Abstract
The impact of temperature (0–80°C) on anaerobic biogeochemical processes and prokaryotic communities in marine sediments (tidal flat) was investigated in slurries for up to 100 days. Temperature had a non-linear effect on biogeochemistry and prokaryotes with rapid changes over small temperature intervals. Some activities (e.g. methanogenesis) had multiple ‘windows’ within a large temperature range (∼10 to 80°C). Others, including acetate oxidation, had maximum activities within a temperature zone, which varied with electron acceptor [metal oxide (up to ∼34°C) and sulphate (up to ∼50°C)]. Substrates for sulphate reduction changed from predominantly acetate below, and H2 above, a 43°C critical temperature, along with changes in activation energies and types of sulphate-reducing Bacteria. Above ∼43°C, methylamine metabolism ceased with changes in methanogen types and increased acetate concentrations (>1 mM). Abundances of uncultured Archaea, characteristic of deep marine sediments (e.g. MBGD Euryarchaeota, ‘Bathyarchaeota’) changed, indicating their possible metabolic activity and temperature range. Bacterial cell numbers were consistently higher than archaeal cells and both decreased above ∼15°C. Substrate addition stimulated activities, widened some activity temperature ranges (methanogenesis) and increased bacterial (×10) more than archaeal cell numbers. Hence, additional organic matter input from climate-related eutrophication may amplify the impact of temperature increases on sedimentary biogeochemistry.
Keywords: sediment, temperature, anaerobic processes, chemoorganotrophic, chemolithotrophic, mineralisation, sulphate reduction, methanogenesis, acetogenesis
Temperature increases had a non-linear effect on biogeochemistry and prokaryotes in marine sediments, with rapid changes over small temperature intervals which are relevant to climate change and the deep biosphere.
Graphical Abstract Figure.
Temperature increases had a non-linear effect on biogeochemistry and prokaryotes in marine sediments, with rapid changes over small temperature intervals which are relevant to climate change and the deep biosphere.
INTRODUCTION
Between 5 and 10 billion tons of particulate, organic matter are constantly sinking in the world's oceans and accumulating as sediments (Jørgensen 1983). Anaerobic microbial processes play a major role in the degradation of this organic matter, especially in coastal sediments (Canfield et al. 1993), with metal oxide and sulphate reduction being major degradation processes. The small amount of organic matter that is not degraded builds up over geological time to become the largest global store of organic carbon (Hedges and Keil 1995), which under some circumstances forms oil and gas after maturation and heating. This burial of reduced carbon also has a major impact on the surface environment in terms of the unused oxygen that accumulates in the atmosphere and removal of nutrients and other compounds. Sedimentary anaerobic communities, therefore, have a major impact on both the biosphere and geosphere.
These anaerobic communities involve interdependent, interacting groups of organisms, including hydrolytic/fermenters, heterotrophic acetogens, syntrophs and terminal-oxidizing groups, such as nitrate, metal oxide and sulphate reducers and methanogens. Although the concentration and turnover of metabolic intermediates between these groups (e.g. H2 and acetate) can provide some information about the dominant processes and their interactions (Lovley and Chapelle 1995; Parkes et al. 2007a), as can inhibitor (Parkes et al. 1989) and biodiversity studies (Fry et al. 2008), there is still only limited information about the interactions between a range of different anaerobic processes and how these are influenced by environmental conditions. The response of some anaerobic processes to changing conditions cannot be adequately explained by thermodynamic considerations (Rothfuss and Conrad 1993; Peters and Conrad 1996); hence, other approaches are also needed. Temperature changes have been used to alter the dominant anaerobic pathways and associated prokaryotic populations (Conrad, Klose and Noll 2009) or uncouple key phases in organic matter degradation, to reveal metabolic interactions (Weston and Joye 2005; Finke and Jørgensen 2008). Often, substrates are added to these experiments to ensure elevated microbial activity, to allow changes to be clearly seen and/or to relieve terminal oxidizers from substrate limitation by fermenters/syntrophs, which might have different metabolic/temperature controls. Using these types of experiments with coastal marine sediments, Weston and Joye (2005) showed that hydrolysis/fermentation was enhanced at low temperatures, <25°C, resulting in an accumulation of organic acids, whilst >25°C sulphate reduction was initially enhanced resulting in net removal of organic acids which ultimately limited sulphate reduction.
Similarly, it has been shown that in anoxic rice field soils acetate concentrations increased with decreasing temperatures, from about 5 μM between 17 and 37°C to about 50 μM at 10°C, and acetoclastic methanogenesis becomes increasingly dominant (Fey and Conrad 2000). The activity of psychrotolerant heterotrophic acetogens was suggested as an explanation for this effect. At higher temperatures, methanogenesis changed from a mixture of acetoclastic and hydrogenotrophic methanogenesis to exclusively hydrogenotrophic methanogenesis over a surprisingly narrow temperature range of 42–46°C (Conrad, Klose and Noll 2009). These studies suggested that temperature defined the structure and function of the methanogenic community in anoxic rice field soils. At temperatures above ∼50°C, in rice field soils (Rui, Qiu and Lu 2011), anaerobic digesters (Ho, Jensen and Batstone 2013) and oil reservoir fluids (Dolfing, Larter and Head 2008; Mayumi et al. 2011) data also suggests dominance of hydrogenotrophic methanogenesis, but that this is coupled to acetate oxidation by syntrophs producing H2.
In contrast to the above, Finke and Jørgensen (2008) concluded that in temperate marine sediments fermentative bacteria tolerated higher temperatures than terminal oxidizing, sulphate-reducing bacteria, and hence, above a critical temperature of ∼30°C concentrations of organic acids and H2 increased. Subsequent removal of H2 in these experiments suggested that methanogens also tolerated higher temperatures than sulphate-reducing bacteria. This is surprising as several studies have shown that thermophilic, spore-forming sulphate-reducing bacteria are present in coastal sediments and become active at these higher temperatures (Isaksen, Bak and Jørgensen 1994; Muller et al. 2014; O'Sullivan et al. 2015).
Despite differences between studies, the above clearly shows that varying temperatures can result in the dominance of different anaerobic prokaryotic processes and can help determine the nature of the interactions between both competing and complementary processes. There is also a suggestion of critical temperatures, where changes in function and community structure occur over a surprisingly narrow temperature range (e.g. Conrad, Klose and Noll 2009). To explore this further, we investigated the impact of incubating temperate estuarine sediments at a wide range of different temperatures (0–80°C) for up to 100 days, either with (1) a small addition of H2 to stimulate activity and interactions (e.g. via the impact of sulphate depletion) or (2) a significant substrate addition to potentially highlight the temperature impact on terminal oxidizers separate from their substrate suppliers, and to increase their population size, and hence, their interaction and detection. Geochemical and direct radiotracer analyses were used to measure biogeochemical activities, and community composition was determined by 16S rRNA gene analysis. This was complemented by thermodynamic and kinetic analysis.
MATERIALS AND METHODS
Sampling and sediment slurries
Sediment slurries were prepared from sediment cores, depths to 49–58 cm, collected at low tide from tidal flats of the Severn Estuary, Woodhill Bay, Portishead, UK (51°29′31.66″ N, 2°46′27.95″ W) on 12 February 2010 using Plexiglas core tubes. At high tide, the sediment was covered by ∼1.5 m of water. The winter in situ sediment temperature was low, 6.8°C compared to average local sea surface temperature (mean: 12.6°C, range 3.6–22.6°C; Joyce 2006). After sampling, cores were sealed with rubber bungs and brought back to the laboratory within two hours for rapid geochemical processing. To assess the effect of seasonality on the geochemical profiles, data were compared to a summer (June) geochemical depth profile analysed under identical conditions (in situ temperature 18.7°C).
To avoid high sulphate concentrations and thus the potential dominance of sulphate reduction, and inhibition of methanogenesis and/or acetogenesis, only sediment below 30 cm, which also contained methane, was slurried (Fig. S1, Supporting Information). All sediments were thoroughly homogenized in a gas-tight plastic bag under oxygen-free nitrogen, and then added to modified 2 L screw-capped bottles (1:4, v/v) containing anoxic mineral salts medium reduced with 1 mM sodium sulphide (Wellsbury, Herbert and Parkes 1994), and the gas headspace replaced with N2:CO2 (80:20, v/v). Slurries were incubated at 10°C (∼in situ annual average temperature) on an orbital shaker (100 rpm) in the dark until sulphate concentration reached steady state and sediment was homogeneously slurried. Replicate slurries were then distributed in an anaerobic cabinet into either 20 mL (10 mL slurry) or 60 mL serum vials (20 mL slurry), and sealed with butyl rubber septa. Half of the slurries were amended with 2 mM acetate and 2 mM methylamine prior to dispensing and then had their headspace gas replaced by H2:CO2 (80:20, v/v); these were the substrate-amended vials. No substrates were added to the other slurries, but they were left with a small amount of H2 (∼45 μM) from the anaerobic cabinet to slightly stimulate prokaryotic activity and interactions; these were termed unamended slurries. Five series of 24 substrate-amended and unamended 20 mL slurry vials were incubated upside down between 0 and 80°C in a Thermal Gradient System (Parkes et al. 2007b). Each series was then sacrificed at different time points, up to 100 days, for analysis. The 60 mL serum vials, which contained only unamended slurry, were incubated upside down in the dark at 10, 25, 38, 46, 55, 66 and 77°C for 14C-activity measurements. Additional aliquots of both slurry types (20 mL) were incubated at the same temperatures, as ‘indicator vials’, whose headspace gases were repeatedly sampled over time to help determine the appropriate sampling times for the thermal gradient vials and 14C-activity measurement times.
Pore water and gas analysis
Sediment and slurry headspace gases were analysed by a natural gas analyser (PerkinElmer Clarus® 500) as previously described (Webster et al. 2010). Anion and cation concentrations from sediment and slurry pore waters were determined by ion chromatography (Dionex ICS-2000 and DX-120, Camberley UK; Webster et al. 2010). Dissolved metals in pore waters were analysed by Inductively-Coupled Plasma Mass Spectrometer (ICP-MS) as previously described (Moreno et al. 2007).
Activity rate measurements
Hydrogenotrophic methanogenesis, hydrogenotrophic acetogenesis, acetoclastic methanogenesis, methylotrophic methanogenesis and acetate oxidation rates were measured using 14C radiolabelled substrates ([1,2-14C]acetic acid, [14C]bicarbonate and [1,2–14C]dimethylamine; Parkes et al. 2007a, 2012). Each activity was measured at three increasing time periods in triplicate; data are expressed as an average of the three time-point means. Acetogenesis was measured by collection of the 14C-acetate fraction using a Dionex ICS-2000 Ion Chromatography System equipped with a Foxy Jr.® fraction collector (Teledyne Isco) followed by liquid scintillation counting. Rates of acetate oxidation to carbon dioxide were calculated by multiplying rates by 2 to account for the two carbon dioxide molecules generated from each acetate molecule. Sulphate removal rates were calculated from the difference in sulphate concentrations between each sampling time point.
Thermodynamic calculations and Arrhenius parameters
The Gibbs free energy (ΔG’r) under non-standard conditions was calculated as previously described (Conrad and Wetter 1990). An estimation of the temperature dependence of each studied anaerobic process was obtained by calculating the activation energy (Ea) and the Q10 factor from Arrhenius plots (Aller and Yingst 1980). The Arrhenius profiles were obtained by plotting the natural logarithm of each maximum rate for each incubation temperature versus the inverse of temperature. The activation energy for each metabolic process was calculated from the following equation:
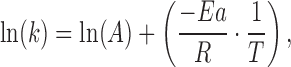 |
where Ea is the activation energy (kJ mol−1), k is the reaction rate (nmol cm−3 day−1), A is the Arrhenius constant, R is the gas constant (8.314 × J K−1 mol−1) and T is the absolute temperature (K). Q10 is the factor by which the rate of reaction increases with a temperature increase of 10°C. The selected temperature range in this study was between 10 and 20°C. Q10 was calculated using the following equation:
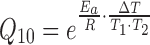 |
Carbon dioxide balance
In order to compare chemoorganotrophic and chemolithotrophic processes, the total carbon dioxide generation rates were determined from the net production or consumption of carbon dioxide by each measured metabolic process. The carbon dioxide generation rate for each metabolic process studied was calculated by multiplying the metabolic rate for each incubation time and temperature by the factor described in Table S1 (Supporting Information). A standardizing factor of 2 was used for putative acetoclastic metal reduction.
DNA extraction
Genomic DNA was extracted from sediment slurries using the FastDNA® Spin Kit for Soil (MP Biomedicals) as described (Webster et al. 2003). Essentially, 3 mL (1.5 mL × 2) of sediment slurry was placed in a lysing matrix E tube (MP Biomedicals) and centrifuged at 15 000 × g for 1 min to pellet cells and sediment. Pellets were then resuspended in 800 μl of sodium phosphate buffer and 120 μl MT buffer (MP Biomedicals) before lysis in a FastPrep® 24 instrument (MP Biomedicals) for 2 × 30 s at speed 5.5 m s−1. All remaining steps were as per the manufacturer's protocol, except that some spin and incubation times were extended. DNA was eluted in 100 μl molecular grade water (Severn Biotech Ltd.) and stored at –80°C until required.
PCR-DGGE analysis of 16S rRNA genes
Bacterial and archaeal 16S rRNA genes were amplified by either direct or nested PCR from all sediment slurry DNA extracts using DreamTaq DNA polymerase (Thermo Fisher Scientific Inc.) with primers 357FGC/518R for Bacteria and 109F/958R followed by SAFGC/PARCH519R for Archaea as previously described (Webster et al. 2006; O'Sullivan et al. 2013). All 16S rRNA gene PCR products (ca. 200 ng of each product) were separated by DGGE on 6–12% gradient (w/v) polyacrylamide DGGE gels with a 30–60% denaturant gradient (Webster et al. 2006; O'Sullivan et al. 2013). DGGE gels were stained with SYBR Gold nucleic acid stain (Invitrogen), viewed under UV and images captured using a Gene Genius Bio Imaging System (Syngene). DGGE bands, representative of all major phylotypes, were excised, reamplified by PCR, sequenced (O'Sullivan et al. 2008) and band identity determined using the NCBI nucleotide BLAST program (http://www.ncbi.nlm.nih.gov/). All 16S rRNA gene sequences reported here have been submitted to the GenBank database under accession numbers KR632942–KR632979.
Quantitative real-time PCR (qPCR)
qPCR was used to quantify 16S rRNA gene copy numbers of Bacteria and Archaea in sediment slurries. SybrGreen chemistry was used for all protocols. All qPCR reactions for standards, no template controls and sediment DNA samples were conducted in triplicate and run on an Agilent Mx3000P QPCR System (Agilent Technologies UK Ltd). For standard curves and calibration, serial dilutions of full length 16S rRNA gene PCR products from Anaerolinea thermophila DSM 14523 and Methanococcoides methylutens DSM 2657 were used as standards for Bacteria and Archaea (Webster et al. 2015). To ensure good quantification data, qPCR results were rejected if the R2 value of the standard curve was below 0.95 or the efficiency of the reaction was below 80%. The qPCR mixtures for all reactions (standards, controls and samples) were contained in a total volume of 20 μl with 400 nM of each primer (Eurofins MWG Operon), 2 μg bovine serum albumin (BSA; Promega) and 1 μl of DNA in 1× qPCRBIO SyGreen Lo-ROX Mix (PCR Biosystems Ltd) made up with molecular grade water (Severn Biotech Ltd). 16S rRNA gene primers 534F/907R and S-D-Arch-0025-a-S-17F/S-D-Arch-0344-a-S-20R were used to target the Bacteria and Archaea, respectively (Webster et al. 2015). The protocol was 95°C for 7 min, 40 cycles of 95°C for 30 s, 52°C for 30 s, 72°C for 60 s, followed by a melting curve from 55 to 95°C. Each cycle was followed by data acquisition at the elongation step.
To estimate the number of bacterial and archaeal cells, the 16S rRNA gene copy numbers were divided by the average 16S rRNA gene copy number for each taxa (4.19 and 1.71, respectively), deduced using the rrnDB database v3.1.225 (https://rrndb.umms.med.umich.edu/); (Stoddard et al. 2015). Average cell numbers and their standard deviations were calculated from each replicate.
V3–V5 16S rRNA gene tag sequencing
Variable regions 3 to 5 (V4–V5) of the 16S rRNA gene from Bacteria and Archaea were amplified from a selected number of unamended slurry DNA samples covering a range of temperatures (25, 35, 38, 46, 66°C) and time points (0, 15, 35, 62 days) using barcoded fusion primers 357F/907R (Muyzer, Dewaal and Uitterlinden 1993; Muyzer et al. 1998) and 341F/958R (Delong 1992; Ovreas et al. 1997), respectively. All PCR reactions (in triplicate) and 454 pyrosequencing were performed by the Research and Testing Laboratory (Lubbock, TX, USA; http://www.researchandtesting.com/index.html) on a Roche 454 GS FLX Titanium system. A total of 126 608 Bacteria and 70 281 Archaea sequences were obtained. Analysis of sequencing data was performed in QIIME version 1.6.0 (Caporaso et al. 2010) using a pipeline developed ‘in house’ at Cardiff University. Essentially, all sequence files were checked using Acacia software release 1.53 (Bragg et al. 2012) for quality, sequence errors and to reduce noise. Chimeras were detected and removed using the USEARCH61 algorithm and each sample randomly subsampled to the lowest number of sequences in each library (2919 for Bacteria and 1617 for Archaea). Representative OTUs were picked with UCLUST (Edgar 2010) at 97% similarity and taxonomy assigned using BLAST (Altschul et al. 1990) with the Greengenes database (DeSantis et al. 2006). Singletons and non-specific sequences were then removed and diversity estimates were calculated in QIIME.
RESULTS AND DISCUSSION
Sediment geochemistry and the effect of different temperatures on anaerobic processes in unamended sediment slurries
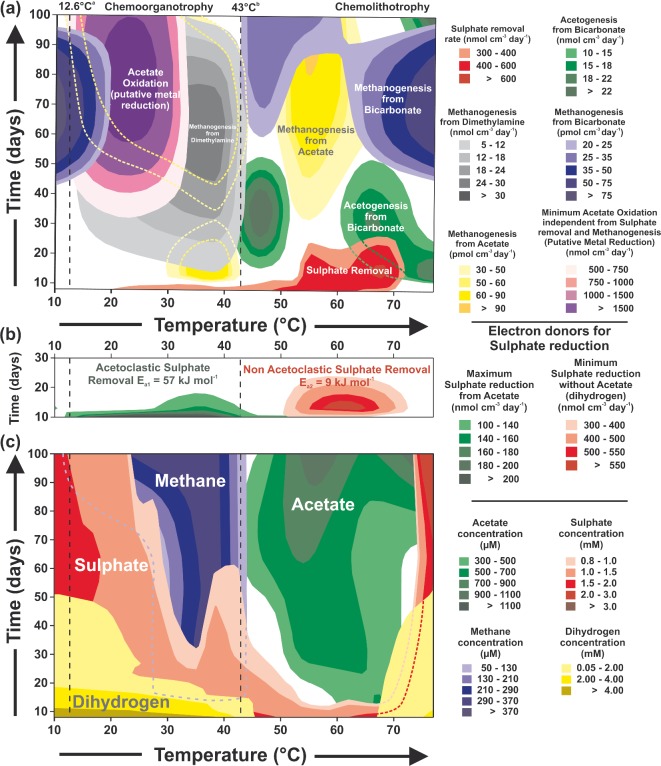
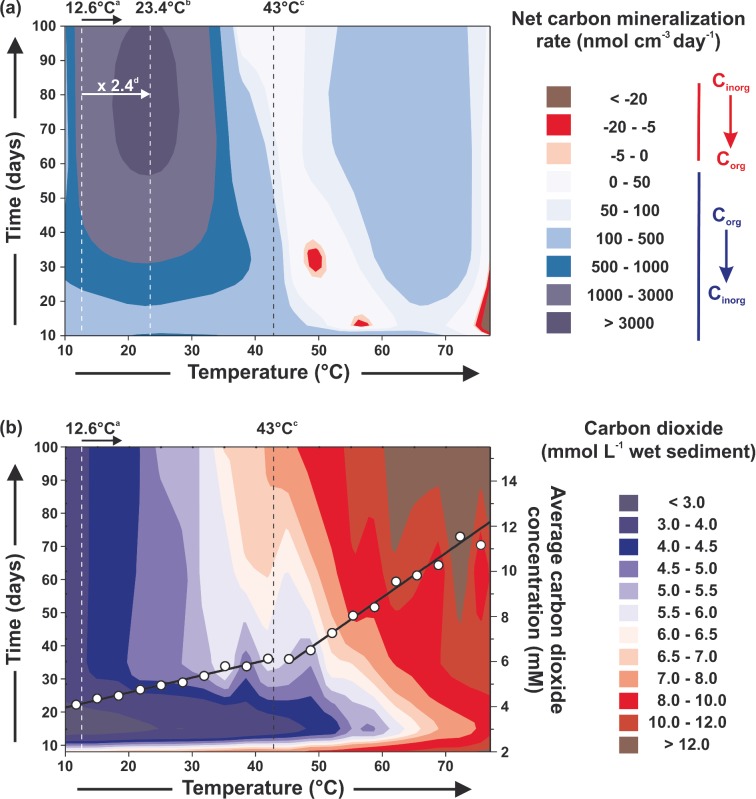
Geochemical analysis of Portishead sediment cores showed that a range of anaerobic prokaryotic activities were cooccurring in situ: metal oxide and sulphate reduction; methanogenesis; acetate, H2 and ammonia formation (Fig. S1, Supporting Information). Interestingly, acetate and H2 concentrations were overall slightly higher in summer (19°C) than winter (7°C) (summer average acetate and H2 concentrations were 9 and 7 μM higher, respectively), which suggests increased organic matter hydrolysis/fermentation at the higher summer temperatures. This is consistent with organic matter reactivity increasing with increasing temperatures (Parkes et al. 2007b; Burdige 2011), but different to the experimental results of Weston and Joye (2005) who suggested that low molecular weight dissolved organic carbon should accumulate at low temperatures. Acetate concentrations also increased over time in the unamended sediment slurries at higher temperatures, above ∼40°C (Fig. 1). Below ∼40°C, however, there was a clear sequence of activities that occurred with increasing temperatures and time as indicated by geochemical changes (Fig. 1). From ∼10°C, both sulphate and H2 were slowly removed, and additionally above ∼23°C up to ∼45°C CH4 began to accumulate. Also at ∼45°C both H2 and sulphate became rapidly depleted and acetate accumulation began. Above ∼70°C, both H2 and sulphate removal slowed and eventually stopped with increasing temperature (Fig. 1). Acetate accumulation of >1 mM occurred above 50°C, as has been shown previously (Parkes et al. 2014).
Figure 1.
Effect of temperature and incubation time on sediment slurries (∼45 μM H2). (a) Specific metabolic activities (very low metabolic rates are not shown). Yellow dashed lines show acetoclastic methanogenic rates when overlaid by other activities. Green dashed lines show rates of autotrophic acetogenesis. (b) Putative substrates for sulphate reduction. (c) Main metabolic substrates and products. Blue dashed lines represent methane concentrations when overlaid by other compounds. Red dashed lines represent sulphate concentrations. aAnnual average in situ temperature. bTemperature at which at least 50% of sulphate reduction was hydrogenotrophic.
Radiotracer activity measurements, however, demonstrated that changes in prokaryotic activity with temperature were more complex than reflected by geochemical changes, there being additional zones of different bacterial activities and changes in substrate utilization (Fig. 1). After about 40 days and at temperatures up to ∼20°C, hydrogenotrophic methanogenesis occurred. Although this result differs from the finding of acetoclastic methanogenesis being dominant at low temperatures in rice field soils (Fey and Conrad 2000), it is consistent with the presence of hydrogenotrophic methanogenesis in some low temperature, near-surface, marine sediments (e.g. Parkes et al. 2007a; Webster et al. 2009). This difference between rice field soils and marine sediments may reflect that generally freshwater systems tend to be dominated by acetoclastic methanogenesis, whilst marine sediments are dominated by hydrogenotrophic methanogenesis, as anaerobic metabolism is thought to be more focused on H2 as an intermediate (Whiticar 1999). The stimulation of hydrogenotrophic methanogenesis occurred in the presence of significant sulphate concentrations, which may be a reflection of acetate being the main substrate for sulphate reduction at low temperatures (Fig. 1, and subsequent discussion), and hence, not competing for H2.
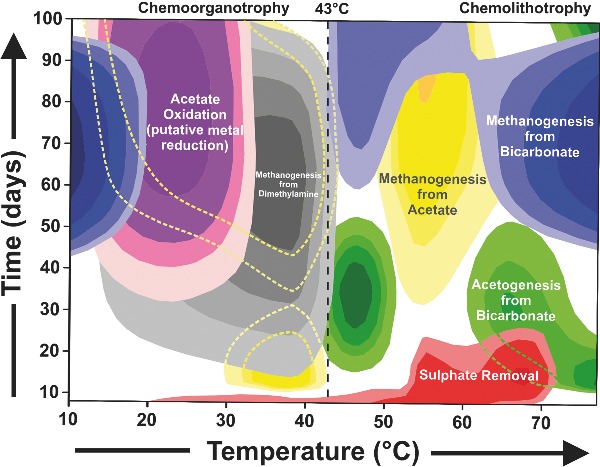
Above about 12°C, with increasing temperatures, first acetate oxidation and then increasingly sulphate removal, and methylotrophic and acetoclastic methanogenesis became significant, followed above ∼30°C by an additional discrete zone of acetoclastic methanogenesis for up to ∼30 days (Fig. 1). Above 43°C there was a marked change in processes, with a clear decrease in CH4 formation and an increase in acetate concentrations. Below 43°C the maximum rates of acetate oxidation to carbon dioxide exceeded sulphate reduction rates (≤122 times) and also occurred ∼50 days after the sulphate reduction rates had peaked; hence, total acetate oxidation must have involved an electron acceptor in addition to sulphate (Fig. 1). As this ‘additional’ acetate oxidation was also 3 × 104 times higher than the maximum rate of acetoclastic methanogenesis, this process could not have been responsible for the large ‘non-sulphate reduction’ acetate oxidation. It seems most likely that the use of metal oxides (e.g. manganese or iron oxides) for acetate oxidation was responsible for this ‘non-sulphate reduction, acetate-oxidation’, especially as other electron acceptors were limited in these anoxic sediments (e.g. nitrate concentrations <1 μM; Webster et al. 2010). Moreover, the in situ presence of dissolved pore water manganese and iron (Fig. S1, Supporting Information), significant concentrations of sedimentary metal oxides (iron and manganese oxides 5.3 and 0.1% of dry sediment, respectively) and phylogenetic analysis of active communities (Webster et al. 2010), all indicate that chemoorganotrophic metal reduction probably represents a significant process in these sediments. Hence, in agreement with in situ results for other coastal sediments (Canfield et al. 1993; Finke et al. 2007), in these slurries, sulphate reduction and putative metal oxide reduction likely accounted for most of the organic matter mineralization (26.5 and 73.2% respectively), despite the heating and H2 addition (Fig. 1).
The occurrence of methylotrophic methanogenesis above 12°C, even in the presence of significant sulphate concentrations (Fig. 1), is understandable because methylamines are not direct substrates for sulphate-reducing bacteria, and these ‘non-competitive’ substrates preferentially stimulate methanogenesis (Oremland, Marsh and Polcin 1982). Similarly, methylotrophic methanogenesis in the sediment in situ could explain the presence of CH4 alongside high sulphate concentrations (>15 mM, Fig. S1, Supporting Information). However, methylated amine concentrations were always low in Portishead sediments (below ∼120 μM detection limits), implying a rapid turnover in situ controlled by the initial depolymerization/hydrolysis of organic matter (Arnosti 2004; Parkes et al. 2012). Significant rates of methylotrophic methanogenesis above ∼12°C in the slurries, therefore, suggests that the supply of methylated substrates was increased by heating, making precursor substrates more bioavailable (Burdige 2011). This increased bioavailability may also have been responsible for the stimulation of methylotrophic acetogenesis (Fig. 2) and acetoclastic methanogenesis above ∼30°C (Fig. 1). Only the high rates of methylotrophic methanogenesis coincided with considerable CH4 accumulation (Fig. 1); this suggests that some of the CH4 produced from low rates of hydrogenotrophic methanogenesis at lower temperatures was anaerobically oxidized in the slurries (ANME 2a and 2b capable of anaerobic oxidation of methane were detected). This situation also occurs in situ, for example, in the coastal sediments of the Danish Skagerrak; there were low levels of both hydrogenotrophic methanogenesis and anaerobic oxidation of methane in the shallow subsurface, and no CH4 was present (Parkes et al. 2007a).
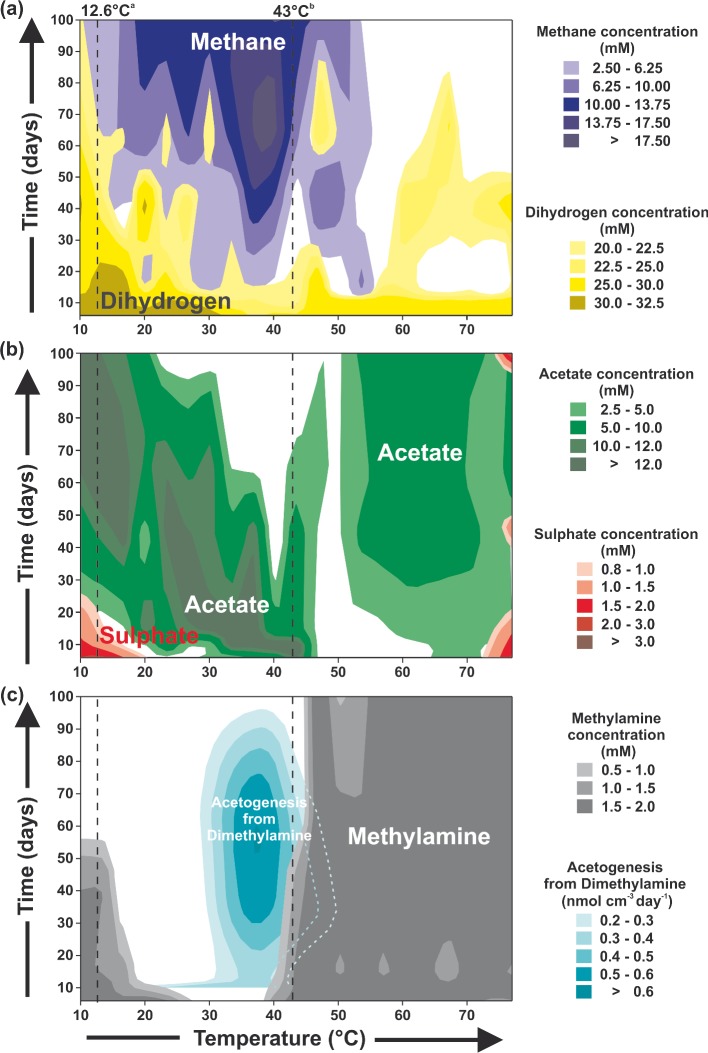
Figure 2.
Effect of temperature and incubation time on the main substrate and product concentrations in substrate-amended sediment slurries (acetate, methylamine, H2). (a) Methane and H2 concentrations. (b) Acetate and sulphate concentrations. (c) Methylamine concentrations (between 10 and 100 days) and rates of methylotrophic acetogenesis in substrate-unamended slurries (∼45 μM H2). aAnnual average in situ temperature. bTemperature at which at least 50% of sulphate reduction could have been hydrogenotrophic.
Abruptly, above 43°C, there was almost no methylotrophic methanogenesis and no CH4 accumulation (Fig. 1). In addition, methylotrophic acetogenesis slowed dramatically (Fig. 2). However, sulphate reduction continued and at faster rates, and first autotrophic acetogenesis developed and then, after about 60 days, this was replaced by hydrogenotrophic methanogenesis (Fig. 1). Also, H2 was removed to below detection limits. Between 37 and 75°C, sulphate reduction rates exceeded total acetate oxidation rates (Fig. S2a, Supporting Information). As this coincides with the zone of decreasing and zero H2 concentrations (Fig. 1), this strongly suggests that H2 became a major substrate for sulphate reduction at higher temperatures. Calculations indicate that at 60°C, at least 71% of sulphate reduction occurred without acetate as a substrate (Fig. S2a, Supporting Information). Interestingly, at 43°C at least 50% of sulphate reduction was using H2 as a substrate, although below this temperature, acetate was the main sulphate reduction substrate (Fig. S2 c, d and f, Supporting Information). Therefore, 43°C is also an important critical temperature for sulphate reduction, marking a switch from predominantly organotrophic to lithotrophic metabolism. Sulphate was only completely removed at temperatures where H2 was the main substrate for sulphate reduction (Fig. 1). This was probably due to organic substrate limitation because in the replicate substrate-amended slurries (2 mM acetate and methylamine), in addition to H2 (∼36 mM gas equivalent), sulphate was completely removed between 3.2 and 73.8°C (Fig. 2b).
As sulphate removal rates increased to maximal above ∼53°C, abruptly autotrophic acetogenesis ceased, and when sulphate was depleted, a zone of acetoclastic methanogenesis developed with maximum rates at ∼55°C and after 80 days (Fig. 1). This change in metabolism occurs at the same temperature, >50°C, as that associated with the switch to the dominance of syntrophic acetate oxidation coupled with hydrogenotrophic methanogenesis in other anaerobic environments (e.g. Ho, Jensen and Batstone 2013). Dolfing, Larter and Head (2008) suggested that there are thermodynamic ‘windows of opportunity’ for various anaerobic metabolisms involving methanogenesis, and the ‘window of opportunity’ in these marine sediment slurries above ∼50°C may favour acetoclastic methanogenesis rather than syntrophic acetate oxidation. The developing high acetate concentrations (∼1 mM and higher) combined with zero H2 concentrations may be aspects of this slurry, which provide a window for acetoclastic methanogenesis between about 50 and 65°C (Fig. 1). At higher temperatures sulphate removal still occurred, but in association with autotrophic acetogenesis again. By about 20 days, however, sulphate reduction became sulphate limited and autotrophic acetogenesis occurred on its own. This acetogenesis was replaced by hydrogenotrophic methanogenesis after about 50 days. The temporal sequence of both these H2 utilizing processes continued at increasing temperatures, although with an increasing time gap between them, as the zone for autotrophic acetogenesis shrank faster than the zone for hydrogenotrophic methanogenesis expanded (Fig. 1). Despite these active H2 utilizing processes at elevated temperatures, above ∼67°C, added H2 removal became restricted. However, it is unclear whether H2 consumption at these high temperatures became balanced by H2 formation from sedimentary organic matter (Parkes et al. 2007b). The upper temperature limit for sulphate reduction in these slurries was ∼73°C (Fig. 1). This upper temperature is almost identical to that of a thermophilic, spore-forming, sulphate-reducing bacteria (Desulfotomaculum sp. C1A60, phylum Firmicutes) previously isolated from Portishead sediments, when growing on H2 (72°C, O'Sullivan et al. 2015). Thermophilic, spore-forming sulphate-reducing bacteria are widespread in marine sediments (Muller et al. 2014). As the highest acetate concentrations occur separately from the main zones of autotrophic acetogenesis (Fig. 1), an additional source of acetate formation must occur, which is presumably, heterotrophic, associated with the temperature activation of organic matter (Parkes et al. 2007b).
The effect of different temperatures on anaerobic metabolism in substrate-amended sediment slurries
Similar results to the above also occurred in the substrate-amended slurries, with the higher substrate concentrations allowing their utilization to be directly analysed (Fig. 2). The total sulphate reduction temperature range remained unchanged with substrate addition (<3 to 73°C), but rates were faster and sulphate depletion more extensive. These results suggest that the syntrophs supplying sulphate-reducing bacteria with substrates may have the same temperature range as the sulphate-reducing bacteria, or that sulphate-reducing bacteria were independent of syntrophs, which seems unlikely as close coupling between fermenters and sulphate reducers has been previously shown (Finke and Jørgensen 2008). In contrast, addition of substrates did extend the temperature range for CH4 formation, especially at lower temperatures, from 23–44°C to 7–55°C. Hence, methanogens had a wider temperature range than their syntrophs and appeared to be substrate limited in the unamended slurries at temperatures below ∼20°C. However, curiously, in both slurry conditions the highest H2 concentrations occurred below ∼30°C (Figs 1 and 2), and in the non-substrate-amended slurries there was also H2 formation at these temperatures (>45 μM) and stimulation of hydrogenotrophic methanogenesis. Perhaps at these temperatures and conditions, there is uncoupling between H2 formation and consumption, H2 leakage (Finke et al. 2007) or other limitations on CH4 production.
Methylamine degradation did occur at low temperatures, but was much slower below than above the average in situ sediment temperature (12.6°C, Fig. 2) and was associated with acetate formation. For example, complete methylamine removal took ∼55 days at ∼10°C, compared to a few days above 20°C. As maximum methylamine degradation did not occur around the average in situ temperatures, other factors than just temperature adaptation must control optimal methylamine degradation. Methylamine degradation also resulted in ammonium accumulation, with maximum concentrations being reached between 10 and 15 days and at 20–40°C (4 mM). However, with continued incubation maximum ammonium concentrations decreased (reaching a minimum of ∼2 mM between 20 and 28°C), suggesting that anaerobic ammonium oxidation was occurring. Above 43°C, methylamine degradation stopped abruptly. This was consistent with the cessation of 14C-dimethylamine metabolism (methanogenesis and acetogenesis) in the non-substrate-amended slurries (Figs 1 and 2). Also similar to the non-substrate-amended slurries, acetate accumulated above 43°C, but concentrations were overall lower than below 43°C, in part probably due to methylamine degradation to acetate below 43°C. Complete H2 removal over time above ∼43°C broadly reflected the two hydrogenotrophic methanogenic zones in the non-substrate-amended slurries (Fig. 1). Despite this indication of hydrogenotrophic methanogenesis above ∼55°C and its direct measurement in non-substrate-amended slurries, no cooccurring CH4 was present in either slurry. This might be due to CH4 oxidation at higher temperatures, above ∼45°C (Kallmeyer and Boetius 2004; Holler et al. 2011).
The effect of different temperatures on bacterial and archaeal cell numbers in sediment slurries
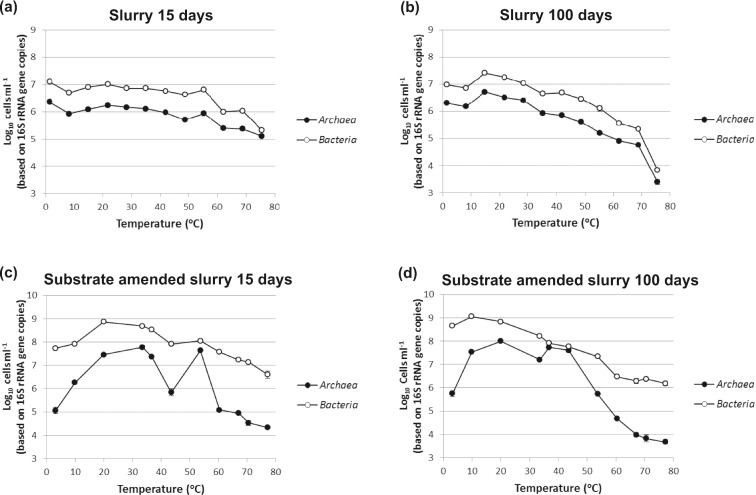
Considerable numbers of bacterial and archaeal cells were in the non-substrate-amended slurries at 15 days (∼106–107 cells based on 16S rRNA gene copies mL−1, Fig. 3), with bacterial cell numbers being consistently higher than archaeal cell numbers. Both archaeal and bacterial cell numbers decreased markedly above about 55°C. After 100 days, bacterial and archaeal cell numbers had increased with a peak at ∼15°C, slightly above the annual average in situ temperature (12.6°C), and close to the zone of maximum acetate oxidation (Fig. 1). Cell numbers subsequently decreased with increasing temperature, for Bacteria, in a bimodal pattern with relatively low numbers at ∼35°C (∼4.5 × 106 ml−1), and overall, decreasing slightly faster than archaeal cells. Above 70°C for both bacterial and archaeal cells, there was a more dramatic decrease. These differences between bacterial and archaeal cell distributions between ∼15 and 70°C perhaps may be due to a combination of the inherently greater thermal tolerance of archaeal membranes (Koga 2012) and germination of thermophilic bacterial spores at higher temperatures (corresponding with an intense zone of thermophilic sulphate reduction [Fig. 1]). However, for both cell types the 100-day cell numbers were lower above ∼50°C compared to the 15 day counts; hence, prokaryotic populations overall decreased due to prolonged incubation at thermophilic temperatures.
Figure 3.
The effect of temperature and time (15 and 100 days) on Bacteria and Archaea cell numbers (16S rRNA gene copies) in unamended (a and b) and substrate amended (c and d) sediment slurries. Cell numbers were calculated from 16S rRNA gene copy numbers by using the average 16S rRNA gene copy number for each taxa (4.19 and 1.71 copies for Bacteria and Archaea, respectively) deduced using the rrnDB database (Stoddard et al. 2015). Standard deviations are plotted but are mostly within the size of the symbols.
The cell distributions with temperature in the substrate-amended slurries were similar to the above for Bacteria, except numbers were over a factor of 10 higher and did not decrease so markedly with temperature, especially above 60°C (Fig. 3). This difference may reflect that bacterial populations with adequate substrate supply were able to respond more effectively to increasing temperatures, such as by membrane lipid changes, and greater spore germination/growth of thermophiles [e.g. increased detection of Firmicutes (Fig. 4) and more complete sulphate removal (Fig. 2)]. Archaeal cell distributions in the substrate-amended slurries were very different, with two peaks: a broad peak around ∼20 to 30°C and a sharper peak at ∼40 to 50°C, present at both 15 and 100 days. This also indicates growth of archaeal cells, although restricted to around meso and lower thermophilic temperatures. Surprisingly, at 100 days archaeal cell numbers decreased more rapidly at temperatures above ∼43°C than did bacterial cells, which is opposite to what occurred in the non-substrate added slurries. This decrease in archaeal cells coincides with the cessation in methylamine degradation (Figs 1 and 2), suggesting that some Archaea may have a role in anaerobic degradation of methylamines, and is another impact of the 43°C critical temperature. In addition, above ∼57°C archaeal cell numbers in the substrate-amended slurry at both 15 and 100 (except 76°C at 100 days) days were actually lower than in the non-substrate-amended slurry. These data suggest that at elevated temperatures, the presence of significant substrate concentrations is detrimental to many archaeal cells, whilst stimulating bacterial populations (Fig. 3). Surprisingly, bacteria dominate the total prokaryotic population at all temperatures, at both 15 and 100 days, and with and without added organic substrates. The theoretically more thermally robust archaeal cells (Valentine 2007; Koga 2012) were thought to be dominant in subsurface sediments (Biddle et al. 2006; Lipp et al. 2008) and some high-temperature environments, but here they only became significant around the two peaks in their cell numbers at ∼30 and 50°C (10 and 40%, respectively of the total population).
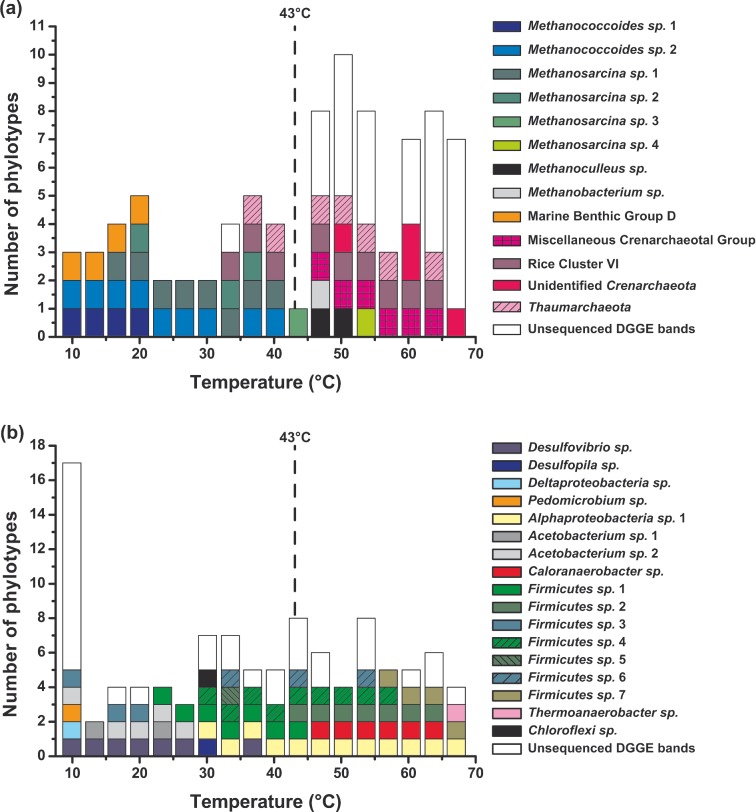
Figure 4.
The effect of temperature on 16S rRNA gene diversity (PCR-DGGE) in substrate-amended slurries incubated for 100 days at different temperatures. (a) Archaea and (b) Bacteria.
The effect of different temperatures on prokaryotic community composition in sediment slurries
Cluster analysis of PCR-DGGE (Fig. S3, Supporting Information) and ARISA data (not shown) of the unamended slurries showed a distinct bacterial community change at temperatures above 40°C already at 15 days. These temperature effects were expanded by 100 days with distinct bacterial communities at 8–22°C, 25–35°C, 39°C, 42–62°C and 69–75°C (Fig. S3, Supporting Information), and for Archaea at 5°C, 8–15°C, 18–45°C, 49°C, 39–65, 52–59°C, 62°C, 69–72°C and 75–79°C (Fig. S4, Supporting Information). Overall, at both 15 and 100 days there was a decrease in the number of phylotypes above ∼40°C. In addition, the 100-day slurry incubated at 1°C clustered with the original slurry, indicating that low temperature incubation in the Thermal Gradient did not change the original slurry community and observed changes were, therefore, a result of the elevated incubation temperatures. The prokaryotic composition of the original slurry was also similar to the fresh sediment (based on 454 pyrosequencing of 16S rRNA genes, unpublished results), so the Thermal Gradient community changes should be representative of the response of the original sediment community. The large number of faint DGGE bands in the unamended slurries made excision and sequencing of bands difficult; however, successful sequencing of some intensely stained bands for Bacteria showed the presence of Deltaproteobacteria below ∼30°C, Deltaproteobacteria and Firmicutes between ∼30 and 35°C and then a mixture of different Firmicutes, including Clostridia-like Bacteria, above ∼40°C.
The presence of Deltaproteobacteria and Firmicutes was also confirmed by pyrosequencing of 16S rRNA gene amplicons and these together with Chloroflexi, Actinobacteria, Bacteroidetes, Acidobacteria and candidate division OP8 were the most abundant bacterial phyla (90%; Fig. S5, Supporting Information). Proteobacteria, including sequences related to the psychrophilic and heterotrophic sulphate-reducing bacteria, Desulfotalea spp. and Chloroflexi were most abundant at low temperatures, but decreased with increasing temperature significantly as Firmicutes, including sequences related to thermophilic, spore-forming and H2-utilizing sulphate-reducing bacteria, Desulfotomaculum spp. (Hubert et al. 2010; de Rezende et al. 2013; O'Sullivan et al. 2015) proliferated toward a peak abundance at 46°C (80% of bacterial phylotypes). Acidobacteria, Bacteriodetes and candidate division OP8 also decreased with increasing temperature, whereas Actinobacteria slightly increased (Fig. S5, Supporting Information). Members of the phylum ‘Bathyarchaeota’ (formerly MCG; Meng et al. 2014) were the most abundant archaeal phylum at all times and temperatures based on pyrosequencing (consistently >45% of Archaea). However, different bathyarchaeotal phylotypes were abundant at different temperatures suggesting considerable physiological diversity within the group. Thaumarchaeota (putative ammonium oxidizers), Euryarchaeota [including the H2-utilizing methanogenic Methanomassiliicocaceae (Dridi et al. 2012); Methanobacteriales and Methanomicrobiales; the substrate versatile methanogens Methanosarcinales, which were most abundant; and sequences related to the anaerobic methane oxidising clade ANME 2a-2b, most abundant at 35°C] and Parvarchaeota (Rinke et al. 2013) were also present but in much lower proportions (Fig. S6, Supporting Information).
Consistent with the significant growth in both bacterial and archaeal populations at a range of temperatures (Fig. 3), PCR-DGGE profiles of the substrate-amended slurries had more intensely stained bands compared to those of the unamended slurries, and this allowed robust sequence analysis. These sequences presumably represent prokaryotes that had grown under their optimum geochemical and temperature conditions, and predominated over organisms just surviving or dying slowly; hence, the community composition of the substrate-amended slurries may provide a clearer link to the changes in metabolism at different temperatures. However, there were still considerable similarities between the community composition changes with temperature in both slurry conditions. In the substrate-amended slurries below ∼20°C where there was both active sulphate reduction and methanogenesis, but neither sulphate or H2 was depleted (Fig. 2), the bacterial community was dominated by Deltaproteobacteria, including organotrophic, incomplete oxidizing sulphate-reducing bacteria (Desulfovibrio), Acetobacterium (acetogens) and different Firmicutes (Fig. 4), consistent with sulphate removal and acetate formation. The Archaea were dominated by the methylotrophic Methanococcoides methanogens (Fig. 4), presumably responsible for the limited non-competitive methylamine utilization, and hence, CH4 production in the presence of sulphate. Marine Benthic Group D/Thermoplasmatales sequences were also present and although these are presently uncultured, genome data indicates that they are capable of exogenous protein degradation in cold anoxic environments (Lloyd et al. 2013), whilst phylogenetic analysis suggests that some clades may even be methanogenic (Paul et al. 2012). Their presence, however, was restricted to temperatures below ∼20°C. Around 20°C substrate versatile Methanosarcina methanogens appeared, in addition to Methanococcoides, and were present up to the 43°C critical temperature, where they became the sole archaeal phylotype present. This temperature range corresponds with maximum CH4 concentrations and methylamine utilization (Fig. 2). Above ∼20°C the bacterial community also changed with additional types of Firmicutes present, this corresponded with more rapid methylamine and sulphate removal, and acetate production (Fig. 2). Above 30°C Alphaproteobacteria appear along with different Firmicutes. Alphaproteobacteria were then consistently present up to 70°C, members of this highly diverse group have been detected in a subsurface oil reservoir (Kryachko et al. 2012) and hydrothermal vents (Takai et al. 2009).
At 43°C the composition of Firmicutes again changed (Firmicutes sp. 2 appear, Fig. 4), and at slightly higher temperatures this was augmented by Caloranaerobacter, a thermophilic, anaerobic, organotrophic bacterium, species of which have been isolated from deep-sea hydrothermal vents (Wery et al. 2001; Jiang et al. 2014), and which can produce acetate as a fermentation product. Caloranaerobacter was present up to ∼65°C, which is the upper temperature limit for cultured isolates (Wery et al. 2001), and acetate concentrations again increased (Fig. 2). Above ∼55°C other Firmicutes groups appeared (Firmicutes sp. 7, Fig. 4), including above ∼65°C Thermoanaerobacter, a heterotrophic, thermophilic anaerobe, the metabolism of which involves both acetate and H2, and some species have been isolated from the deep subsurface (Fardeau et al. 2000; Roh et al. 2002). The composition of Archaea changes even more markedly above 43°C than did Bacteria (Fig. 4), from the single Methanosarcina phylotype to the first appearance of Methanoculleus and then its presence up to ∼50°C, which is consistent with a temperature window for hydrogenotrophic methanogenesis documented in the unamended slurries (Fig. 1) and continued CH4 formation in the amended slurries (Fig. 2). This upper temperature limit is the same as for Methanoculleus submarinus isolated from deep-sea sediments (247 m; Mikucki et al. 2003). The hydrogenotrophic methanogen, Methanobacterium was also present but only at ∼45°C. This genus has thermophilic species (Zeikus and Wolfe 1972) and has also been detected in subsurface environments (Kotelnikova and Pedersen 1997; Kryachko et al. 2012). In addition, Thaumarchaeota originally first present above 35°C became a consistent member of the archaeal population (Fig. 4). All known members of Thaumarchaeota are chemolithoautotrophic ammonia oxidizers and this is consistent with ammonium removal between ∼20 and 40°C. However, they have not so far been shown to be capable of anaerobic growth (Stahl and de la Torre 2012) despite being common in anoxic, subseafloor sediments (Parkes et al. 2014). Our results suggest that some Thaumarchaeota may be capable of anaerobic metabolism. In addition, the continued presence of Thaumarchaeota above 40°C indicates metabolism other than ammonia oxidization, as suggested in some other environments (Mussmann et al. 2011; Stahl and de la Torre 2012). Above 45°C the common subseafloor ‘Bathyarchaeota’/MCG appears and is present up to ∼65°C. These Archaea have been detected in sediments up to 95°C (Biddle et al. 2012) and are thought to be heterotrophic (Lloyd et al. 2013), and perhaps activation of recalcitrant organic matter at higher temperatures stimulated their growth (Parkes et al. 2007b). The temperature distribution of the Bathyarchaeota/MCG was similar to another uncultured archaeal group, the probably non-methanogenic Rice cluster VI (Fey and Conrad 2000) which has also been found in deep, thermophilic, subseafloor sediments (Roussel et al. 2008).
These biodiversity results demonstrate that there are clear changes in community composition with temperature, often over narrow temperature intervals, and that these changes match those in biogeochemical activity, and the temperature ranges of cultivated representatives. The distribution and activity of methanogens, sulphate-reducing and acetogenic bacteria was supported by the distribution of their functional genes (mcrA, dsrB and fths, respectively, unpublished results).
The dominant anaerobic metabolic processes in substrate-unamended slurries
Regardless of the temperature, sulphate reduction and acetate oxidation were major metabolic activities in these coastal sediment slurries (Fig. 5), with highest activity rates being carbon dioxide from acetate oxidation (1644 nmol cm−3 day−1 at 25°C) and sulphate reduction (602 nmol cm−3 day−1 at 69°C) (Table S2, Supporting Information). Acetate was an important compound over the whole temperature range (10–77°C), both as a substrate for some metabolic processes and product of others. Total acetate oxidation to carbon dioxide increased with temperature up to a maximum at 25°C, with rates 7.5 times greater below 40°C than above this temperature. As previously described, metal oxide reduction must have been responsible for the majority of acetate oxidation, as often occurs in coastal marine sediments (Canfield et al. 1993).
Figure 5.
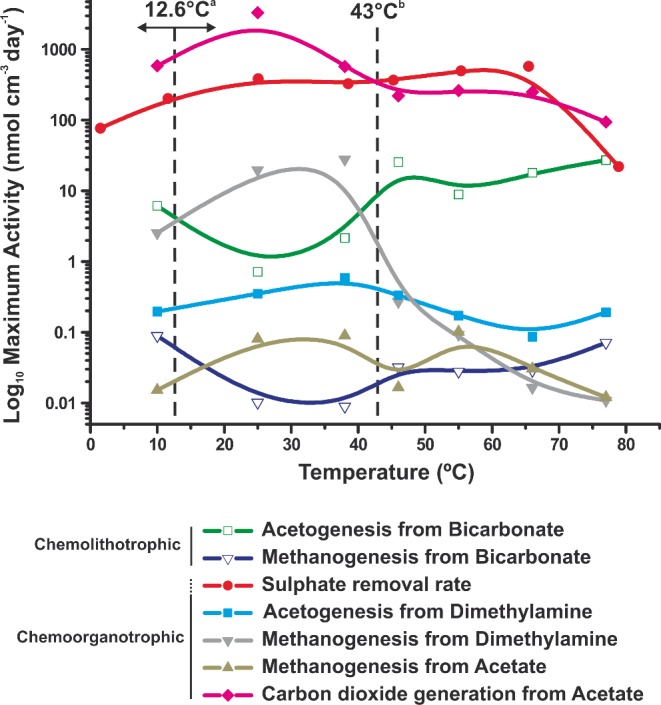
Maximum activity rates for studied metabolic processes at different temperatures in sediment slurries (H2 ∼45 μM). aAnnual average in situ temperature (Arrows indicate range between minimum and maximum in situ temperature). bTemperature at which at least 50% of sulphate removal was hydrogenotrophic.
The effect of temperature on sulphate reduction rates was also variable with a bimodal peak (R2 = 0.78, Fig. 1 and S2, Supporting Information), suggesting that total sulphate removal was due to two sulphate reduction processes with different optimal temperatures (29 and 61°C), as found in other sediments (Isaksen, Bak and Jørgensen 1994; Hubert et al. 2009). Here we also show that the two sulphate reduction processes were probably utilizing different substrates (Fig. 1b), and likely involved different bacteria (Fig. 4). Sulphate reduction was the second most important chemoorganotrophic process (Fig. 5) and below 43°C was most likely coupled to acetate oxidation. That this activity was due to substrates other than acetate (e.g. lactate, propionate, butyrate and valerate; Parkes et al. 1989) was unlikely, as these compounds were either below detection limit (<1 μM) or their concentration profiles were not correlated with sulphate reduction (data not shown). Acetate is also the major in situ substrate for sulphate reduction in marine sediments (Parkes et al. 1989). Between 37 and 75°C, however, sulphate reduction rates exceeded total acetate oxidation rates (Fig. S2, Supporting Information) and coincided with complete removal of H2 (Fig. 1), which strongly suggests that H2 had become the major substrate for sulphate reduction (e.g. 71% of sulphate reduction at 60°C, Fig. S2, Supporting Information). Interestingly, at the 43°C critical temperature H2 was probably driving at least 50% of sulphate reduction, with acetate responsible for the rest (Fig. S2, Supporting Information).
A switch from acetoclastic to hydrogenotrophic anaerobic terminal oxidizing processes has been reported previously but for methanogenesis in rice fields and lake sediments at temperatures higher than about 30–40°C (Fey and Conrad 2000; Nozhevnikova et al. 2007). The temperature-driven transition between substrates for sulphate reduction in these slurries might be a consequence of (1) acetoclastic sulphate and metal oxide reducers being restricted by temperature, as suggested by the change in bacterial diversity (Fig. 4); (2) acetoclastic processes being outcompeted for electron acceptors by hydrogenotrophic processes at high temperatures; and/or (3) specific syntrophic relationships between chemoorganotrophic acetogens and acetate oxidizers up to ∼40°C might become disrupted at higher temperatures, resulting in H2 becoming the dominant anaerobic intermediate and acetate accumulation (Fig. 1c). Assuming that this also occurs in situ, it would provide an additional mechanism for acetate accumulation in deep sediments above ∼40°C (Parkes et al. 2007b), including petroleum reservoirs (Seewald 2003).
Carbon mineralization controls and temperature
Although the Portishead sediment depth profiles had very low sulphate reduction activity (Fig. S1, Supporting Information), sulphate removal rates in unamended Portishead sediment slurries (plus H2) were in the range for active coastal sediments (Jørgensen 1982), demonstrating that some anaerobic metabolic processes in these tidal flat sediments were strongly substrate limited. Anaerobic processes were also strongly controlled by temperature, as has been previously shown (e.g. Middelburg et al. 1996); however, changes were not linear and the response of individual processes to temperature increases was variable (Fig. 5). In addition, average net total carbon dioxide production rates (calculated from overall carbon dioxide production and consumption rates of each metabolic activity) were actually greater below 43°C (8.7 times, 781 nmol cm−3 day−1) than above this temperature. Hence, processes oxidizing organic carbon dominated below 43°C (Fig. 6). Marked changes in archaeal and bacterial diversity also occurred with temperature, particularly around this 43°C critical temperature (Fig. 4, S3–6, Supporting Information), demonstrating that distinct prokaryotic communities with specific metabolisms for each temperature ‘window of opportunity’ (Dolfing, Larter and Head 2008) were driving the temperature-related biogeochemical changes (Figs 1 and 2). Rates of chemoorganotrophic methanogenesis, acetogenesis, sulphate and putative metal oxide reduction all decreased rapidly above 43°C, whilst chemolithotrophic sulphate reduction, acetogenesis and methanogenesis were all stimulated (Figs 1 and 2), suggesting that a common factor might control the changes in both types of processes. Carbon dioxide partial pressures increased with temperature and time, particularly above 43°C where rates of increase were three times greater (Fig. 6b). Since biotic organic matter oxidation rates decreased above 43°C (Fig. 6a), this carbon dioxide increase must have been due to other mechanisms, such as decreased carbon dioxide solubility (Fig. S7, Supporting Information), and slurry acidification from organic acid accumulation (e.g. acetate; Fig. S7, Supporting Information). However, the main driving processes were probably related to the type of catagenic reactions occurring above ∼50°C during organic matter burial and maturation (Horsfield et al. 2006) responsible for increasing carbon dioxide concentrations in petroleum formations (Seewald 2003).
Figure 6.
(a) Effect of temperature and incubation time on the net carbon mineralization balance of sediment slurries (∼45 μM H2). Net carbon mineralization balance was calculated by applying the standardizing factor in Table S1 (Supporting Information) to each specific rate, and then summing these. (b) Effect of temperature and incubation time on carbon dioxide concentrations measured from the slurry headspace (left vertical axis) and on the average carbon dioxide concentrations (open circles and right vertical axis). aAnnual average in situ temperature (Arrow indicates maximum average in situ temperature). bTemperature of maximum mineralization rate. cTemperature at which at least 50% of sulphate reduction was hydrogenotrophic. dFactor of increase of mineralization rate.
In these slurries (Fig. 6b), the increasing carbon dioxide concentrations with temperature may be responsible for enhancing chemolithotrophic metabolism, as increased carbon dioxide concentration has been shown to impact both prokaryotic activities and communities in other marine sediments (Mayumi et al. 2013; Yanagawa et al. 2013). In addition to the direct substrate increase for chemolithotrophic metabolism, carbon dioxide increase may cause product inhibition of organic carbon oxidation by chemoorganotrophic processes above ∼43°C. A potential causal relationship between the beginning of more rapid increases in carbon dioxide concentrations (Fig. 6b) at the 43°C critical temperature window and fundamental biogeochemical changes is suggested by the following: (1) the abrupt cessation of methylotrophic methanogenesis and acetogenesis, (2) the end of complete methylamine removal and (3) the reduction in biogenic carbon dioxide production, being combined with, (4) the start of hydrogenotrophic acetogenesis, (5) increasing dominance of hydrogenotrophic sulphate reduction, (6) prokaryotic diversity changes and (7) start of the rapid decrease in archaeal cell numbers in the substrate-amended slurries; all occurring around 43°C.
Different metabolic windows of opportunity are reflected in different optimal temperatures and incubation times for different anaerobic processes (Table S2, Supporting Information). Interestingly, the two autotrophic processes of acetogenesis and methanogenesis both had two temperature optima and these were the same (≤10°C, ≥77°C), whilst heterotrophic processes only had one temperature optimum and these varied from 25 to 55°C. Sulphate reduction, which had acetate as a substrate at lower temperatures but H2 as a substrate at higher temperatures (Fig. 1), had optimum activities during the zone of predominantly H2 utilization (∼95% H2 utilization, 69°C; Table S2 and Fig. S2, Supporting Information). Sulphate reduction also had two activation energies, the values during high-temperature H2 utilization being ∼6 times lower than during lower temperature acetate utilization (8.9 and 56.8 kJ mol−1), and hence, H2 utilization was a highly favoured reaction at higher temperatures which resulted in maximum activities and complete H2 removal (Fig. 1). The response of activities to a 10°C temperature increase (Q10 between 10 and 20°C, Table S2, Supporting Information) varied considerably from 1.1 (hydrogenotrophic sulphate reduction, acetoclastic methanogenesis) to 2.5 (hydrogenotrophic acetogenesis, methylotrophic methanogenesis), suggesting considerable differences in their response to climate warming and temperature increases during sediment burial.
SUMMARY
It is generally considered that the impact of temperature on anaerobic biogeochemical processes in marine sediments, and other environments, would be controlled by the temperature characteristics of the prokaryotic community (e.g. Robador, Bruchert and Jørgensen 2009). Hence, there should be a progression from psychrophilic, mesophilic to thermophilic etc. populations for the dominant metabolism and generally increasing activities as temperatures increase. However, the results presented here, which greatly extend previous findings from a range of different environments (e.g. rice paddies, oil reservoirs, marine sediments, and unpublished mud volcano sediments studies), demonstrate that temperature increases have a direct, non-linear effect on the dominant biogeochemical processes and causative prokaryotes in a series of temperature windows of ‘opportunity’ with surprisingly rapid metabolic changes over small temperature increases (Figs 1 and 2). Here a critical 43°C temperature window was related to a more rapid increase in carbon dioxide concentrations (Fig. 6b) and this may also occur in other environments, particularly in subsurface sediments with increasing temperatures and carbon dioxide concentrations with depth (Seewald 2003). Deep hot sediments also have elevated acetate concentrations (Parkes et al. 2007b), as occurred in these slurries. Some activities such as hydrogenotrophic methanogenesis had multiple windows of opportunity within a large temperature range (∼10 to 80°C). Others including acetate oxidation had a more restrictive temperature range for maximum activities and this was also dependent on availability of electron acceptors [metal oxide (up to ∼34°C) and sulphate (up to ∼50°C), Fig. 1]. In addition, some activities switched substrates with temperature, this occurred for sulphate reduction, with acetate being the main substrate below 43°C and H2 being the main substrate above 43°C (Fig. 1, both with different energy of activations). This is consistent with (1) the widespread occurrence of spores of thermophilic sulphate-reducing bacteria, Firmicutes—Desulfotomaculum and other types, in near-surface, marine sediments (Muller et al. 2014) and their widespread activity in deep, hot sediments (Aullo et al. 2013) and (2) the prevalence of Firmicutes in our slurries and their species changes above 43°C (Fig. 4). A close relationship between functional biogeochemical changes and structural community changes occurred across the whole temperature range (∼0 to 80°C); for example, the presence of methylotrophic Methanococcoides methanogens was related to methylamine removal, methylotrophic methanogenic activity and methane formation (Fig. 1 and 2, and when methanogenic substrates changed above 43°C so did the types of methanogens present (Fig. 4). Several of the uncultured Archaea, characteristic of deep marine sediments, such as Marine Benthic Group D and Miscellaneous Crenarchaeatol Group/’Bathyarchaeota’, developed in the slurries and this indicated their possible metabolisms and temperature ranges.
Bacterial cell numbers were always higher than archaeal cells (Fig. 3), although in the non-substrate-amended slurries above ∼15°C bacterial cell numbers decreased rather more rapidly than the more robust archaeal cells with increasing temperatures. This situation has been suggested to occur in deep marine sediments (Schouten et al. 2010). Both bacterial and archaeal cell numbers decreased markedly above ∼70°C which could provide substrates in the form of necromass for prokaryotes able to grow at these temperatures (Parkes et al. 2014). Although both archaeal and bacterial cell numbers increased due to substrate addition, the pattern of their response was very different (Fig. 3). Archaeal cells increased around mesophilic and lower thermophilic temperatures (reaching ∼10 and 40% of the total prokaryotic population, respectively) and then decreased rapidly above the 43°C critical temperature. This coincided with the abrupt cessation in methylamine degradation, suggesting that some Archaea were degrading methylamines. Surprisingly, above ∼57°C archaeal cell numbers were actually lower than in the non-substrate-amended slurries (except 76°C at 100 days). This suggests that at these high temperatures, the presence of significant substrate concentrations is detrimental to some archaeal cells. In contrast, bacterial cell numbers were stimulated at all temperatures by substrate addition, including above 70°C (∼10 times higher). This differential response to substrate addition by Bacteria and Archaea with increasing temperatures may be significant in some high temperature and high substrate environments, such as, oil reservoirs.
In addition to temperature windows changing the structure and function of prokaryotic communities across a wide temperature range, the data presented here shows that even small temperature increases can have a significant impact on anaerobic organic matter degradation. For example, a temperature increase of only 2°C above the average annual in situ temperature of Portishead tidal flat sediments would increase organic carbon mineralization by 40% (Q10, 10–20°C, 1.1 to 2.5), further potentially contributing to total greenhouse gas emissions. Furthermore, the effect of temperature increase on organic carbon mineralization is also enhanced by substrate addition (Fig. 2). Hence, in addition to direct temperature effects of global warming, potential-associated eutrophication of coastal environments and elevated organic matter input would further increase the intensity of anaerobic activity and deleterious environmental impacts.
Supplementary Material
Acknowledgments
The authors thank Miriam Olivier for assisting with the summer core sampling, and Ian McDonald for the ICP-MS analyses. This work was supported by NERC UK grant numbers NE/F00477X/1, NE/F018983/1, NE/H021531/1, NE/H02042X/1 and European Community's Seventh Framework Program (FP7/2007-2013) under the HERMIONE project, grant agreement n° 226354.
Conflict of interest. None declared.
Footnotes
Present address: Laboratoire de Microbiologie des Environnements Extremes, UMR6197, IFREMER, CNRS, UBO, Technopôle Brest Iroise, Plouzané, France.
SUPPLEMENTARY DATA
REFERENCES
- Aller RC, Yingst JY. Relationships between microbial distributions and the anaerobic decomposition of organic-matter in surface sediments of long-island sound, USA. Mar Biol. 1980;56:29–42. [Google Scholar]
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnosti C. Speed bumps and barricades in the carbon cycle: substrate structural effects on carbon cycling. Mar Chem. 2004;92:263–73. [Google Scholar]
- Aullo T, Ranchou-Peyruse A, Ollivier B, et al. Desulfotomaculum spp. and related Gram-positive sulfate-reducing bacteria in deep subsurface environments. Front Microbiol. 2013;4:362. doi: 10.3389/fmicb.2013.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle JF, Cardman Z, Mendlovitz H, et al. Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 2012;6:1018–31. doi: 10.1038/ismej.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle JF, Lipp JS, Lever MA, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. P Natl Acad Sci USA. 2006;103:3846–51. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg L, Stone G, Imelfort M, et al. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods. 2012;9:425–6. doi: 10.1038/nmeth.1990. [DOI] [PubMed] [Google Scholar]
- Burdige DJ. Temperature dependence of organic matter remineralization in deeply-buried marine sediments. Earth Planet Sc Lett. 2011;311:396–410. [Google Scholar]
- Canfield DE, Jørgensen BB, Fossing H, et al. Pathways of organic-carbon oxidation in three continental-margin sediments. Mar Geol. 1993;113:27–40. doi: 10.1016/0025-3227(93)90147-n. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Klose M, Noll M. Functional and structural response of the methanogenic microbial community in rice field soil to temperature change. Environ Microbiol. 2009;11:1844–53. doi: 10.1111/j.1462-2920.2009.01909.x. [DOI] [PubMed] [Google Scholar]
- Conrad R, Wetter B. Influence of temperature on energetics of hydrogen metabolism in homoacetogenic, methanogenic, and other anaerobic bacteria. Arch Microbiol. 1990;155:94–8. [Google Scholar]
- de Rezende JR, Kjeldsen KU, Hubert CRJ, et al. Dispersal of thermophilic Desulfotomaculum endospores into Baltic Sea sediments over thousands of years. ISME J. 2013;7:72–84. doi: 10.1038/ismej.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delong EF. Archaea in coastal marine environments. PNatl Acad Sci USA. 1992;89:5685–9. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfing J, Larter SR, Head IM. Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J. 2008;2:442–52. doi: 10.1038/ismej.2007.111. [DOI] [PubMed] [Google Scholar]
- Dridi B, Fardeau ML, Ollivier B, et al. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Micr. 2012;62:1902–7. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fardeau ML, Magot M, Patel BKC, et al. Thermoanaerobacter subterraneus sp nov., a novel thermophile isolated from oilfield water. Int J Syst Evol Micr. 2000;50:2141–9. doi: 10.1099/00207713-50-6-2141. [DOI] [PubMed] [Google Scholar]
- Fey A, Conrad R. Effect of temperature on carbon and electron flow and on the archaeal community in methanogenic rice field soil. Appl Environ Microb. 2000;66:4790–7. doi: 10.1128/aem.66.11.4790-4797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke N, Hoehler T, Jørgensen B. Hydrogen ‘leakage’ during methanogenesis from methanol and methylamine: implications for anaerobic carbon degradation pathways in aquatic sediments. Environ Microbiol. 2007;9:1060–71. doi: 10.1111/j.1462-2920.2007.01248.x. [DOI] [PubMed] [Google Scholar]
- Finke N, Jørgensen BB. Response of fermentation and sulfate reduction to experimental temperature changes in temperate and Arctic marine sediments. ISME J. 2008;2:815–29. doi: 10.1038/ISMEJ.2008.20. [DOI] [PubMed] [Google Scholar]
- Finke N, Vandieken V, Jørgensen BB. Acetate, lactate, propionate, and isobutyrate as electron donors for iron and sulfate reduction in Arctic marine sediments, Svalbard. FEMS Microbiol Ecol. 2007;59:10–22. doi: 10.1111/j.1574-6941.2006.00214.x. [DOI] [PubMed] [Google Scholar]
- Fry JC, Parkes RJ, Cragg BA, et al. Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol Ecol. 2008;66:181–96. doi: 10.1111/j.1574-6941.2008.00566.x. [DOI] [PubMed] [Google Scholar]
- Hedges JI, Keil RG. Marine Chemistry Discussion Paper. Sedimentary organic matter preservation: an assessment and speculative synthesis. Mar Chem. 1995;4:81–115. [Google Scholar]
- Ho DP, Jensen PD, Batstone DJ. Methanosarcinaceae and acetate-oxidizing pathways dominate in high-rate thermophilic anaerobic digestion of waste-activated sludge. Appl Environ Microb. 2013;79:6491–500. doi: 10.1128/AEM.01730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler T, Widdel F, Knittel K, et al. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 2011;5:1946–56. doi: 10.1038/ismej.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield B, Schenk HJ, Zink K, et al. Living microbial ecosystems within the active zone of catagenesis: implications for feeding the deep biosphere. Earth Planet Sci Lett. 2006;246:55–69. [Google Scholar]
- Hubert C, Arnosti C, Bruchert V, et al. Thermophilic anaerobes in Arctic marine sediments induced to mineralize complex organic matter at high temperature. Environ Microbiol. 2010;12:1089–104. doi: 10.1111/j.1462-2920.2010.02161.x. [DOI] [PubMed] [Google Scholar]
- Hubert C, Loy A, Nickel M, et al. A constant flux of diverse thermophilic Bacteria into the cold Arctic seabed. Science. 2009;325:1541–4. doi: 10.1126/science.1174012. [DOI] [PubMed] [Google Scholar]
- Isaksen MF, Bak F, Jørgensen BB. Thermophilic sulfate-reducing bacteria in cold marine sediments. FEMS Microbiol Ecol. 1994;14:1–8. [Google Scholar]
- Jiang L, Long C, Wu X, et al. Optimization of thermophilic fermentative hydrogen production by the newly isolated Caloranaerobacter azorensis H53214 from deep-sea hydrothermal vent environment. Int J Hydrogen Energ. 2014;39:14154–60. [Google Scholar]
- Jørgensen BB. Mineralization of organic-matter in the sea bed—the role of sulfate reduction. Nature. 1982;296:643–5. [Google Scholar]
- Jørgensen BB. Processes at the sediment-water interface. In: Bolin B, Cook RB, editors. The Major Biogeochemical Cycles and Their Interactions. New York: Wiley; 1983. pp. 477–509. [Google Scholar]
- Joyce AE. The coastal temperature network and ferry route programme: long-term temperature and salinity observations. CEFAS Sci Ser. 2006;43:1–129. [Google Scholar]
- Kallmeyer J, Boetius A. Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl Environ Microbiol. 2004;70:1231–3. doi: 10.1128/AEM.70.2.1231-1233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y. Thermal adaptation of the Archaeal and Bacterial lipid membranes. Archaea. 2012;2012:789652. doi: 10.1155/2012/789652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelnikova S, Pedersen K. Evidence for methanogenic Archaea and homoacetogenic Bacteria in deep granitic rock aquifers. FEMS Microbiol Rev. 1997;20:339–49. [Google Scholar]
- Kryachko Y, Dong XL, Sensen CW, et al. Compositions of microbial communities associated with oil and water in a mesothermic oil field. Anton Leeuw. 2012;101:493–506. doi: 10.1007/s10482-011-9658-y. [DOI] [PubMed] [Google Scholar]
- Lipp JS, Morono Y, Inagaki F, et al. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature. 2008;454:991–4. doi: 10.1038/nature07174. [DOI] [PubMed] [Google Scholar]
- Lloyd KG, Schreiber L, Petersen DG, et al. Predominant archaea in marine sediments degrade detrital proteins. Nature. 2013;496:215–8. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Chapelle FH. Deep subsurface microbial processes. Rev Geophys. 1995;33:365–81. [Google Scholar]
- Mayumi D, Dolfing J, Sakata S, et al. Carbon dioxide concentration dictates alternative methanogenic pathways in oil reservoirs. Nat Commun. 2013;4:1998. doi: 10.1038/ncomms2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayumi D, Mochimaru H, Yoshioka H, et al. Evidence for syntrophic acetate oxidation coupled to hydrogenotrophic methanogenesis in the high-temperature petroleum reservoir of Yabase oil field Japan. Environ Microbiol. 2011;13:1995–2006. doi: 10.1111/j.1462-2920.2010.02338.x. [DOI] [PubMed] [Google Scholar]
- Meng J, Xu J, Qin D, et al. Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J. 2014;8:650–9. doi: 10.1038/ismej.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelburg JJ, Klaver G, Nieuwenhuize J, et al. Organic matter mineralization in intertidal sediments along an estuarine gradient. Mar Ecol-Prog Ser. 1996;132:157–68. [Google Scholar]
- Mikucki JA, Liu YT, Delwiche M, et al. Isolation of a methanogen from deep marine sediments that contain methane hydrates, and description of Methanoculleus submarinus sp. nov. Appl Environ Microb. 2003;69:3311–6. doi: 10.1128/AEM.69.6.3311-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno T, Oldroyd A, McDonald I, et al. Preferential fractionation of trace metals-metalloids into PM10 resuspended from contaminated gold mine tailings at Rodalquilar, Spain. Water Air Soil Poll. 2007;179:93–105. [Google Scholar]
- Muller AL, de Rezende JR, Hubert CRJ, et al. Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J. 2014;86:1153–65. doi: 10.1038/ismej.2013.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussmann M, Brito I, Pitcher A, et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. P Natl Acad Sci USA. 2011;108:16771–6. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Brinkoff T, Nübel U, et al. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans ADL, Van Elsas JD, De Bruijn FJ, et al., editors. Molecular Microbial Ecology Manual. Dordrecht: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- Muyzer G, Dewaal EC, Uitterlinden AG. Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozhevnikova AN, Nekrasova V, Ammann A, et al. Influence of temperature and high acetate concentrations on methanogenensis in lake sediment slurries. FEMS Microbiol Ecol. 2007;62:336–44. doi: 10.1111/j.1574-6941.2007.00389.x. [DOI] [PubMed] [Google Scholar]
- O'Sullivan LA, Roussel EG, Weightman AJ, et al. Survival of Desulfotomaculum spores from estuarine sediments after serial autoclaving and high-temperature exposure. ISME J. 2015;9:922–33. doi: 10.1038/ismej.2014.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan LA, Sass AM, Webster G, et al. Contrasting relationships between biogeochemistry and prokaryotic diversity depth profiles along an estuarine sediment gradient. FEMS Microbiol Ecol. 2013;85:143–57. doi: 10.1111/1574-6941.12106. [DOI] [PubMed] [Google Scholar]
- O'Sullivan LA, Webster G, Fry JC, et al. Modified linker-PCR primers facilitate complete sequencing of DGGE DNA fragments. J Microbiol Meth. 2008;75:579–81. doi: 10.1016/j.mimet.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Marsh LM, Polcin S. Methane production and simultaneous sulphate reduction in anoxic, salt marsh sediments. Nature. 1982;296:143–5. [Google Scholar]
- Ovreas L, Forney L, Daae FL, et al. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–73. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes RJ, Brock F, Banning N, et al. Changes in methanogenic substrate utilization and communities with depth in a salt-marsh, creek sediment in southern England. Estuar Coast Shelf S. 2012;96:170–8. [Google Scholar]
- Parkes RJ, Cragg B, Roussel E, et al. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar Geol. 2014;352:409–25. [Google Scholar]
- Parkes RJ, Cragg BA, Banning N, et al. Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments Skagerrak, Denmark. Environ Microbiol. 2007a;9:1146–61. doi: 10.1111/j.1462-2920.2006.01237.x. [DOI] [PubMed] [Google Scholar]
- Parkes RJ, Gibson GR, Mueller-Harvey I, et al. Determination of the substrates for sulfate-reducing bacteria within marine and estuarine sediments with different rates of sulfate reduction. J Gen Microbiol. 1989;135:175–87. [Google Scholar]
- Parkes RJ, Wellsbury P, Mather ID, et al. Temperature activation of organic matter and minerals during burial has the potential to sustain the deep biosphere over geological time scales. Org Geochem. 2007b;38:845–52. [Google Scholar]
- Paul K, Nonoh JO, Mikulski L, et al. ‘Methanoplasmatales,’ Thermoplasmatales-related Archaea in termite guts and other environments, are the seventh order of methanogens. Appl Environ Microbiol. 2012;78:8245–53. doi: 10.1128/AEM.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters V, Conrad R. Sequential reduction processes and initiation of CH4 poduction upon flooding of oxic upland soils. Soil Biol Biochem. 1996;28:371–82. [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Robador A, Bruchert V, Jørgensen BB. The impact of temperature change on the activity and community composition of sulfate-reducing bacteria in arctic versus temperate marine sediments. Environ Microbiol. 2009;11:1692–703. doi: 10.1111/j.1462-2920.2009.01896.x. [DOI] [PubMed] [Google Scholar]
- Roh Y, Liu SV, Li GS, et al. Isolation and characterization of metal-reducing Thermoanaerobacter strains from deep subsurface environments of the Piceance Basin, Colorado. Appl Environ Microbiol. 2002;68:6013–20. doi: 10.1128/AEM.68.12.6013-6020.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfuss F, Conrad R. Thermodynamics of methanogenic intermediary metabolism in littoral sediment of Lake Constance. FEMS Microbiol Ecol. 1993;12:265–76. [Google Scholar]
- Roussel EG, Cambon-Bonavita MA, Querellou J, et al. Extending the sub-sea-floor biosphere. Science. 2008;320:1046. doi: 10.1126/science.1154545. [DOI] [PubMed] [Google Scholar]
- Rui J, Qiu Q, Lu Y. Syntrophic acetate oxidation under thermophilic methanogenic condition in Chinese paddy field soil. FEMS Microbiol Ecol. 2011;77:264–73. doi: 10.1111/j.1574-6941.2011.01104.x. [DOI] [PubMed] [Google Scholar]
- Schouten S, Middelburg JJ, Hopmans EC, et al. Fossilization and degradation of intact polar lipids in deep subsurface sediments: a theoretical approach. Geochim Cosmochim Acta. 2010;74:3806–14. [Google Scholar]
- Seewald JS. Organic-inorganic interactions in petroleum-producing sedimentary basins. Nature. 2003;426:327–33. doi: 10.1038/nature02132. [DOI] [PubMed] [Google Scholar]
- Stahl DA, de la Torre JR. Physiology and diversity of ammonia-oxidizing Archaea. Annu Rev Microbiol. 2012;66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- Stoddard SF, Smith BJ, Hein R, et al. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593–8. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Miyazaki M, Hirayama H, et al. Isolation and physiological characterization of two novel, piezophilic, thermophilic chemolithoautotrophs from a deep-sea hydrothermal vent chimney. Environ Microbiol. 2009;11:1983–97. doi: 10.1111/j.1462-2920.2009.01921.x. [DOI] [PubMed] [Google Scholar]
- Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nature Rev Microbiol. 2007;5:316–23. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- Webster G, Blazejak A, Cragg BA, et al. Subsurface microbiology and biogeochemistry of a deep, cold-water carbonate mound from the Porcupine Seabight IODP Expedition 307. Environ Microbiol. 2009;11:239–57. doi: 10.1111/j.1462-2920.2008.01759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster G, Newberry CJ, Fry JC, et al. Assessment of bacterial community structure in the deep sub- seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J Microbiol Meth. 2003;55:155–64. doi: 10.1016/s0167-7012(03)00140-4. [DOI] [PubMed] [Google Scholar]
- Webster G, Parkes RJ, Cragg BA, et al. Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol. 2006;58:65–85. doi: 10.1111/j.1574-6941.2006.00147.x. [DOI] [PubMed] [Google Scholar]
- Webster G, Rinna J, Roussel EG, et al. Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol Ecol. 2010;72:179–97. doi: 10.1111/j.1574-6941.2010.00848.x. [DOI] [PubMed] [Google Scholar]
- Webster G, Sullivan LA, Meng Y, et al. Archaeal community diversity and abundance changes along a natural salinity gradient in estuarine sediments. FEMS Microbiol Ecol. 2015;91:1–18. doi: 10.1093/femsec/fiu025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellsbury P, Herbert RA, Parkes RJ. Bacterial [methyl-H-3]thymidine incorporation in substrate-amended estuarine sediment slurries. FEMS Microbiol Ecol. 1994;15:237–48. [Google Scholar]
- Wery N, Moricet JM, Cueff V, et al. Caloranaerobacter azorensis gen. nov., sp. nov., an anaerobic thermophilic bacterium isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2001;51:1789–96. doi: 10.1099/00207713-51-5-1789. [DOI] [PubMed] [Google Scholar]
- Weston NB, Joye SB. Temperature-driven decoupling of key phases of organic matter degradation in marine sediments. P Natl Acad Sci USA. 2005;102:17036–40. doi: 10.1073/pnas.0508798102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiticar MJ. Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol. 1999;161:291–314. [Google Scholar]
- Yanagawa K, Morono Y, de Beer D, et al. Metabolically active microbial communities in marine sediment under high-CO2 and low-pH extremes. ISME J. 2013;7:555–67. doi: 10.1038/ismej.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeikus JG, Wolfe RS. Methanobacterium thermoautotrophicus sp n, an anaerobic, autotrophic, extreme thermophile. J Bacteriol. 1972;109:707–13. doi: 10.1128/jb.109.2.707-713.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.