Abstract
By convention, CD4+ T lymphocytes recognize foreign and self peptides derived from internalized antigens in combination with MHC class II molecules. Alternative pathways of epitope production have been identified but their contributions to host defense have not been established. We show here in a mouse infection model that the CD4+ T cell response to influenza, critical for durable protection from the virus, is driven principally by unconventional processing of antigen synthesized within the infected antigen-presenting cell, not by classical processing of endocytosed virions or material from infected cells. Investigation of the cellular components involved, including the H2-M molecular chaperone, the proteasome, and gamma-interferon inducible lysosomal thiol reductase revealed considerable heterogeneity in the generation of individual epitopes, an arrangement that ensures peptide diversity and broad CD4+ T cell engagement. These results could fundamentally revise strategies for rational vaccine design and may lead to key insights into the induction of autoimmune and anti-tumor responses.
The classical MHCII processing pathway, developed chiefly through work with stable, globular proteins, entails: 1) engulfment of extracellular material, 2) delivery of nascent MHC class II (MHCII) molecules to a late endosomal compartment via its transient partner invariant chain (Ii), 3) catabolism of both Ii and internalized material in the endocytic compartment. 4) exchange of the remaining class II-associated invariant chain peptide (CLIP) portion of Ii for high affinity peptides and 5) trafficking of peptide/MHCII complexes to the cell surface where they can trigger cognate CD4+ T cells1. MHCII molecules are highly polymorphic and in most cases CLIP-MHCII affinity is sufficiently high that CLIP-peptide exchange requires participation of a heterodimeric chaperone termed HLA-DM in humans and H-2M in mice2.
Viral proteins are distinct from nominal exogenous antigens in accessing intracellular compartments beyond the endosomal network and in interacting far more dynamically with cellular machinery. Indeed, studies of MHCII processing with such proteins have revealed several alternatives that diverge to greater or lesser extents from the classical scheme. Examples include: 1) a “recycling” pathway in which partially or completely disordered peptides derived from exogenous antigen load onto MHCII in the early endosome without H2-M participation3, 2) macroautophagy, which delivers cytosolic contents to the late endosomal network for conventional proteolysis and loading4, and 3) a pathway that depends upon delivery to the cytosol and participation of both the proteasome and the transporter associated with antigen processing (TAP)5, well established components of the conventional MHC class I (MHCI) processing pathway but rarely implicated in MHCII processing.
Because MHCII processing studies have traditionally focused on individual epitopes that are largely derived from exogenously provided antigens, the relative contributions of alternative pathways have remained unknown. In an initial attempt to address this issue, we previously carried out ex vivo analysis of a polyclonal influenza-specific CD4+ T cell population, estimating that 30–40% of the responding T cells were specific for proteasome-dependent epitopes5. This figure is consistent with a significant contribution from non-classical processing; however there were limitations to the indirect ELISpot-based approach that we utilized. First was the use of proteasome inhibitor at concentrations that, in retrospect, may have reduced protein (endogenous antigen) synthesis6. Second was the inability to determine whether the 30–40% fraction lay with a few dominant epitopes or reflected 30–40% of all the specificities involved in the response. In addition, given the existence of several alternative processing pathways, the other 60–70% of the response may or may not have been driven by classical processing. These are fundamental issues considering the importance of CD4+ T cells in potentiating humoral and CD8+ (cytolytic) T cell responses1 and the predictive power of a broad CD4+ T cell response for protection against several human pathogens, including the hepatitis B, hepatitis C and influenza viruses7–9. Greater processing complexity will enhance epitope diversity and, consequently, CD4+ T cell participation in establishing protection. Vaccine strategies that assume sufficient CD4+ T cell activation via the classical pathway may engender suboptimal protection.
In order to explore both the prevalence and complexity of alternative MHCII processing, we turned to a mouse model of influenza infection that has provided numerous fundamental insights into defense against the virus10. We were guided by the principle that definitive information would be gained only by accounting for each of the MHCII-restricted epitopes that drive the influenza-specific CD4+ T cell response, subsequently exploring the processing requirements of each epitope through complementary in vivo- and in vitro-based approaches. Although our previous efforts5 led us to suspect that alternative processing would make more than a minor contribution, we show here that the vast majority of the influenza-specific CD4+ T cell response in infected mice is driven by bona fide endogenous processing of antigen by the infected antigen-presenting cell (APC).
RESULTS
Influenza virions are poor MHCII processing substrates and induce weak CD4+ T cell responses
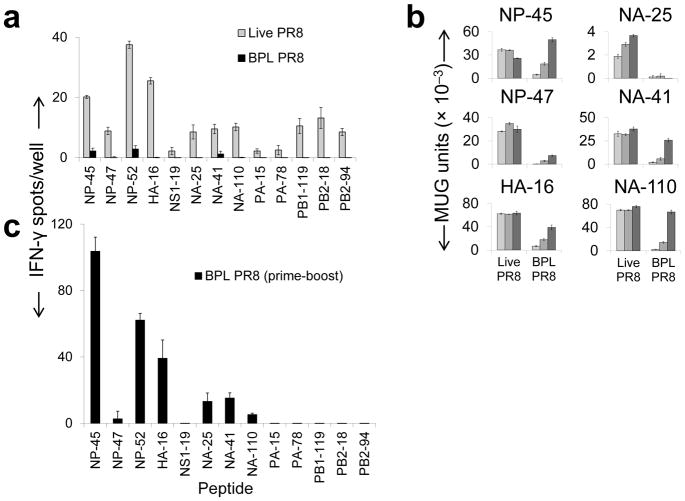
We reasoned that if the CD4+ T cell response to influenza is driven primarily by the classical pathway (defined here as conversion of internalized virions, infectious or not, to peptides that load onto nascent MHCII), live and inactivated influenza should elicit comparable responses since most encoded proteins are assembled into the virion11. Wildtype (WT) C57BL/6 (B6) mice, which express only the Ab MHCII molecule, were inoculated intranasally (i.n.) with a low dose of mouse-adapted influenza A virus, A/Puerto Rico/8/1934 (PR8) or β-propiolactone (BPL) inactivated PR8 at a much higher dose (~4 × 106 fold) to compensate for absence of replication12. The responding CD4+ T cells were analyzed by ELISpot assay with a comprehensive overlapping peptide library. Live PR8 elicited thirteen distinct specificities, six established and seven novel (Fig. 1a, Supplementary Fig. 1 and Supplementary Table 1). Seven mapped to three major structural proteins: nucleoprotein (NP), hemagglutinin (HA), and neuraminidase (NA). Five mapped to the polymerase subunits (PA, PB1 and PB2) that are less prevalent in the virion11. One mapped to the nonstructural protein NS1, absent from the virion. In contrast, BPL PR8 elicited only three specificities and quite weakly. No specificities unique to BPL PR8 were elicited (not shown), eliminating the possibility of a redistributed response. The disparity was not attributable to limited access to inert virions by APC nor to destruction of processable antigen by the inactivation procedure; intraperitoneal (i.p.), intramuscular (i.m.), intradermal (i.d.) and even intravenous (i.v.) delivery produced similar results as did UV and hydrogen peroxide inactivation of the virus (Supplementary Fig. 2–4).
Figure 1.
Impact of virus inactivation on in vivo and in vitro epitope-specific CD4+ T cell responses to PR8. (a) B6 (WT) mice were primed i.n. with live PR8 (0.001 hemagglutination units [HAU], gray bars) or BPL-inactivated PR8 (BPL PR8, 3800 HAU, black bars). 12 days later, IFN-γ-secreting CD4+ T cell specific for the indicated peptides (sequences in Supplementary Table 1) were quantified by ELISpot (b) BMDCs were pulsed with either live PR8 or BPL PR8 and co-cultured with CD4+ T cell hybridomas of the indicated specificities Virus amounts from light to dark, respectively: 1, 16, 256 HAU. (c) B6 mice were primed intraperitoneally with 3800 HAU BPL PR8 on day (d) 0 and boosted with the same dose on d21. On day 28, CD4+ T cells were purified from harvested spleens and responses to individual influenza peptides were quantified by ELISpot assay. Results were similar with prime/boost of UV-inactivated PR8 (data not shown).100,000 CD4+ T cells/well were cultured in all ELISpot assays shown here and elsewhere. Female mice were used in all experiments. (a, b, c) (representative of three independent experiments performed in triplicate). Data are presented as mean ± s.d. Background [(a, c) mean DMSO ± (2 × standard deviation (s.d.)); (b) mean uninfected APC] was subtracted from the experimental group results. MUG, methyl-umbelliferyl-β-D-galactoside
In opposing the receptor binding activity of HA, NA might contribute to the low potency of inactivated virus by reducing antigen capture by APC11. However, elimination of NA activity did not enhance the response to BPL inactivated virus (Supplementary Fig. 5).
Response patterns of CD4+ T cell hybridomas to six of the specificities were consistent with the in vivo results; presentation by bone marrow derived dendritic cells (BMDCs) (Fig. 1b) and MHCII-expressing fibroblasts (Supplementary Fig. 6) was markedly, and in some cases absolutely, more efficient from infectious virus. Multiple independent clones for three of the hybridomas demonstrated comparable reactivity patterns (Supplementary Fig. 7), discounting individual TCR bias toward possible conformations that are unique to endogenously produced MHC/peptide complexes.
The data indicate that influenza virions are inherently poor processing substrates. To reinforce this notion, we primed and boosted WT B6 mice i.p. with high-dose BPL PR8. The specificities elicited in the primary response were amplified to widely varying degrees and four additional specificities were elicited at uniformly low levels (Fig. 1c). Five specificities mapping to structural proteins remained undetectable. Results were similar with prime/boost of UV-inactivated PR8 (data not shown). Collectively, these results indicate that processing of exogenous virus is capable of producing only some of the epitopes and at efficiencies that range from low to marginally detectable compared to processing of biosynthesized antigen.
Inactivated virus elicits low antibody titers that poorly control a lung infection
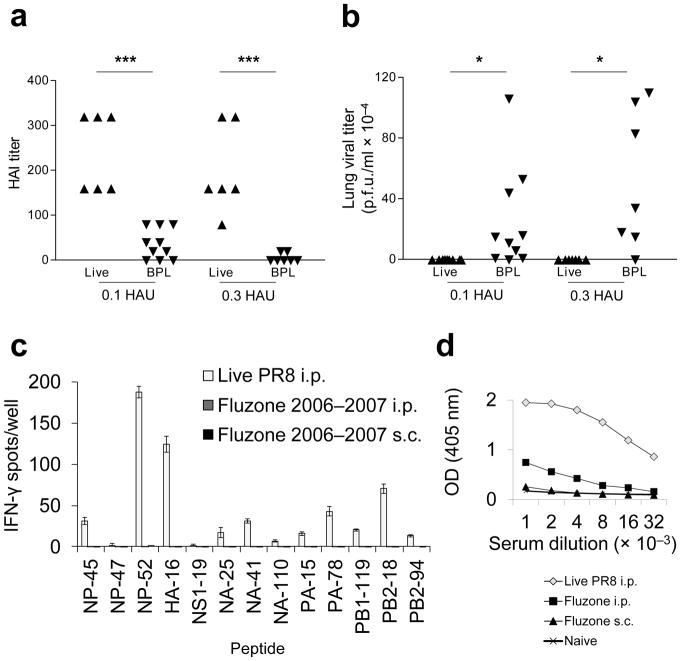
The low CD4+ T cell response induced by inactivated virus is predicted to compromise induction of durable, high-titer neutralizing antibodies13,14, the hallmark of protection from influenza15, since “help” to B cells will be considerably diminished. We investigated this by immunizing WT B6 mice with two different doses of live and inactivated virus i.m., the standard inoculation route for the licensed subunit vaccine. Identical doses of virus were compared since, as we confirmed (Supplementary Fig. 8), extrapulmonary replication of influenza is negligible16. Induction of neutralizing antibodies, measured by hemagglutination inhibition (HAI) assay, was severely compromised by virus inactivation (Fig. 2a) and, consequently, so was ability to control a live virus challenge, with clear, statistically significant differences in viral lung titers three days after inoculation (Fig. 2b).
Figure 2.
Vaccination with inactivated PR8 induces low levels of influenza-specific neutralizing antibodies, which poorly control virus replication. The licensed split subunit vaccine similarly displays poor CD4+ T cell and B cell immunogenicitty. (a) Mice were primed i.m with 0.1 HAU live PR8 (▲) (n = 7), 0.1 HAU BPL PR8 (▼) (n = 10), 0.3 HAU live PR8 (▲) (n = 7), or 0.3 HAU BPL PR8 (▼) (n = 7) and serum HAI antibody titers were quantified on d26 (left graph). (b)They were then challenged i.n with 20 HAU live homologous PR8. Live virus was titered by plaque assay from lung homogenates 3 days post immunization (d.p.i.). (c) B6 mice were primed i.p. with 4 HAU live PR8 (white bars), 22.5 μg H1 Fluzone 2006–2007 formula i.p. (dark gray bars, not visible), or 22.5 μg H1 Fluzone 2006–2007 formula s.c. (black bars, not visible). This is higher than the dose administered to humans (15 μg HA each). 12 days later, cytokine ELISpot assays were performed for detection of IFN-γ-secreting CD4+ T cells in response to individual influenza peptides. (d) Serum IgG titer to PR8 was determined by ELISA. Sera were collected on d12 from the same mouse groups - live PR8 i.p. (◇), Fluzone i.p. (■), Fluzone s.c. (▲) - inoculated in panel (c) and sera from naive (×) mice were used as controls. (c) (representative of two independent experiments performed in triplicate) (d) (representative of two independent experiments performed in duplicate, error bars covered by symbols). Data are presented as mean ± s.d. Background (mean DMSO + 2s.d.) was subtracted from the experimental group results. Statistical significance was tested (for a and b) by one-tailed Student’s t-test and (for d) P < 0.0001 by two-way ANOVA and post-hoc pairwise comparisons with Bonferroni correction.***P < 0.005, * P < 0.05.
Consistent with the observed low immunogenicity of BPL PR8 and previous work17, i.p. or subcutaneous (s.q.) inoculation with copious quantities of a licensed split subunit vaccine, which cross-reacts strongly with PR8 yielded undetectable CD4+ T cell responses (Fig. 2c) and antibody titers (Fig. 2d) were significantly lower than those elicited by live PR8 i.p.
Active infection does not enhance virion presentation
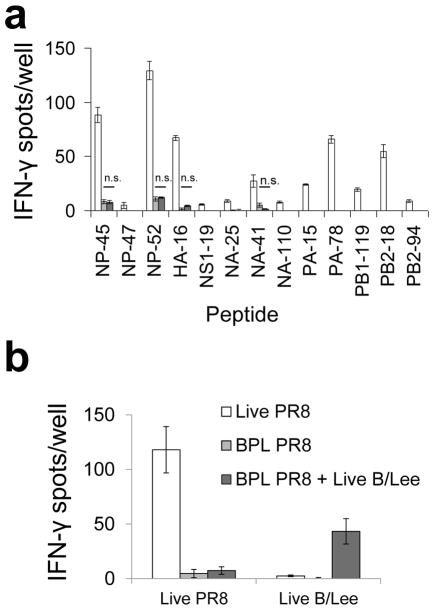
The superior immunogenicity of infectious virus could have been due to upregulation of antigen processing via the inflammatory signals triggered by viral replication18. To test this possibility, we inoculated WT B6 mice with a mixture of inactivated PR8 and infectious non-cross-reactive B/Lee/1940 (B/Lee) influenza. Despite inducing a robust autologous response, infectious B/Lee did not enhance the anti-PR8 response (Fig. 3). Similar results were obtained following co-inoculation i.m., i.d. (Supplementary Figs. 2b and c) and i.n. (not shown). We observed the same outcome in vitro using BMDCs and the hybridoma panel where we were able to ensure, through the use of sufficiently high multiplicities of infection, internalization of both viruses by the same APC. In no case was presentation from BPL PR8 enhanced by the presence of infectious B/Lee and in several cases presentation was diminished (Supplementary Fig. 9).
Figure 3.
Co-inoculation with infectious B/Lee does not enhance the CD4+ T cell response to BPL PR8. (a) B6 mice were primed intraperitoneally with 4 HAU live PR8 (white bars), 4800 HAU BPL PR8 (light gray bars), or 4800 HAU BPL PR8 and 400 HAU live B/Lee (dark gray bars). 12 days later, cytokine ELISpot assays were performed for detection of IFN-γ-secreting CD4+ T cells in response to individual influenza peptides. (b) To verify efficient B/Lee priming, BMDCs were pulsed with either 4 HAU live PR8 or 4 HAU live B/Lee (x-axis) and co-cultured with purified CD4+ T cell from each priming condition. Data are representative of two independent experiments performed in triplicate. Data are presented as mean ± s.d. Background (DMSO ± 2s.d.) was subtracted from the experimental group results. Statistical significance (for a) was tested by one-tailed Student’s t-test.
Bona fide endogenous processing is the main driver of CD4+ T cell stimulation
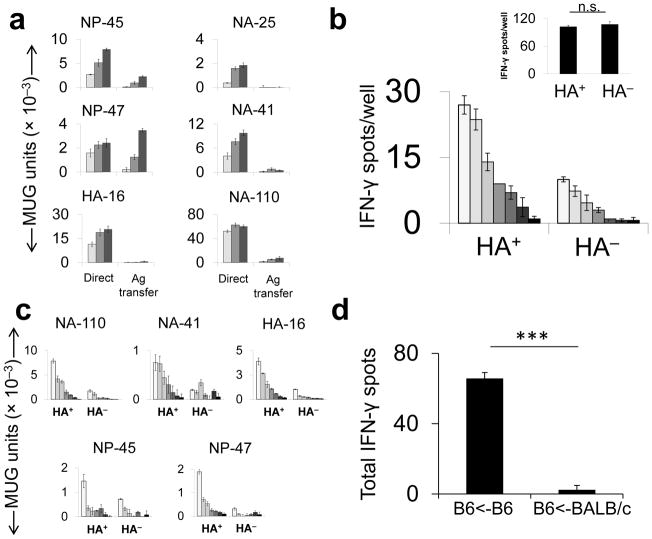
Skewing toward presentation of biosynthesized antigen could be due to bona fide endogenous processing by the infected cell or transfer to an uninfected APC of subviral material that is more easily processed via the exogenous pathway than whole virions. To assess the relative contributions of these two general mechanisms in vitro, we infected MHCII-negative (MHCII−)donor cells and combined them with uninfected BMDCs, the T hybridoma panel and neutralizing antibody to prevent infection of the recipient APCs. Only one epitope (NP-47) demonstrated relatively robust presentation via transfer (Fig. 4a), and this was at least 16-fold less efficient than presentation by directly infected BMDCs. Use of BMDCs from H2-M-deficient mice as recipients demonstrated that presentation via transfer of all six epitopes is H-2M-dependent (Supplementary Fig. 10), suggesting that processing of transferred antigen is typically via the classical pathway.
Figure 4.
Relative contributions of direct endogenous presentation and antigen transfer in vitro and in vivo. (a) MHCII− L929 cells were infected with live PR8 and co-cultured with uninfected BMDCs and T hybridomas (“Antigen [Ag] Transfer”). For the comparator (“Direct”), BMDC were pulsed with live PR8 under the same conditions as in Fig. 1b. (b) B6 mice were infected with PR8 i.n. (128 HAU) and 3 d.p.i., MHCII+ cells were isolated from homogenized lungs, flow-sorted into HA+ and HA− cell pools and combined in an ELISpot assay with PR8-immune (main panel) or vaccinia virus (VACV)-immune (inset) CD4+ T cells. Inset: The HA+ and HA− pools were pulsed with a VACV-derived peptide and co-incubated with a VACV-specific polyclonal CD4+ T cell population (c) MHCII+HA+ and MHCII+HA− cells, prepared as described in b were co-cultured with T hybridomas overnight. (d) Naive splenocytes from B6 or BALB/c donor mice were infected with 50 HAU live PR8 per 1×106 cells, cultured overnight and then UV irradiated to prevent transmission of infectious virus. B6 mice were immunized i.p with 5 ×106 infected cells. 11d later, IFN-γ-producing CD4+ T cells were quantified by ELISpot assay.
(a,c & d) representative of three and (b) two independent experiments performed in triplicate. Data are presented as mean ± s.d. Background [(a) mean uninfected APC; (b & c) MHC-II+ cells from B/Lee inoculated mice, (d) mean DMSO] was subtracted from the experimental group results.(b & d). Statistical significance was tested by one-tailed Student’s t-test.*** P < 0.0001.(d) Results with individual peptides were summed and mean standard deviation for the total was determined by calculating the square root of the averaged variances. MUG, methyl-umbelliferyl-β-D-galactoside
To assess the relative contributions of direct presentation and transfer in vivo we utilized three independent approaches. First, magnetic bead-isolated MHCII+ cells from the lungs of infected mice, which migrate to regional lymph nodes to initiate T cell responses19, were separated by FACS into surface HA+ (infected) and HA− pools. Twofold serial dilutions of the otherwise unmanipulated pools were co-incubated with polyclonal flu-specific CD4+ T cells (Fig. 4b) or individual T hybridomas (Fig. 4c). If antigen transfer were a robust mechanism in vivo, then the MHCII+HA− pool should demonstrate strong stimulatory capacity. However, consistent with the in vitro assay (Fig. 1b), the HA+ pool was substantially more potent (8–16-fold) while the pools demonstrated similar presentation abilities when pulsed with exogenously provided synthetic peptide. Results were similar for cells collected 3 d (shown) and 1 d (not shown) after infection.19 This approach likely overestimates the contribution of antigen transfer since most flu infected cells do not express the full complement of viral proteins20 and, indeed, we confirmed that a fraction of the HA− pool stains positive for NP and NS1 (not shown). The deleterious impact of cell permeabilization on antigen presentation precluded us from staining for internal proteins during the sorting procedure.
In our second approach, we infected BALB/c and B6 splenocytes with influenza, UV irradiated them to prevent subsequent release of infectious progeny, verifying inactivation by plaque assay (not shown), and adoptively transferred them by i.p. injection into naive BALB/c and B6 recipients. The B6→B6 transfer resulted in an influenza-specific CD4+ T cell response as detected by peptide-based ELISpot assay while the BALB/c→B6 configuration, which precludes direct presentation, did not (Fig. 4d).
In the third approach, i.n. or i.p injection of a PR8-infected cell lysate yielded no detectable CD4+ T cell response even when co-inoculated with live B/Lee (not shown). All three outcomes support our hypothesis that directly infected APCs carrying out bona fide endogenous MHCII antigen processing are the main drivers of the CD4+ T cell response.
PR8 presentation in vivo involves H2-M-dependent and -independent mechanisms
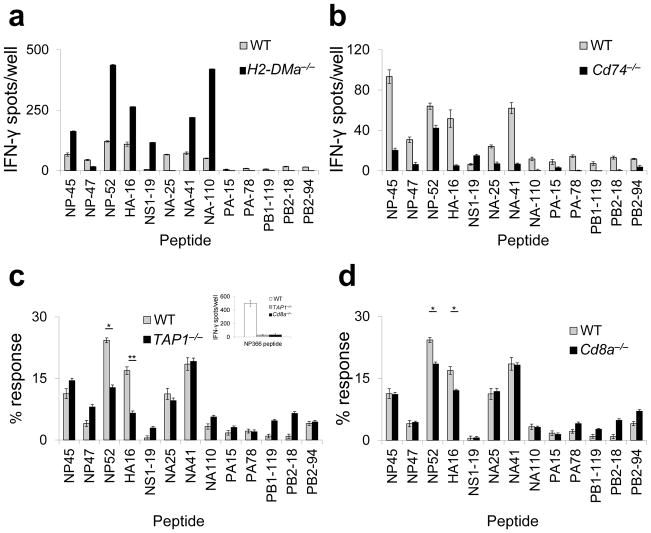
We initially explored the endogenous processing mechanisms underlying presentation of the thirteen PR8 epitopes with a panel of knockout mice on the H-2b background. In earlier studies, we had identified an Ed-restricted epitope within HA that is presented by recycling MHCII independent of H-2M expression. This behavior was attributable to location of the epitope in the stalk region of HA that unfolds in response to acidification shortly after internalization, rendering the epitope available for MHCII binding within the early endosome. We anticipated that none of the thirteen Ab-restricted epitopes would be presentable in the absence of H-2M because the one epitope within HA (HA-16) is not located in the stalk region and none of the other flu proteins is known to undergo unfolding in the early endosome. In addition Ab has a high affinity for CLIP21 and participation of H-2M in classical, late endosomal loading appears to be mandatory22. Nevertheless, six specificities were robustly elicited in H2-M-deficient mice (Fig. 5a). Indeed, all responses were substantially higher than in WT mice, possibly due to H2-M-mediated suppression as described for an HLA-DQ2-restricted epitope23. A similar peptide-based analysis of the CD4+ T cell response to ectromelia virus has revealed a much lower frequency of H2-M-independent presentation (M. Mendonca and B. DeHaven, unpublished), discounting anomalous hyperreactivity of H2-M-deficient mice to viral infection. The results suggest processing schemes for these six epitopes that do not entail peptide loading in the late endosome. In support of this, nearly the same reactivity pattern was elicited in (Cd74 encodes Ii) mice although, compared to H2-M-deficient mice, T cell expansions were lower (Fig. 5b). Of particular note, all three of the epitopes that were presentable in vivo from inactivated virus (NP-45, NP-52 and NA-41) fall into this H2-M-independent category. Therefore, none of the thirteen epitopes is processed in vivo solely, or even predominantly, via the classical pathway as defined, and the alternatives involve both H2-M-dependent and -independent mechanisms.
Figure 5.
Selective H-2M-, and Ii-independent presentation in vivo. WT (gray bars), H2-DMa−/−(a), Cd74−/−(b), TAP1−/−(c) and CD8a−/−(d) (all black bars) mice were primed with live PR8 and analyzed by IFN-γbased ELISpot assay as described for Fig. 1a. For c and d, the data were normalized as % total response.) (c, inset) In the same ELISpot assay, whole splenocytes from WT (white bar), TAP-deficient (gray bar), or CD8-deficient (black bar) mice were pulsed with TAP-dependent MHC-I restricted influenza epitope, NP (366–374). (a) (representative of three independent experiments performed in triplicate). (b,c,d)(representative of two independent experiments performed in triplicate). Data are presented as mean ± s.d. Background (DMSO + 2s.d.) was subtracted from the experimental group results. Statistical significance was tested by one-tailed Student’s t-test. *P<0.05; **P<0.01.
We also previously described two Ed-restricted epitopes whose endogenous presentation depended upon TAP function5. Immunization of TAP-deficient vs. WT mice in the H-2Ab background and normalization of the ELISpot data revealed partial reduction of the responses to two epitopes responses to two epitopes (NP-52 and HA-16, Fig. 5c). However, CD8-deficient mice also displayed selectively diminished responses to the same two epitopes (Fig. 5d), suggesting that TAP-dependent CD8+ T cell activity in some way enhances responses to these two epitopes. A lack of TAP dependence for any of the Ab-restricted epitopes suggests that TAP participation in MHCII processing may be allele specific. In addition, our findings are in contrast with the recent report that absence of TAP profoundly alters the Ab-bound peptidome24.
In vitro analyses reveal additional endogenous processing heterogeneities
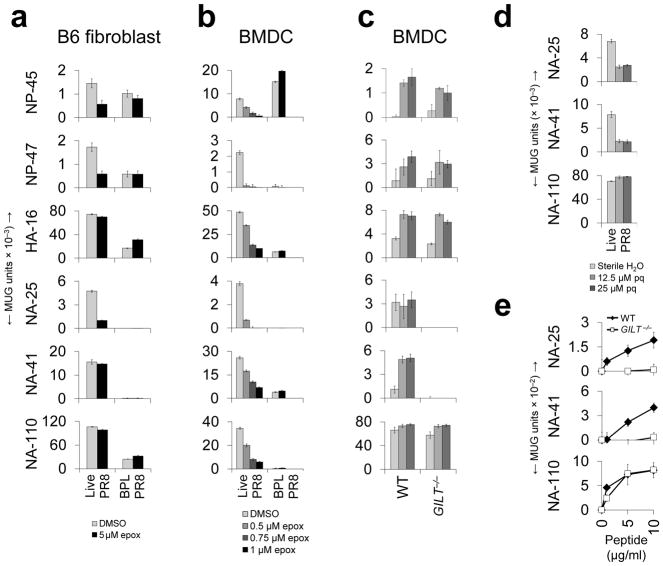
Further exploring underlying processing mechanisms, we utilized the hybridoma panel to investigate participation by the proteasome. In both BMDCs and a MHCII-expressing fibroblast line, endogenous presentation from infectious virus of two epitopes (NP-47 and NA-25) was potently reduced by treatment versus mock treatment with the proteasome inhibitor exoxomicin under conditions of persisting protein synthesis (Supplementary Fig. 11), with only stimulation from input (exogenous) virus remaining (the stimulating activities of BPL PR8 providing these reference points). Presentation from inactivated virus was uniformly unaffected (Figs. 6a and b). In the case of the NP-45 epitope, inhibition of endogenous presentation was again complete in the fibroblast line but only partial in BMDCs, suggesting both dependent and independent endogenous pathways in BMDCs. Thus, for half of the six epitopes, derived from both cytosolic and glycoprotein antigens, and including one that can also be generated via exogenous presentation in vivo (NP-45), robust presentation is proteasome-dependent. This frequency is in line with our previous analysis of a polyclonal influenza-specific CD4+ T cell population5 but clearly additional study is needed to determine the actual fraction. Since macroautophagy does not appear to be operative in the processing of PR825,26, novel mechanisms are likely involved in the endogenous processing of the remaining three epitopes (HA-16, NA-41 and NA-110).
Figure 6.
Involvement of the proteasome and GILT in endogenous processing. B6 fibroblasts (a) and BMDCs (b) were pre-treated with the indicated concentrations of epoxomicin (epox) or DMSO, pulsed with 10 HAU of live PR8 or BPL PR8 (fibroblasts) or 1 HAU of live PR8, 1 HAU BPL PR8 or 10 HAU BPL PR8 (BMDCs), and co-cultured with the indicated CD4+ T cell hybridomas. For BMDC data, responses to 1 HAU BPL PR8 (negligible for all but the NP-47 hybridoma) were subtracted from responses to 1 HAU live PR8, which facilitated comparison between groups. Because proteasome inhibition can compromise protein synthesis6, particularly in the case of BMDCs, complicating interpretation of results, we first identified appropriate concentrations of epoxomicin by flow cytometry (Supplementary Fig. 12). (c) BMDCs (WT or GILT−/−) were pulsed with increasing amounts of live PR8 (1, 50, 250 HAU; light to dark columns) and co-cultured with the indicated CD4+ T cell hybridomas. (d) B6 fibroblasts were pre-treated with the indicated concentrations of primaquine (pq) and pulsed with 1 HAU live PR8. APC were fixed after 18 hours and co-cultured with the indicated CD4+ T cell hybridomas. (e) BMDCs (WT or GILT−/−) were pulsed with increasing amounts of disulfide-constrained synthetic peptides and co-cultured with the indicated CD4+ T cell hybridomas. (a,b,c,e) (representative of three independent experiments performed in triplicate). (d)(representative of two independent experiments performed in triplicate).Data are presented as mean ± s.d. Background [(a,b,c,d) mean uninfected APC; (e) mean DMSO-pulsed APC] was subtracted from the experimental group results.
Seeking additional mechanistic insight, we investigated generation of the NA-25 and NA-41 epitopes, both being constrained within native NA by disulfide bonds. The NA-110 epitope, which lacks a cysteine residue, provided an internal control. Gamma-interferon induced lysosomal thiol reductase (GILT) can be critical for processing of exogenously obtained, disulfide bonded antigens27 but has not been implicated in endogenous processing. Indeed, endogenous processing of both epitopes by BMDCs was strongly GILT-dependent (Fig. 6c). Expansion of these two specificities was also selectively reduced in GILT-deficient mice (Supplementary Fig. 12a), but only partially, suggesting that GILT-independent processing mechanisms are operative in vivo. The relevant GILT activity appears to reside in the endosomal compartment since endogenous presentation of both epitopes was strongly inhibited by primaquine, a compound that disrupts endosomal trafficking3, while presentation of the NA-110 epitope was unaffected (Fig. 6d). Furthermore, presentation of exogenously provided disulfide-constrained synthetic peptides was also GILT-dependent (Fig. 6e and Supplementary Fig. 13). A second potential site of GILT action we considered was the endoplasmic reticulum, where GILT might mediate ERAD-driven reduction of full-length NA in order to facilitate retrograde translocation to the cytosol for conventional MHCI-like processing. However, this function should impact NA presentation globally and presentation of NA-110 is not GILT-dependent. Furthermore, responses to several MHCI-restricted epitopes within NA are not compromised in GILT-deficient mice (indeed, they are enhanced, Supplementary Fig. 12b). The results suggest novel pathways that pair proteasome-dependent and -independent processing in the cytosol with GILT-dependent processing in the endosome (Supplementary Fig. 14) via an as yet unidentified transport mechanism since neither TAP (Fig. 5c) nor macroautophagy25,26 appear to be involved. This analysis expands the striking heterogeneity in endogenous antigen processing evidenced in earlier figures. Indeed, when the various parameters examined here are compiled, each of the six epitopes is unique. Collectively, results suggest the existence of an endogenous processing network that is uniquely traversed in the generation of each epitope. Factors such as proteolytic susceptibility of the epitope and subcellular location of the parent protein may determine the route that leads to productive processing. The open-ended peptide binding groove of MHCII, which obviates the need for precise processing, would facilitate such an arrangement.
DISCUSSION
The outcomes reported here are strongly at odds with the prevailing model of MHCII processing. We attribute this to: 1) historical focus by the field on purified proteins, originally developed to study delayed-type hypersensitivity responses28, and having restricted access to extra-endosomal compartments, 2) the greater complexity of viruses that renders them less amenable to classical processing and more disposed to alternatives, owing to production of antigen within the APC, 3) the common practice of studying epitopes in isolation, which precludes the “accounting” approach that was taken here, and 4) the generation of many viral epitopes by more than one pathway, particularly appreciable in vitro (Fig. 1b), which could potentially confine scope of investigation to exogenous processing.
Live influenza vaccines are generally more protective than inactivated preparations, particularly in first time vaccinees29,30, and live vaccines in general require fewer immunizations to attain durable protection1. The engagement of multiple, non-redundant processing routes, would serve to maximize peptide diversity, thereby ensuring a broadly reactive CD4+ T cell response that is important for protection against several human pathogens, including influenza7–9, and perhaps many others, through the potentiation of both cellular and humoral immunity. It should therefore be of benefit to incorporate this principle into rational vaccine design, bearing in mind that processing pathway distribution will likely differ for each pathogen based upon structure, replication strategy and tropism. Finally, in light of findings reported here, and with autoimmune disorders and cancer immunotherapy in mind, greater exploration of non-classical MHCII processing of self-antigens seems warranted.
Methods
Peptides
The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, National Institute of Allergy and Infectious diseases (NIAID), NIH: Peptide Array, Influenza Virus A/Puerto Rico/8/1934 (H1N1) Hemagglutinin Protein, NR-18973; Peptide Array, Influenza Virus A/Puerto Rico/8/1934 (H1N1) Neuraminidase Protein, NR-19257; Peptide Array, Influenza Virus A/New York/348/03 (H1N1) PB1 Protein, NR-2617; Peptide Array, Influenza Virus A/New York/348/03 (H1N1) PB2 Protein, NR-2616; Peptide Array, Influenza Virus A/New York/348/03 (H1N1) Nucleocapsid Protein, NR-2611; Peptide Array, Influenza Virus A/New York/444/01 (H1N1) Nonstructural Protein 1, NR-2612; Peptide Array, Influenza Virus A/New York/348/03 (H1N1) Nonstructural Protein 2, NR-2615; Peptide Array, Influenza Virus A/New York/348/03 (H1N1) Matrix Protein 1, NR-2613; Peptide Array, Influenza Virus A/New York/348/03 (H1N1) Matrix Protein 2, NR-2614; Peptide Array, Influenza Virus A/New York/348/03 (H1N1) PA Protein, NR-2618, Peptide Array, Influenza Virus A/Brisbane/59/2007 (H1N1) Hemagglutinin Protein, NR-18970. Lyophilized peptides were reconstituted in dimethyl sulfoxide (DMSO; Sigma) and stored at −20 °C. Synthetic peptides (NA-25, NA-41, NA-110) were pre-treated with 0.5 mM 2-Mercaptoethanol (2-ME) for 30 minutes at 37 °C to allow for reduction of disulfide bonds.
Influenza viruses
Influenza A virus, A/Puerto Rico/8/1934 (PR8), subtype H1N1, and Influenza B virus, B/Lee/1940 (B/Lee) were grown, harvested, titered, and plaqued from isolates as described previously31,32. One hemagglutinating unit (HAU) of live PR8 virus ≈1.5 ×105 plaque forming units, titered as previously described32. Viruses were inactivated using either β-propiolactone33 (BPL; Sigma) or UV light34. In the BPL protocol, allantoic fluid containing virus was buffered in HEPES (0.1 M) and treated with BPL (0.1%) at 4 °C overnight. The next day, the prep was incubated at 37 °C for two hours to inactivate BPL. Hemagglutinin activity was determined by HAU titer using chicken red blood cells35. Heat inactivation of NA and measurement of NA activity using 4-MUNANA (Sigma) were as previously described36. The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: Influenza A Virus, A/Brisbane/59/2007 (H1N1), NR-12282; Fluzone® Influenza Virus Vaccine, 2006–2007 Formula, NR-10483; Fluzone® Influenza Virus Vaccine, 2008–2009 Formula, NR-17423.
Primary APCs and cell lines
Immature bone marrow-derived dendritic cells (BMDCs) were generated as previously described31 with the following modifications: media changes were performed every 3 days and BMDCs were harvested on day 8. The DC2.4 cell line was maintained in RPMI (Corning Cellgro) supplemented with 10% FBS (Gibco), 1× penicillin/streptomycin(Fisher Scientific), 1× L-glutamine (Fisher Scientific), and 0.05 mM 2-mercaptoethanol (2-ME; Gibco). The following reagent was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: Mouse Interferon Gamma (MuIFN-γ), NR-3081. For ELISpot assays, DC2.4 were pretreated with 2 International Units (IU)/ml MuIFN-γ for 48 hours to upregulate MHCII expression. B6 fibroblast cell line has been described previously37. Class II, major histocompatibility complex, transactivator (CIITA)-transduced B6 fibroblasts, MC57G (ATCC), L929 (ATCC), and I-Ab transduced L929 (L-IAb) cell lines were maintained in DMEM (Corning Cellgro) supplemented with 10% FBS and 0.05 mM 2-ME. All cell lines were periodically surveyed for mycoplasma using a commercial detection kit (Agilent technologies, catalog no. 302109)
T cell hybridomas
LacZ-inducible CD4+ T cell hybridomas, which express β-galactosidase upon recognition of peptide-MHCII complexes, were generated for NP-45, NP-47, HA-16, NA-25, NA-41, and NA-110 epitope specificities following previously described methods31,38. T cell hybridomas were maintained in RPMI supplemented with 10% FBS, 1× Penicillin/Streptomycin, 1× L-glutamine and 0.05 mM 2-ME. Activation was measured by detection of fluorometric β-galactosidase substrate methyl-umbelliferyl-β-D-galactoside (MUG; Sigma) as previously described39.
Mice
6–8 week old female C57BL/6, BALB/c, CD8−/− Tap1−/− (B6.129S2-Tap1tm1Arp/J), (B6.129S2-Tap1tm1Arp/J) (Jackson Laboratories), C57BL/6 H2-DMa−/− (Jackson Laboratories and provided by P. Roche), Ifi30−/− (provided by K. Hastings,) and Cd74−/−40 (provided by Guo-Ping Shi,) mice were utilized in all experiments. Colonies were maintained by Thomas Jefferson University Office of Laboratory Animal Services. Animal numbers were empirically determined to optimize numbers necessary for statistical significance. Exclusion criteria for mouse studies were weight loss in excess of 20% and no mice were excluded from this study. Animal studies were not randomized and investigators were not blinded for this study. All experimental protocols were preapproved by the Thomas Jefferson University Institutional Animal Care and Use Committee (IACUC). Numbers of mice used in each experiment were the minimums needed to obtain sufficient cell numbers for in vitro assays, statistical significance in in vivo assays and to demonstrate reproducibility; these were consistent with usage outlined in the IACUC-approved protocols.
Peptide-based ELISpot assays
Mice were primed intranasally with the following viruses at indicated doses: live PR8 (0.001 HAU), BPL-inactivated PR8 (3800 HAU). Groups of 3 mice were primed per condition. Mouse spleens were harvested 10–12 days post-priming, pooled, and CD4+ or CD8+ T cells were purified using Dynal CD4 or CD8 Negative Isolation Kits according to manufacturer’s instructions (Invitrogen). BMDC or MuIFN-γ-activated DC2.4 were used as APC and co-cultured overnight with purified CD4+ T cells (105 per well in all cases) in the presence of indicated peptides and screened for production of IFN-γ as a readout of T-cell activation. Overlapping peptide libraries were screened in triplicate as either matrixed peptide pools (containing 8–12 peptides/well)41 or individual peptides at a final concentration of 10 μg/ml. For all ELISpot assays, baselines were calculated as 2s.d. above background.
Fluzone immunogenicity
B6 mice were primed i.p. with 4 HAU of live PR8, 22.5 μg of H1 Fluzone 2006–2007 formula i.p., or 22.5 μg of H1 Fluzone 2006 -2007 formula s.c. 12 d later, cytokine ELISpot assays were performed for detection of IFN-γ -secreting CD4+ T cells in response to individual influenza peptides. CD4+ T cell immunogenicity of the H1 HA molecule contained in the Fluzone 2006–2007 preparation was confirmed (Supplementary Fig. 15).
In vivo cross priming studies
Adapted from Norbury et al42. Splenocytes from two naive female B6 or BALB/c mice were first infected with live PR8 virus for 1 hour at 37 °C in serum-free medium. To allow for expression of viral proteins, infected cells were cultured overnight at 37 °C and cells were UVB irradiated for 15 minutes. Cells were harvested and washed with PBS. Groups of three female B6 mice were primed intraperitoneally with 5×106 PR8 infected B6 or BALB/c splenocytes in a total volume of 200 μL per mouse. Ten days post-immunization, spleens were harvested and CD4+T cells were isolated to perform ELISpot assays. MuIFN-γ-activated DC2.4 were used as APC and pulsed with individual peptides at a final concentration of 10 μg/ml. Results with individual peptides were summed and mean standard deviation for the total was determined by calculating the square root of the averaged variances.
Protection studies
Sets of 7 female B6 mice were primed intramuscularly either with PBS or 0.1 or 0.3 HAU live or BPL inactivated PR8. Mice were bled at days 14 and 26 to perform serum HAI antibody titers using chicken red blood cells (RBCs). Mice were challenged intranasally with 20 HAU live PR8 at day 28 and lungs were harvested 3 days post challenge. Harvested lungs were processed using gentleMACSdissociator and live virus was titered by plaque assay from homogenates. Control of a heterologous challenge, indicating breadth of neutralization capability, was not investigated in these studied.
i.p. lavage and virus titer assay
B6 mice were injected i.p. with 1 HAU of live PR8 on days 0, 1, 2, or 3. Mice were subsequently sacrificed and peritoneal washes were performed by i.p. injection of 1 ml cold PBS. Lavage fluid was harvested on ice, pre-cleared of cells by centrifugation and directly assessed for infectious virus (plaque-forming units, p.f.u.) in a viral plaque assay32.
ELISA
Serum was collected from primed mice by retro-orbital eye bleeding and analyzed for IgG titer by ELISA. Briefly, serum was serially diluted in PBS supplemented with 1% low-IgG bovine serum albumin (BSA; Gemini Bio-Products), ranging from 1:500 to 1:32,000, and incubated in high binding EIA/RIA plates (Corning) pre-coated with 2 HAU of PR8. Plates were then washed with PBS + 0.01% Tween (PBST) and incubated with peroxidase labeled anti-mouse IgG (H+L) (Vector Laboratories, catalog no. PI-2000) at 1:1500 dilution in PBS/BSA (1%). Plates were developed using ABST Peroxidase Substrate (KPL) and read at detection wavelength of 405 nm.
In vitro antigen presentation assays
Epitope-specific LacZ-inducible T cell hybridomas were co-cultured in the presence of various antigen presenting cells (APC) pulsed with infectious or inactivated virus as described previously31. For antigen transfer assays, L929 cells were infected with live PR8 (1, 16 and 256 HAU) for 45 minutes and then washed twice to remove remaining virus. They were subsequently co-cultured overnight (18–20 h) with uninfected BMDC and T cell hybridomas. For proteasome inhibition assays, APCs were pre-treated with epoxomicin (Boston Biochem) for 15 minutes and washed once before infection with virus. To assess the effect of epoxomicin on protein synthesis, treated APCs were cultured for 18–20 h, stained with primary anti-HA IgG (H28-E23) antibody sup and secondary anti-mouse IgG FITC at 0.5 μg per test (eBioscience, catalog no. 11–4011), and analyzed by flow cytometry for surface expression of PR8 HA. Acquisition was performed on a FACS calibur. Baseline MFIs, calculated as 2s.d.above background, were subtracted from individual conditions. Amino acid supplementation43 did not alter the dose response curve in BMDCs (not shown). In assays involving primaquine-treated APCs, B6 fibroblasts were pre-treated with the compound for 30 minutes before infection with live virus. APCs were maintained overnight in the presence of primaquine, then fixed with 0.3% paraformaldehyde in 1.5 × HBSS for 15 minutes at room temperature and washed twice with complete medium before co-culturing with T cell hybridomas. For disulfide-bonded peptide assays, synthetic peptides (New England Peptides) were stored in DMSO (an oxidizing agent) allowing for disulfide-homodimerization to form between the single cysteine residues in each peptide. APC were then pulsed with varying amounts of disulfide-constrained peptides before co-culturing with T cell hybridomas.
Ex vivo cross-presentation assay
Groups of 5 female B6 mice were immunized i.n. with 128 HAU infectious PR8 or B/Lee. 3 d.p.i., mouse lungs were perfused with 5 ml cold PBS and harvested. Lung tissue fragments were collected to gentleMACS C tubes (MiltenyiBiotec) and enzymatically digested with Dispase II (9.4 U/ml) (Roche) and Collagenase A (50 mg/ml) (Roche) in serum-free RPMI media at 37 °C for 45 min under slow mixing. Tubes were transferred to gentleMACSDissociator (MiltenyiBiotec) and the protocol was run according to the manufacturer’s recommendations. Cell clumps were removed with a cell strainer. ACK lysis buffer (Life Technologies) was used to lyse any remaining RBCs. Lung cells were enriched for MHCII+ cells using anti-MHCII bead separation (MiltenyiBiotec, catalog no. 130-052-401). Cells were blocked with mouse Fc block (BD Biosciences) and stained with Alexa Fluor 647-conjugated anti-HA (mouse MAb H36-26) at 1:100 dilution. Surface HA cells were sorted using FACS (BD FACSAria II). Doubling serial dilutions of APC were then co-incubated with a polyclonal flu-specific CD4+ T cell in an ELISpot assay (96 well plate format, starting at 105 APC per well and a constant 1 × 105 CD4+ T cells per well) or individual T cell hybridomas in a MUG assay (384 well plate format, starting at 5 × 104 APC per well and a constant 1 × 104 T cell hybridomas per well). Polyclonal flu-specific CD4+ T cell were isolated using Dynal CD4 Negative Isolation Kits according to manufacturer’s instructions (Invitrogen) from mice primed with 0.003 HAU live PR8 i.n. 12 d.p.i.. To verify if surface-HA sorted cells have equal antigen presentation, processed/purified lung cells (105 per well) were pulsed with a VACV-specific peptide identified as an H-2Ab-restricted epitope by M. Mendonca and B. DeHaven (unpublished, peptide sequence LTGYAPVSPIVIART, 0.02 μg/ml) and co-incubated with VACV-primed CD4+ T cells (1 × 105 per well) in an ELISpot assay. VACV-specific CD4+ T cell were isolated 12d after priming with 1 × 105 p.f.u. live VACV-WR i.p. using Dynal CD4 Negative Isolation Kits. For all ELISpot assays, baselines were calculated as 2s.d. above background.
Statistical Analyses
All samples represent technical replicates, n = 3, unless noted. Data are presented as mean ± s.d. Reported P values were calculated using a one-tailed Student’s t-test or two-way ANOVA and post-hoc pairwise comparisons with Bonferroni correction (GraphPad Prism Software), and the data met the assumptions of the test. P values are indicated by the following symbols: *P< 0.05, **P< 0.01, ***P< 0.0005 and n.s.= not significant.
Supplementary Material
Acknowledgments
We thank M. Kinnarney for logistical support, S. Hensley (Wistar Institute, Philadelphia, PA) for technical advice on virus inactivation, and J. Comber for assistance with some of the ELISpot assays. We also thank the Flow Cytometry Facility and Division of Biostatistics (in particular B. Leiby and S. Keith) at Thomas Jefferson University. Mouse-adapted PR8 was provided by C. Lopez (University of Pennsylvania School of Veterinary Medicine, Philadelphia, PA). The H2-DMa−/− mice were provided by P. Roche (National Institute of Health/ National Cancer Institute, Bethesda, MD). Ifi30−/− mice were provided by K. Hastings (University of Arizona College of Medicine). Cd74−/− mice were provided by G. Shi at Brigham and Women’s Hospital (Harvard University, Boston, MA.). We thank C. Norbury, P. Roche, B. DeHaven and E. Wold for critical reading of this manuscript and P. Cresswell, S. Hensley and R. Langlois for helpful discussions. This study was funded by NIH grants AI036331 and AI101134.
Footnotes
Contributions
M. Miller carried out most of the in vivo and in vitro experiments included in this manuscript and some data analysis. A. Veerappan Ganesan conducted the in vivo cross priming, epoxomicin, primaquine experiments and most primary data analyses. M. Mendonca carried out the ex vivo cross presentation experiments. A. Veerappan Ganesan & M. Mendonca conducted the HAI titer and protection experiments. M. Miller and N. Lucashenak carried out pilot experiments. L. Eisenlohr provided overall supervision and wrote most of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Murphy K, Travers P, Walport M, Janeway C. Janeway’s immunobiology. Garland Science; New York: 2012. [Google Scholar]
- 2.Busch R, et al. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol Rev. 2005;207:242–260. doi: 10.1111/j.0105-2896.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- 3.Sinnathamby G, Eisenlohr LC. Presentation by recycling MHC class II molecules of an influenza hemagglutinin-derived epitope that is revealed in the early endosome by acidification. J Immunol. 2003;170:3504–3513. doi: 10.4049/jimmunol.170.7.3504. [DOI] [PubMed] [Google Scholar]
- 4.Gannage M, Munz C. Autophagy in MHC class II presentation of endogenous antigens. Curr Top Microbiol Immunol. 2009;335:123–140. doi: 10.1007/978-3-642-00302-8_6. [DOI] [PubMed] [Google Scholar]
- 5.Tewari MK, Sinnathamby G, Rajagopal D, Eisenlohr LC. A cytosolic pathway for MHC class II-restricted antigen processing that is proteasome and TAP dependent. Nat Immunol. 2005;6:287–294. doi: 10.1038/ni1171. [DOI] [PubMed] [Google Scholar]
- 6.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, et al. Dominance of the CD4+ T helper cell response during acute resolving hepatitis A virus infection. J Exp Med. 2012 doi: 10.1084/jem.20111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol. 2004;58:391–424. doi: 10.1146/annurev.micro.58.030603.123836. [DOI] [PubMed] [Google Scholar]
- 10.O’Donnell CD, Subbarao K. The contribution of animal models to the understanding of the host range and virulence of influenza A viruses. Microbes Infect. 2011;13:502–515. doi: 10.1016/j.micinf.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb RA. The influenza virus RNA segments and their encoded proteins. In: Palese P, Kingsbury DW, editors. Genetics of Influenza Viruses. Springer-Verlag; New York: 1983. pp. 21–69. [Google Scholar]
- 12.Moltedo B, et al. Cutting edge: stealth influenza virus replication precedes the initiation of adaptive immunity. J Immunol. 2009;183:3569–3573. doi: 10.4049/jimmunol.0900091. [DOI] [PubMed] [Google Scholar]
- 13.Sangster MY, et al. An early CD4+ T cell-dependent immunoglobulin A response to influenza infection in the absence of key cognate T-B interactions. J Exp Med. 2003;198:1011–1021. doi: 10.1084/jem.20021745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee BO, et al. CD4 T cell-independent antibody response promotes resolution of primary influenza infection and helps to prevent reinfection. J Immunol. 2005;175:5827–5838. doi: 10.4049/jimmunol.175.9.5827. [DOI] [PubMed] [Google Scholar]
- 15.Boyden AW, Frickman AM, Legge KL, Waldschmidt TJ. Primary and long-term B-cell responses in the upper airway and lung after influenza A virus infection. Immunol Res. 2014;59:73–80. doi: 10.1007/s12026-014-8541-0. [DOI] [PubMed] [Google Scholar]
- 16.Baz M, et al. Replication and immunogenicity of swine, equine, and avian h3 subtype influenza viruses in mice and ferrets. J Virol. 2013;87:6901–6910. doi: 10.1128/JVI.03520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lay M, et al. Cationic lipid/DNA complexes (JVRS-100) combined with influenza vaccine (Fluzone) increases antibody response, cellular immunity, and antigenically drifted protection. Vaccine. 2009;27:3811–3820. doi: 10.1016/j.vaccine.2009.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair P, Amsen D, Blander JM. Co-ordination of incoming and outgoing traffic in antigen-presenting cells by pattern recognition receptors and T cells. Traffic. 2011;12:1669–1676. doi: 10.1111/j.1600-0854.2011.01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legge KL, Braciale TJ. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity. 2003;18:265–277. doi: 10.1016/s1074-7613(03)00023-2. [DOI] [PubMed] [Google Scholar]
- 20.Brooke CB, et al. Most influenza a virions fail to express at least one essential viral protein. J Virol. 2013;87:3155–3162. doi: 10.1128/JVI.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sette A, Southwood S, Miller J, Apella E. Binding of major histocompatibility complex class II to the invariant chain-derived peptide, CLIP, is regulated by allelic polymorphism in class II. J Exp Med. 1995;181:677–683. doi: 10.1084/jem.181.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bikoff EK, Wutz G, Kenty GA, Koonce CH, Robertson EJ. Relaxed DM requirements during class II peptide loading and CD4+ T cell maturation in BALB/c mice. J Immunol. 2001;166:5087–5098. doi: 10.4049/jimmunol.166.8.5087. [DOI] [PubMed] [Google Scholar]
- 23.Hou T, et al. An insertion mutant in DQA1*0501 restores susceptibility to HLA-DM: implications for disease associations. J Immunol. 2011;187:2442–2452. doi: 10.4049/jimmunol.1100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spencer CT, et al. Sculpting MHC class II-restricted self and non-self peptidome by the class I Ag-processing machinery and its impact on Th-cell responses. Eur J Immunol. 2013;43:1162–1172. doi: 10.1002/eji.201243087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comber JD, Robinson TM, Siciliano NA, Snook AE, Eisenlohr LC. Functional macroautophagy induction by influenza A virus without a contribution to major histocompatibility complex class II-restricted presentation. J Virol. 2011;85:6453–6463. doi: 10.1128/JVI.02122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gannage M, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maric M, et al. Defective Antigen Processing in GILT-Free Mice. Science. 2001;294:1361–1365. doi: 10.1126/science.1065500. [DOI] [PubMed] [Google Scholar]
- 28.Silverstein AM. A history of immunology. Academic Press; San Diego: 1989. [Google Scholar]
- 29.Johnson PR, Jr, Feldman S, Thompson JM, Mahoney JD, Wright PF. Comparison of long-term systemic and secretory antibody responses in children given live attenuated or inactivated influenza A vaccine. J Med Virol. 1985;17:325–335. doi: 10.1002/jmv.1890170405. [DOI] [PubMed] [Google Scholar]
- 30.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 31.Tewari MK, Sinnathamby G, Rajagopal D, Eisenlohr LC. A cytosolic pathway for MHC class II-restricted antigen processing that is proteasome and TAP dependent. Nat Immunol. 2005;6:287–294. doi: 10.1038/ni1171. [DOI] [PubMed] [Google Scholar]
- 32.Langlois RA, Varble A, Chua MA, Garcia-Sastre A, tenOever BR. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proceedings of the National Academy of Sciences. 2012;109:12117–12122. doi: 10.1073/pnas.1206039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstein MA, Tauraso NM. Effect of formalin, beta-propiolactone, merthiolate, and ultraviolet light upon influenza virus infectivity chicken cell agglutination, hemagglutination, and antigenicity. Appl Microbiol. 1970;19:290–294. doi: 10.1128/am.19.2.290-294.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comber JD, Robinson TM, Siciliano NA, Snook AE, Eisenlohr LC. Functional macroautophagy induction by influenza A virus without a contribution to major histocompatibility complex class II-restricted presentation. J Virol. 2011;85:6453–6463. doi: 10.1128/JVI.02122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fazekas de St Groth S, Webster RG. Disquisition on original antigenic sin. I. Evidence in man. J Exp Med. 1966;124:331–345. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosaka Y, Sasao F, Yamanaka K, Bennink JR, Yewdell JW. Recognition of noninfectious influenza virus by class I-restricted murine cytotoxic T lymphocytes. J Immunol. 1988;140:606–610. [PubMed] [Google Scholar]
- 37.Testa JS, Apcher GS, Comber JD, Eisenlohr LC. Exosome-driven antigen transfer for MHC class II presentation facilitated by the receptor binding activity of influenza hemagglutinin. J Immunol. 2010;185:6608–6616. doi: 10.4049/jimmunol.1001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanderson S, Shastri N. LacZ inducible, antigen/MHC-specific T cell hybrids. Int Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 39.Sinnathamby G, Eisenlohr LC. Presentation by recycling MHC class II molecules of an influenza hemagglutinin-derived epitope that is revealed in the early endosome by acidification. J Immunol. 2003;170:3504–3513. doi: 10.4049/jimmunol.170.7.3504. [DOI] [PubMed] [Google Scholar]
- 40.Bikoff EK, et al. Defective major histocompatibility complex class II assembly, transport, peptide acquisition, and CD4+ T cell selection in mice lacking invariant chain expression. J Exp Med. 1993;177:1699–1212. doi: 10.1084/jem.177.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roederer M, Koup RA. Optimized determination of T cell epitope responses. J Immunol Methods. 2003;274:221–228. doi: 10.1016/s0022-1759(02)00423-4. [DOI] [PubMed] [Google Scholar]
- 42.Norbury CC, et al. CD8+ T cell cross-priming via transfer of proteasome substrates. Science. 2004;304:1318–1321. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 43.Suraweera A, Munch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 2012;48:242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.