tubes on ice, centrifuged and washed twice in cold PBS (Life Technologies). After the last washing step the supernatant was removed and the pellets were snap-frozen in liquid N2 and stored at −80 °C until SDS lysis. MCF-7 cells were plated 1 day before the experiment at a concentration of 1′ 106 cells per well in 6 well plates in growth medium (MEM + 10% FBS + 1% NEAA + sodium pyruvate) to let them recover and adhere. Medium was replaced with 1.5 ml fresh growth medium, and 500 μl of a 4′ compound solution prepared in growth medium (DMSO, active or inactive ERRα-PROTAC) was added and the cells were treated for the indicated time points (4, 8 or 24 h) at 37 °C, 5% CO2. After treatment medium was removed and the cells were scraped in 1 ml ice cold PBS and collected into 1.5 ml tubes. Cells were washed with PBS and the supernatant was removed completely before cells were lysed in 2% SDS for 3 min at 95 °C in a thermomixer (Thermo Fisher Scientific), followed by digestion of DNA with Benzonase at 37 °C for 1.5 h. Lysate was cleared by centrifugation and protein concentration in the supernatant was determined by BCA assay. Proteins were reduced by DTT and alkylated with iodacetamide, separated on 4–12% NuPAGE (Invitrogen) gels and stained with colloidal Coomassie41 before trypsin digestion and mass spectrometric analysis (see below).
Ternary complex formation
THP-1 cells were harvested and lysed in lysis buffer (50 mM Tris-HCl, 5% Glycerol, 1.5 mM MgCl2, 150 mM NaCl, 1 mM Na3VO4, 0.008% NP40 (Igepal), with Complete EDTA-free protease inhibitor cocktail). Mouse anti-VHL antibody (IgG1k, BD Biosciences) was immobilized at 0.125 μg antibody per μl agarose beads (AminoLink Plus, Thermo Fisher Scientific), and separately, mouse IgG1κ at 0.125 μg antibody per μl agarose beads was immobilized as control antibody. PROTAC_RIPK2, PROTAC_RIPK2_epi and RIPK2-binding ligand were prepared in DMSO at 200 times the final assay concentration. THP-1 lysate was diluted to 5 mg ml−1 total protein concentration with IP buffer (50 mM Tris-HCl, 5% Glycerol, 1.5 mM MgCl2, 150 mM NaCl, 1 mM Na3VO4, 0.008% NP40 (Igepal), Complete EDTA-free protease inhibitor cocktail tablet), and 10 mg total protein was incubated with 300 nM, 30 nM and 3 nM of PROTACs or 30 nM RIPK2-binding ligand or DMSO at 4 °C for 2 h. AminoLinked agarose beads were washed and equilibrated in IP buffer, and incubated with lysate compound mixture at 4 °C for 2h. The beads were settled and supernatant was removed. The beads were washed twice with 30 times bed volume of IP buffer and once with 30 times bed volume of IP buffer without detergent. Bound protein was eluted from the agarose beads by 2′ Nupage buffer (Life Technologies) heated at 95 °C for 10 min. The eluate was separated from the agarose beads, and heated at 95 °C for 5 min after addition of DTT (final DTT concentration 50 mM in the sample). The eluate was subjected to Immunoblotting and LC-MS analysis.
Kinobeads assays
Competition binding assays were performed as described previously by using a modified bead matrix39,42,43. Briefly, 1 ml (5 mg protein) cell extract was pre-incubated with test compound or vehicle for 45 min at 4 °C followed by incubation with Kinobeads (Cellzome; 35 ml beads per sample) for 1 h at 4 °C. The nonbound fraction was removed by washing the beads with DP buffer (50 mM Tris-HCl, 0.8% (v/v) Igepal-CA630, 5% (v/v) glycerol, 150 mM NaCl, 1.5 mM MgCl2, 25 mM NaF, 1 mM sodium vanadate, 1 mM dithiothreitol, Complete EDTA-free protease inhibitor tablet (Roche), pH 7.5). Proteins retained were eluted with 50 ml 2× SDS sample buffer. Proteins were alkylated with 200 mg/ml iodoacetamide for 30 min, partially separated on 4–12% NuPAGE (Invitrogen), and stained with colloidal Coomassie. PROTAC_RIPK2, PROTAC_RIPK2_epi and the RIPK2-binding ligand were tested at 3, 0.75, 0.18, 0.046, 0.012, 0.0029, 0.00073, 0.00018, and 0.000046 mM.
Sample preparation for MS
Gel lanes were cut into three slices covering the entire separation range (~2 cm) and subjected to in-gel digestion39. Peptide samples were labeled with 10-plex TMT (TMT10, Thermo Fisher Scientific, Waltham, MA) reagents, enabling relative quantification of a broad range of 10 conditions in a single experiment. The labeling reaction was performed in 40 mM triethylammoniumbicarbonate, pH 8.53, at 22 °C and quenched with glycine. Labeled peptide extracts were combined to a single sample per experiment, and subjected to additional fractionation on an Ultimate3000 (Dionex, Sunnyvale, CA) by using reverse-phase chromatography at pH 12 [1 mm Xbridge column (Waters, Milford, MA)], as previously described44.
LC-MS/MS analysis
Samples were dried in vacuo and resuspended in 0.05% trifluoroacetic acid in water. Of the sample, 50% was injected into an Ultimate3000 nanoRLSC (Dionex, Sunnyvale, CA) coupled to a Q Exactive (Thermo Fisher Scientific). Peptides were trapped on a 5 mm × 300 μM C18 column (Pepmap100, 5 μM, 300 Å, Thermo Fisher Scientific) in water with 0.05% TFA at 60 °C. Separation was performed on custom 50 cm × 100 mM (ID) reverse-phase columns (Reprosil) at 55 °C. Gradient elution was performed from 2% acetonitrile to 40% acetonitrile in 0.1% formic acid and 3.5% DMSO over 2 h. Samples were online injected into Q-Exactive plus mass spectrometers operating with a data-dependent top 10 method. MS spectra were acquired by using 70.000 resolution and an ion target of 3′ 106. Higher energy collisional dissociation (HCD) scans were performed with 35% NCE at 35.000 resolution (at m/z 200), and the ion target settings was set to 2′ 105 so as to avoid coalescence45. The instruments were operated with Tune 2.3 and Xcalibur 3.0.63.
Peptide and protein identification
Mascot 2.4 (Matrix Science, Boston, MA) was used for protein identification by using a 10 p.p.m. mass tolerance for peptide precursors and 20 mD (HCD) mass tolerance for fragment ions. Carbamidomethylation of cysteine residues and TMT modification of lysine residues were set as fixed modifications and methionine oxidation, and N-terminal acetylation of proteins and TMT modification of peptide N termini were set as variable modifications. The search database consisted of a customized version of the International Protein Index protein sequence database combined with a decoy version of this database created by using a script supplied by Matrix Science. Unless stated otherwise, we accepted protein identifications as follows: (i) For single-spectrum to sequence assignments, we required this assignment to be the best match and a minimum Mascot score of 31 and a score difference of 10× the next best assignment. Based on these criteria, the decoy search results indicated <1% false discovery rate (FDR). (ii) For multiple spectrum to sequence assignments and using the same parameters, the decoy search results indicate <0.1% FDR.
Peptide and protein quantification
Reporter ion intensities were read from raw data and multiplied with ion accumulation times (the unit is milliseconds) so as to yield a measure proportional to the number of ions40; this measure is referred to as ion area46. Spectra matching to peptides were filtered according to the following criteria: mascot ion score >15, signal-to-background of the precursor ion >4, and signal-to-interference >0.5 (ref. 47). Fold changes were corrected for isotope purity as described and adjusted for interference caused by co-eluting nearly isobaric peaks as estimated by the signal-to-interference measure48. Protein quantification was derived from individual spectra matching to distinct peptides by using a sum-based bootstrap algorithm; 95% confidence intervals were calculated for all protein fold changes that were quantified with more than three spectra46. Protein fold changes were only reported for proteins with at least 2 quantified unique peptide matches.
Dose-response curves were fitted using R (http://www.r-project.org/) and the drc package (http://www.bioassay.dk), as described previously39. All measured half-maximum inhibitory concentration (IC50) values were corrected for the influence of the immobilized ligand on the binding equilibrium using the Cheng-Prusoff relationship49.
Average relative log2 fold changes for proteins in the ternary complex formation experiment were calculated for proteins quantified with ³2 quantified unique peptide matches in both replicates.
Relative protein abundances were generated based on MS1 abundances41,50,51. XIC peaks were matched to the identified peptides. The apex of the XIC peak was required to be within 30 s from the time of the MS/MS event performed on the peptide precursor. The raw abundances of the XIC peaks of the peptides with identical sequences were summed (i.e., same sequence, but different charge states and/or different modifications), and the resulting single entity was referred to as a sequence. For each protein the 3 sequences with the highest raw XIC peak abundance from a given sample were selected and log10-transformed51. These values were then summed, and the mean was calculated. If less than 3 sequences were identified for a given protein, the mean was calculated on the 2 or 1 log10-transformed MS1 values of the sequences.
Statistical analysis
Quantified proteins were divided into bins. The bins are constructed according to the number of quantified spectrum sequence matches. Each bin consists of at least 300 proteins. Once each bin has been completed, the remaining number of proteins is counted; if this number is below 300, the remaining proteins are added to the last completed bin. This data quality–dependent binning strategy is analogous to the procedure described previously52. The statistical significance of differences in protein fold change was calculated using a z-test with a robust estimation of the standard deviation (using the 15.87, 50 and 84.13 percentiles) and calculating the P values for all measurements for a specific bin exactly as previously described52. Subsequently, an adjustment for multiple hypothesis testing was performed on the full data set by using Benjamini-Hochberg (BH) correction53. Additionally proteins were filtered if the absolute log2 fold change difference between the 2 replicate vehicle controls was ≥ 0.38. Finally, proteins were counted as regulated when they had P ≤ 0.05 and showed a change in expression in both replicates of 50% in the same direction.
Supplementary Material
Acknowledgments
We thank K. Mueller and D. Poeckel for cell culture support; S. Melchert, E. Stonehouse, A. Lachert, J. Cox, M. Leveridge, C. Pancevac and M. Jundt for biochemistry and sample preparation support; M. Boesche, T. Rudi, M. Kloes-Hudak and K. Kammerer for mass spectrometry support; and S. Gade for data analysis support. This research was partially supported by US National Institutes of Health grants AI084140, T32GM067543 and T32GM007223.
Footnotes
Author contributions
I.E.D.S., E.K., S.C., A.H.M., J.D.H., D.L.B., J.L.G., L.N.C. and B.J.V. contributed to the design and synthesis of compounds. D.P.B., A.M., K.E.M., N.R., C.C., D.A.G., R.R.W., J.J.F. and W.d.B. contributed to running of in vitro, cellular and in vivo experiments. N.Z., P.G., S.S., G.B., M.F.-S. and M.B. designed, performed and interpreted proteomic analyses. D.P.B., A.M., M.B., P.G., G.B., J.J.F., K.F., L.K., P.S.C., J.D.H., I.C. and C.M.C. designed studies and interpreted results. D.P.B., C.M.C. and I.C. wrote the manuscript.
Competing financial interests
The authors declare competing financial interests: details accompany the online version of the paper.
Additional information
Supplementary information and chemical compound information is available in the online version of the paper.
References
- 1.Adjei AA. What is the right dose? The elusive optimal biologic dose in phase I clinical trials. J. Clin. Oncol. 2006;24:4054–4055. doi: 10.1200/JCO.2006.07.4658. [DOI] [PubMed] [Google Scholar]
- 2.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 3.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tokatlian T, Segura T. siRNA applications in nanomedicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010;2:305–315. doi: 10.1002/wnan.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley DL, Crews CM. Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system. Angew. Chem. Int. Edn. Engl. 2014;53:2312–2330. doi: 10.1002/anie.201307761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneekloth AR, Pucheault M, Tae HS, Crews CM. Targeted intracellular protein degradation induced by a small molecule: en route to chemical proteomics. Bioorg. Med. Chem. Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneekloth JS, Jr., et al. Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 9.Hines J, Gough JD, Corson TW, Crews CM. Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs. Proc. Natl. Acad. Sci. USA. 2013;110:8942–8947. doi: 10.1073/pnas.1217206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min JH, et al. Structure of an HIF-1α -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 11.Hon WC, et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Gonzalez A, et al. Targeting steroid hormone receptors for ubiquitination and degradation in breast and prostate cancer. Oncogene. 2008;27:7201–7211. doi: 10.1038/onc.2008.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puppala D, Lee H, Kim KB, Swanson HI. Development of an aryl hydrocarbon receptor antagonist using the proteolysis-targeting chimeric molecules approach: a potential tool for chemoprevention. Mol. Pharmacol. 2008;73:1064–1071. doi: 10.1124/mol.107.040840. [DOI] [PubMed] [Google Scholar]
- 14.Itoh Y, et al. Double protein knockdown of cIAP1 and CRABP-II using a hybrid molecule consisting of ATRA and IAPs antagonist. Bioorg. Med. Chem. Lett. 2012;22:4453–4457. doi: 10.1016/j.bmcl.2012.04.134. [DOI] [PubMed] [Google Scholar]
- 15.Okuhira K, et al. Development of hybrid small molecules that induce degradation of estrogen receptor-alpha and necrotic cell death in breast cancer cells. Cancer Sci. 2013;104:1492–1498. doi: 10.1111/cas.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekine K, et al. Small molecules destabilize cIAP1 by activating autoubiquitylation. J. Biol. Chem. 2008;283:8961–8968. doi: 10.1074/jbc.M709525200. [DOI] [PubMed] [Google Scholar]
- 17.Buckley DL, et al. Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1alpha interaction. J. Am. Chem. Soc. 2012;134:4465–4468. doi: 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crews CM, et al. Compounds and methods for the inhibition of vcb e3 ubiquitin ligase. 2013. Patent PCT/US2013/021141.
- 19.Buckley DL, et al. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1alpha. Angew. Chem. Int. Edn Engl. 2012;51:11463–11467. doi: 10.1002/anie.201206231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichner LJ, Giguere V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11:544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- 21.Patch RJ, et al. Identification of diaryl ether-based ligands for estrogen-related receptor α as potential antidiabetic agents. J. Med. Chem. 2011;54:788–808. doi: 10.1021/jm101063h. [DOI] [PubMed] [Google Scholar]
- 22.Humphries F, Yang S, Wang B, Moynagh PN. RIP kinases: key decision makers in cell death and innate immunity. Cell Death Differ. 2015;22:225–236. doi: 10.1038/cdd.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miceli-Richard C, et al. CARD15 mutations in Blau syndrome. Nat. Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 24.Kanazawa N, et al. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood. 2005;105:1195–1197. doi: 10.1182/blood-2004-07-2972. [DOI] [PubMed] [Google Scholar]
- 25.Meng L, et al. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Windheim M, Lang C, Peggie M, Plater LA, Cohen P. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem. J. 2007;404:179–190. doi: 10.1042/BJ20061704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 28.Douglass EF, Jr., Miller CJ, Sparer G, Shapiro H, Spiegel DA. A comprehensive mathematical model for three-body binding equilibria. J. Am. Chem. Soc. 2013;135:6092–6099. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamura T, et al. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwai K, et al. Identification of the von Hippel–Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanvin O, Bianco S, Kersual N, Chalbos D, Vanacker JM. Potentiation of ICI182,780 (Fulvestrant)-induced estrogen receptor-alpha degradation by the estrogen receptor-related receptor-alpha inverse agonist XCT790. J. Biol. Chem. 2007;282:28328–28334. doi: 10.1074/jbc.M704295200. [DOI] [PubMed] [Google Scholar]
- 32.Lee J, Udugamasooriya DG, Lim HS, Kodadek T. Potent and selective photo-inactivation of proteins with peptoid-ruthenium conjugates. Nat. Chem. Biol. 2010;6:258–260. doi: 10.1038/nchembio.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jewell UR. Induction of HIF-1a in response to hypoxia is instantaneous. FASEB J. 2001;15:1312–1314. [PubMed] [Google Scholar]
- 34.Guo Y, et al. Structural basis for hijacking CBF-β and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 35.Spiegel DA. A call to ARMs: the promise of immunomodulatory small molecules. Expert Rev. Clin. Pharmacol. 2013;6:223–225. doi: 10.1586/ecp.13.10. [DOI] [PubMed] [Google Scholar]
- 36.Clark MA, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 37.Noblin DJ, et al. A HaloTag-based small molecule microarray screening methodology with increased sensitivity and multiplex capabilities. ACS Chem. Biol. 2012;7:2055–2063. doi: 10.1021/cb300453k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradner JE, et al. A robust small-molecule microarray platform for screening cell lysates. Chem. Biol. 2006;13:493–504. doi: 10.1016/j.chembiol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Bantscheff M, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 40.Bantscheff M, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 41.Becher I, et al. Chemoproteomics reveals time-dependent binding of histone deacetylase inhibitors to endogenous repressor complexes. ACS Chem. Biol. 2014;9:1736–1746. doi: 10.1021/cb500235n. [DOI] [PubMed] [Google Scholar]
- 42.Bergamini G, et al. A selective inhibitor reveals PI3Kg dependence of TH17 cell differentiation. Nat. Chem. Biol. 2012;8:576–582. doi: 10.1038/nchembio.957. [DOI] [PubMed] [Google Scholar]
- 43.Werner T, et al. High-resolution enabled TMT 8-plexing. Anal. Chem. 2012;84:7188–7194. doi: 10.1021/ac301553x. [DOI] [PubMed] [Google Scholar]
- 44.Kruse U, et al. Chemoproteomics-based kinome profiling and target deconvolution of clinical multi-kinase inhibitors in primary chronic lymphocytic leukemia cells. Leukemia. 2011;25:89–100. doi: 10.1038/leu.2010.233. [DOI] [PubMed] [Google Scholar]
- 45.Werner T, et al. Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal. Chem. 2014;86:3594–3601. doi: 10.1021/ac500140s. [DOI] [PubMed] [Google Scholar]
- 46.Savitski MM, et al. Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on Orbitrap-type mass spectrometers. Anal. Chem. 2011;83:8959–8967. doi: 10.1021/ac201760x. [DOI] [PubMed] [Google Scholar]
- 47.Savitski MM, et al. Targeted data acquisition for improved reproducibility and robustness of proteomic mass spectrometry assays. J. Am. Soc. Mass Spectrom. 2010;21:1668–1679. doi: 10.1016/j.jasms.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Savitski MM, et al. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J. Proteome Res. 2013;12:3586–3598. doi: 10.1021/pr400098r. [DOI] [PubMed] [Google Scholar]
- 49.Sharma K, et al. Proteomics strategy for quantitative protein interaction profiling in cell extracts. Nat. Methods. 2009;6:741–744. doi: 10.1038/nmeth.1373. [DOI] [PubMed] [Google Scholar]
- 50.Grossmann J, et al. Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J. Proteomics. 2010;73:1740–1746. doi: 10.1016/j.jprot.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 51.Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel ms acquisition. Mol. Cell. Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 53.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.