Abstract
The spermidine N-acetyltransferase SpeG is a dodecameric enzyme that catalyzes the transfer of an acetyl group from acetyl-coenzyme A to polyamines such as spermidine and spermine. SpeG has an allosteric polyamine-binding site and acetylating polyamines regulates their intracellular concentrations. The structures of SpeG from Vibrio cholerae in complexes with polyamines and cofactor have been characterized earlier. Here, we present the dodecameric structure of SpeG from V. cholerae in a ligand-free form in three different conformational states: open, intermediate and closed. All structures were crystallized in C2 space group symmetry and contain 6 monomers in the asymmetric unit cell. Two hexamers related by crystallographic twofold symmetry form the SpeG dodecamer. The open and intermediate states have a unique open dodecameric ring. This SpeG dodecamer is asymmetric except for the one twofold axis and is unlike any known dodecameric structure. Using a fluorescence thermal shift assay, size exclusion chromatography with multi-angle light scattering, small angle X-ray scattering analysis, negative stain electron microscopy, and structural analysis we demonstrate that this unique open dodecameric state exists in solution. Our combined results indicate that polyamines trigger conformational changes and induce the symmetric closed dodecameric state of the protein when they bind to their allosteric sites.
Keywords: dodecameric enzyme, asymmetric structure, allosteric site, spermidine/spermine, GNAT acetyltransferase
Graphical abstract
Introduction
Spermidine N-acetyltransferase is an enzyme encoded by the speG gene that regulates polyamine levels in bacteria by acetylating them and converting them to an inert, less toxic form that can be excreted. It has been shown that SpeG from Escherichia coli can acetylate spermidine in response to a variety of environmental stresses [1–5]. Sequence analysis of SpeG shows that the enzyme is a member of the GNAT superfamily of acetyltransferases [6, 7]. The crystal structure of SpeG from V. cholerae has been determined in one ligand-free enzyme form and in binary and ternary complexes with spermidine/spermine and/or acetyl coenzyme A/coenzyme A (acetyl-CoA/CoA) [8]. Unlike all other known GNAT structures, the crystal structure of SpeG displays a unique dodecameric architecture that is formed by two stacked hexamers. Each hexamer consists of 6 monomers that share a conserved GNAT fold [6, 7]. Two monomers from opposite hexamers form the dimer that is typically observed among known GNAT protein structures [6, 7]. Two different conformational states of the SpeG subunits corresponding to a ligand-free form and ligand bound form have been identified [8].
The SpeG dodecameric structure reveals a distinct mechanism of allosteric activation [8]. It has been observed that polyamines (spermidine/spermine) bind in a subunit interface of the dodecamer between adjacent monomers of the hexamers and induce a shift of the active site loop α1–α2 that is necessary for catalysis. The kinetic data show that SpeG from V. cholerae can acetylate only long polyamines, such as spermidine and spermine, in vitro [9] and fit a bireactant random steady-state kinetic mechanism [8].
Here, we report crystal structures of SpeG from V. cholerae in ligand-free form in three distinct conformational states obtained in the same space group symmetry C2: open, intermediate and closed. The open and intermediate dodecamers reveal an unusual asymmetric open arrangement of the dodecameric ring. Negative stain electron microscopy (EM) and small angle X-ray scattering analysis (SAXS) show that SpeG with this unusual open dodecamer exists in solution. The data indicate that an increase in polyamine concentration induces a shift from the asymmetric open state to the closed symmetric dodecameric state.
Results
Conservation of the SpeG dodecamer in different bacterial species
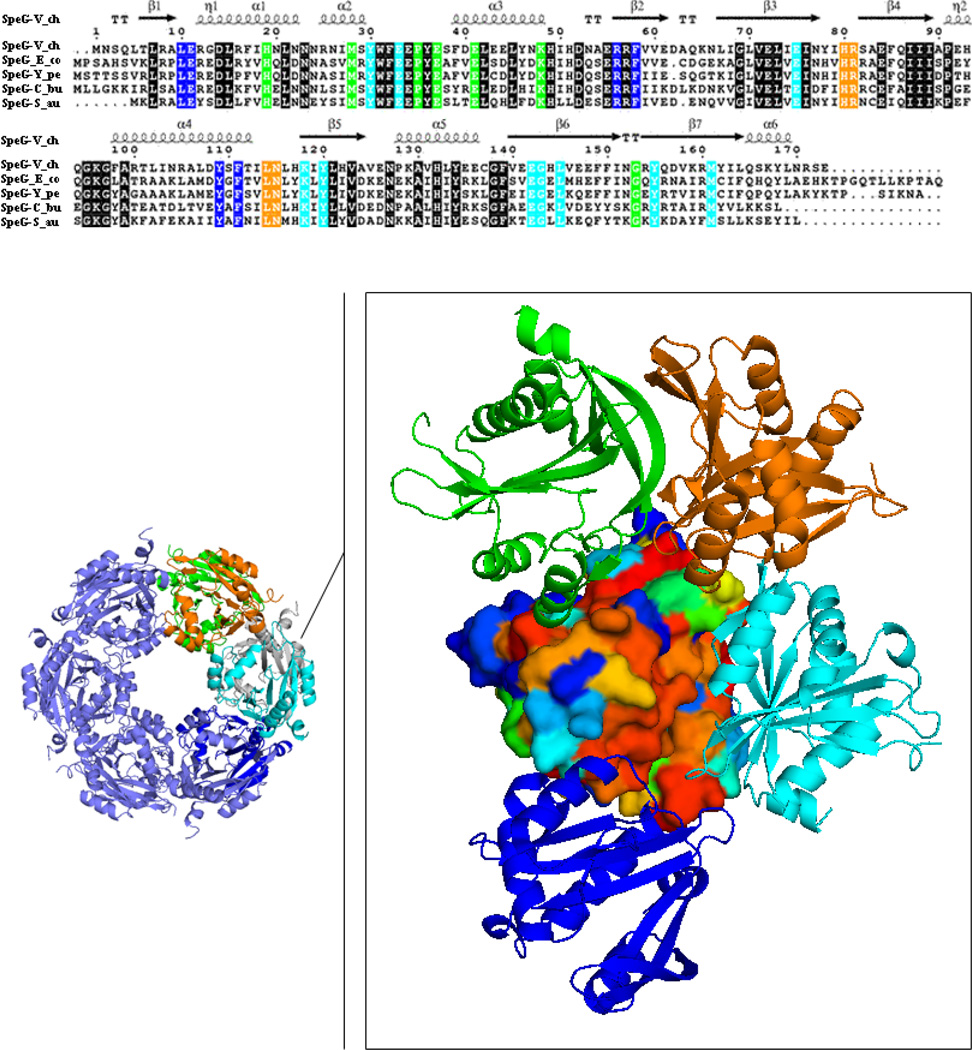
Among the GNAT family of acetyltransferases with known structures, SpeG from V. cholerae forms a unique functional unit, a dodecamer. The dodecameric structure of SpeG from V. cholerae in ligand-free form in two space groups (P42212, I222), in complexes with spermidine/spermine in space group I222 and complexed with AcCoA/CoA in space group P3 have been characterized previously [8]. In all of those structures, the SpeG dodecamer is composed of 12 monomers related by dihedral symmetry D6 that describes two hexamers related by twofold axis (Fig. 1). The six monomers in each hexamer of the SpeG dodecamer contribute to the six allosteric polyamine-binding sites, which cross between protomers. It has already been shown that SpeG binds spermidine/spermine in an allosteric polyamine-binding site, which activates the bireactant random steady-state kinetic mechanism [8]. To verify that homologous SpeG proteins might adopt similar dodecameric architecture and function, we performed a sequence-structure alignment analysis (see Materials and methods section). A list of 60 randomly chosen SpeG sequences was generated. Based on a multisequence alignment, residues of the SpeG monomeric structure were colored by sequence conservation (Fig. 1).
Fig. 1.
SpeG sequence-structure alignment. Surface representation of the SpeG monomer colored by degree of sequence conservation from red for 100 % conserved residues to blue for non-conserved residues. The neighboring monomers interacting with the SpeG monomer within the dodecamer are displayed as ribbon diagram and colored in cyan, green, blue and orange. Conserved residues located on the interface between contacting monomers in the SpeG dodecamer are colored according to the sequence alignment that shown on top of the figure. Sequence alignment of SpeG from V. cholerae (SpeG-V_ch) with other SpeG homologs includes: SpeG from E. coli (SpeG-E_co), SpeG from Y. pestis (SpeG-Y_pe), SpeG from C. burnetii (SpeG-C_bu) and SpeG from S. aureus (SpeG-S_au). In the sequence alignment conserved residues of the protein active site are shown on black background. Secondary structure elements of the SpeG monomer from V. cholerae are presented above the sequence. The sequence alignment of SpeG from V. cholerae with other homologous proteins was generated using CLUSTALW [48] and formatted in ESPript [49].
Each monomer in the SpeG dodecamer interacts with four neighboring monomers: with the monomer (cyan) that forms the GNAT dimer, two adjacent monomers of the same hexamer (green and blue) and with one other monomer (orange) from the opposite hexamer. We observed that residues involved in surface contacts between monomers of the SpeG dodecamer are highly conserved (Fig. 1). The surface area between monomers (grey and cyan) of the typical GNAT dimer has the largest interaction surface area and accounts for ~ 1070 Å2 (see Table 1). Most GNAT proteins are dimers with conserved monomer-monomer interactions that involve the C-terminal strands (β6 and β7 in SpeG) [7]. The interaction surface area between a SpeG monomer and the adjacent monomer (green or blue) of the hexamer includes the allosteric polyamine-binding site and has an area of interaction of ~ 700 Å2. All conserved residues of this motif are involved in polyamine binding as previously described [8]. The interaction between a SpeG monomer and the second monomer (orange) from the opposite hexamer has the smallest surface area, ~ 340 Å2. The residues located on loops between β3–β4 and α4-β5 in the second monomer from the opposite hexamer create the fourth conserved region of interactions. Based on this analysis, we predict that homologous SpeG enzymes from different bacteria will form a similar oligomeric state with allosteric polyamine-binding sites and functions. The homolog structure of SpeG from Coxiella burnetii (PDBID: 3TTH) and recently deposited structure of SpeG from E. coli determined in our CSGID center (http://www.csgid.org) at 2.9 Å resolution (PDBID: 4R9M) with 55 % and 54 % identity, respectively, to V. cholerae represent conserved SpeG dodecamers with the equivalent intersubunit interactions.
Table 1.
Intersubunit surface area# between dimers in SpeG dodecamer.
| Open State | Intermediate State | Closed State | |
|---|---|---|---|
| Subunit Interface |
Interface Area, Å2 | Interface Area, Å2 | Interface Area, Å2 |
| 1–2 | 1641 | 1648 | - |
| 2–3 | 1637 | 1737 | - |
| 3–4 | 1163 | 1233 | - |
| 4–5 | 1638 | 1740 | 1582 |
| 5–6 | 1640 | 1649 | 1598 |
| 6-1 | 96.6 | 581 | 1585 |
| GNAT dimer | |||
| 1 | 1084 | 1120 | 1076 |
| 2 | 1057 | 1090 | - |
| 3 | 1073 | 1084 | - |
| 4 | 1073 | 1081 | - |
| 5 | 1056 | 1091 | 1094 |
| 6 | 1085 | 1123 | 1076 |
Intersubunit surface area differences due to oligomer formation were calculated by QtPISA program from CCP4 Program Suite [29]
Structure and crystal packing of the SpeG dodecamer in ligand-free form
Screening of SpeG crystals from different crystallization conditions allowed us to determine three SpeG structures in ligand-free form (Table 2). These structures exhibit different dodecameric states (closed, intermediate, open) in the same space group C2 (Fig. 2, 3). The closed dodecamer in C2 space group is like the SpeG dodecamer characterized in ligand-free form and in complexes with spermidine/spermine or AcCoA/CoA that crystallized in higher symmetry space groups I222, P42212 or P3 [8]. In contrast, the arrangement of SpeG dodecamers in open and in intermediate state is different and can be described as an asymmetric dodecameric ring with a wide opening (Fig. 2a, 2b). The distance between monomers on each side of the asymmetric opening of the SpeG dodecamer in open and intermediate states is ≈ 17 and 8 Å, respectively. The total surface area calculated by the program AreaIMol [10] of the open and intermediate SpeG dodecamers are higher (89496 and 85294 A2) compared to that of the closed state (81863 A2). The main contribution to the increased total surface area of the open and intermediate dodecamers comes from the monomers adjacent to the opening.
Table 2.
Data collection, structure determination and refinement statistics&.
| SpeG dodecamer in open state |
SpeG dodecamer in intermediate state |
SpeG dodecamer in closed state |
|
|---|---|---|---|
| Crystal parameters | |||
| Resolution (Å) | 40.0-2.38 (2.47-2.38) | 30.0-2.5 (2.54-2.5) | 30.0-2.88 (2.95-2.88) |
| Space group | C2 | C2 | C2 |
| Unit cell parameters | |||
| a, b, c (Å) | 159.9, 134.2, 77.5 | 155.1, 135.8, 72.8 | 194.9, 71.8, 136.7 |
| α, β, γ (°) | 90, 114.4, 90 | 90, 117.1, 90 | 90, 134.5, 90 |
| Matthews coefficient (Å3/Da) | 2.99 | 2.8 | 2.7 |
| Solvent content (%) | 58.8 | 56.8 | 54.6 |
| Data Collection | |||
| Completeness (%) | 99.1 (91.6) | 100 (100) | 99.9 (100) |
| No of unique reflections | 58601 | 46610 | 30456 |
| I/σ(I) | 28.5 (2.1) | 25.4 (2.0) | 22.8 (1.7) |
| Rmerge (%) | 0.07 (0.5) | 0.06 (0.6) | 0.08 (0.7) |
| Redundancy | 4.6 (3.5) | 3.8 (3.8) | 3.8 (3.8) |
| Wilson B-factor (Å2) | 43.9 | 54.8 | 76.9 |
| Refinement | |||
| R (%)/Rfree (%) | 18.5/23.6 | 18/25.3 | 18.5/27.7 |
| RMSD bond length (Å) | 0.012 | 0.015 | 0.012 |
| RMSD bond angle (°) | 1.5 | 1.8 | 1.56 |
| Average B value (Å2) | 50.2 | 61 | 84 |
| No of atoms | |||
| Protein | 8689 | 8764 | 8655 |
| Metal Ions/Other Ligands | 3/8 | 6/2 | −/25 |
| Water molecules | 237 | 144 | 30 |
| Ramachandran analysis# | |||
| Favoured (%)/n | 97.3/994 | 97.2/976 | 93.3/949 |
| Allowed (%)/n | 2.7/28 | 2.8/28 | 6.7/68 |
| Outlier (%)/n | - | - | - |
Data for the highest resolution shell are given in parentheses. The abbreviations RMSD and ASU stand for root-mean-square deviation and asymmetric unit, respectively.
Defined by validation program MolProbity
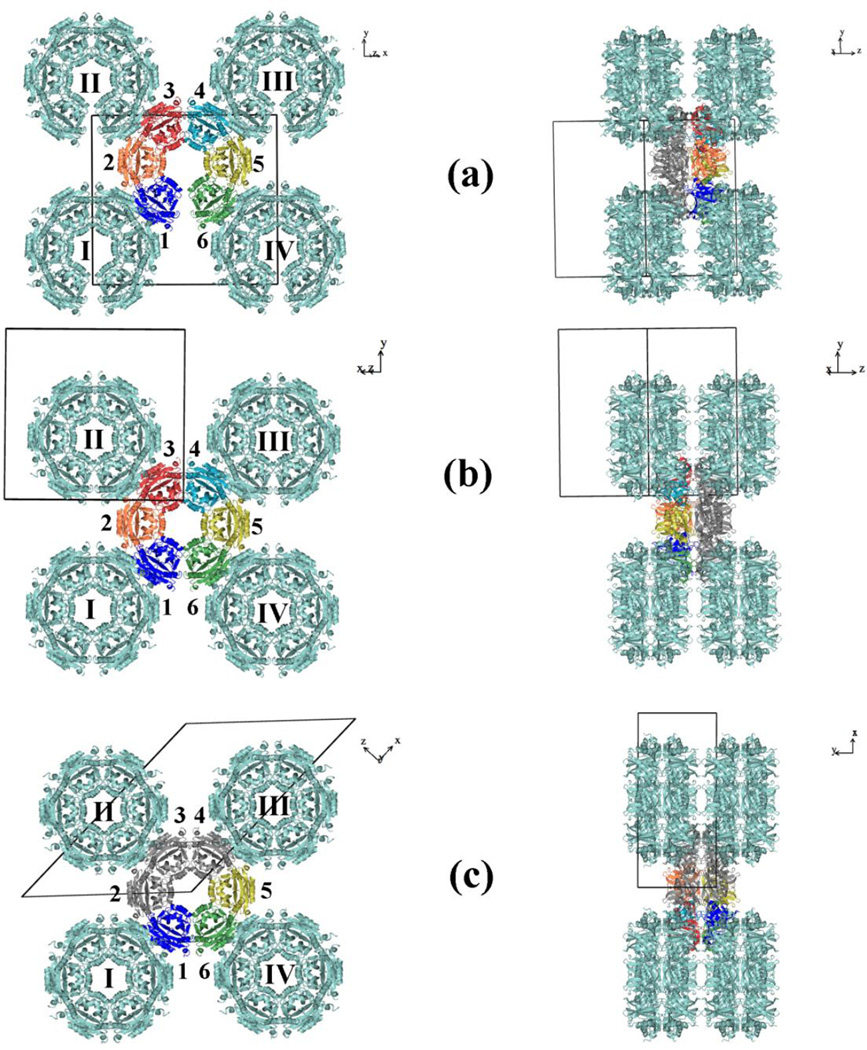
Fig. 2.
Crystal packing. Molecular layers in the structure of SpeG in open dodecameric state (a), intermediate dodecameric state (b) and closed dodecameric state (c). The monomers of the asymmetric unit cell are colored. The 2-fold rotation axis goes along the crystallographic axis Y. Monomers of the SpeG dodecamer that are related by crystallographic symmetry are shown in grey. The GNAT dimers in the dodecameric structure that are composed from monomers related by 2-fold crystallographic or noncrystallographic rotation axis are numbered clockwise from 1 to 6.
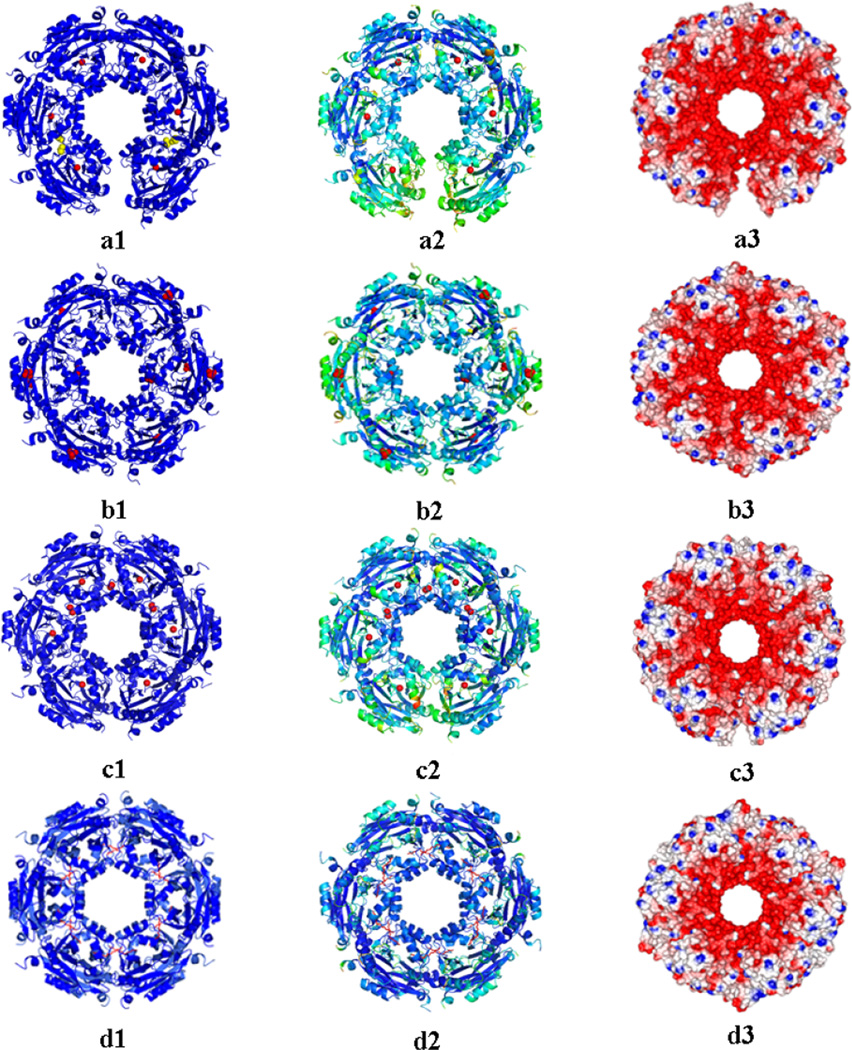
Fig. 3.
Ribbon diagram of the SpeG dodecamer. The SpeG dodecameric structure in open state in C2 space group with magnesium ions and isopropyl alcohol (IPA) molecules (a1), in closed state in C2 space group with sulfate ions (b1), in intermediate state in C2 space group with calcium ions (c1), in closed state in presence of spermine in I222 space group (d1); B-factor distribution of the SpeG dodecamers colored from the least degree of motion (in blue) to highest (in yellow) values (a2–d2); Electrostatic surface of SpeG dodecamers that corresponds to red for the negative charge, blue for the positive and white for the neutral charges (a3–d3); Metal ions are shown as red spheres, spermine shown as a stick model in red, sulfate as red spheres and IPA as yellow spheres.
In the crystal lattice, tightly packed SpeG dodecamers stack on top of each other to make long tunnels. The Mathews coefficients for the open and intermediate SpeG dodecameric structures are slightly higher (Vm = 2.99 and 2.92) then for closed (Vm = 2.71). The asymmetric unit in each of these structures contains 6 protein monomers with essentially the same tertiary structure. In the SpeG structures with open and intermediate dodecameric states, six monomers comprise an open hexameric ring that then forms the SpeG dodecamer by a 2-fold rotation axis (Fig. 2a, 2b). Related monomers from each hexamer form GNAT dimers [7]. In the SpeG structure in the closed dodecameric state, 6 monomers form three GNAT dimers in the asymmetric unit of the crystal cell and comprise a half of the dodecamer donut (Fig. 2c). Therefore, the crystallographic 2-fold rotation axis generates a full SpeG dodecamer in the closed state. The intersubunit interface between monomers related either by crystallographic or a noncrystallographic 2-fold rotation axis comprising the GNAT dimers in all three dodecameric structures is very similar (Table 1). However, the intersubunit surface area between adjacent GNAT dimers within the dodecamer is different in the different dodecameric states. Analysis of interfaces between GNAT dimers in open and intermediate dodecameric structures suggests that the interactions between dimers 1–2, 2–3, 4–5 and 5–6 as shown on Fig. 2a and 2b are the strongest compared to interactions between dimers 3–4 and especially 1–6 (Table 1). In the SpeG structure with the closed dodecameric state all intersubunit interfaces between GNAT dimers inside the dodecamer are equivalent.
It is possible that the different dodecamer structures are a consequence of different crystal packing. To examine packing of structurally different SpeG dodecamers in the crystal cell, we performed a crystal contact analysis. The contacts between a SpeG hexamer and symmetry related hexamers in three different forms (closed, open and intermediate) were calculated by program CONTACT from the CCP4 program suite [11] with the Van der Waals interactions being < 4.6 Å. The contacts between the remaining residues of the N-terminal histidine-tag in the SpeG structures were not included. The analysis indicates that the hexamers in all three forms have similar contact areas with symmetry related hexamers. Each SpeG hexamer forms crystal contacts with four symmetry equivalent dodecamers (Fig. 2). The number of hydrogen bonds between one hexamer and symmetry equivalent hexamers in closed, open and intermediate forms are 36, 43 and 43, respectively. The most extensive interactions of SpeG hexamers with crystallographically related monomers in all three crystal forms were observed between the C-terminal region (residues 170 – 173) and the conserved AcCoA binding motif (residues 94–100) in four monomers (1, 3, 4, and 6 as shown on Fig. 2) of the SpeG hexamer (Table S1). The small differences between open, intermediate and closed state were found in the symmetry contact area of monomer 2 and 5. Compared to the closed SpeG structure these subunits of the SpeG structures with open or intermediate states have additional contacts with symmetry equivalent monomers between residues 170-95, 171-129, 169-129 (only in intermediate state) and 154-136, 169, 170-129, respectively in monomer 2 and 5 (Table S1).
Additionally, to identify stable complexes within the crystal environment, we calculated the Gibbs free energy for the SpeG dodecamer in open, intermediate and closed forms using the Protein Interface, Surfaces and Assemblies (PISA) server that “employs thermodynamic models based on optimal position of the protein assemblies” [12]. The PISA analysis suggests that the SpeG dodecamers in the open and intermediate states are more stable with ΔGdiss = 46.4 and 42.8 kcal/mol, respectively, compared to the closed dodecameric form that gave a ΔGdiss = 31.2 kcal/mol. Therefore, our combined results allow us to conclude that the crystal packing most likely does not induce conformation changes in the SpeG quaternary structure. On the contrary, the differences in the unit cell parameters and solvent content could allow the different dodecameric states of the protein. Each SpeG dodecameric state may occur in solution and crystallization conditions could favor different states of the dodecameric structure in the crystal environment. The SpeG dodecamer in the closed state contains sulfate ions that were localized in 8 monomers near the “P-loop” (conserved AcCoA binding region) and in two monomers on the surface of the protein (Fig. 3b). In the open and intermediate dodecameric states magnesium and calcium ions, respectively, were identified between monomers comprising the GNAT dimers (Fig. 3a, 3c). In the crystal structures magnesium and calcium ions interact with the side chains of E33 and E75 from both monomers of the dimer. Additionally, in 4 SpeG monomers in the intermediate dodecameric state, calcium ions were observed in the interface between adjacent monomers of the hexamer at the location of the allosteric polyamine-binding pocket [8]. In this site calcium ions interact with N53, E34, E41 and P35 from adjacent monomers. Finally, an isopropyl alcohol molecule was found in a hydrophobic pocket near the allosteric polyamine-binding site in 4 monomers of the SpeG structure in open state.
SEC-MALS analysis of SpeG enzyme in ligand-free form, in complex with polyamine and cofactor
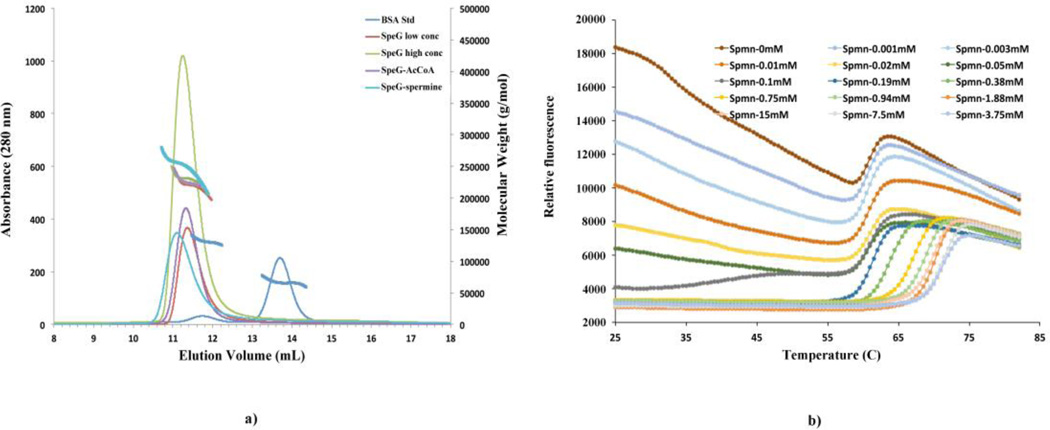
SEC-MALS was employed to measure the average molecular weight (MW) of SpeG in solution in ligand-free form and in presence of ligands required for SpeG activity (spermine and AcCoA) (Fig. 4a). At low concentration of the SpeG in ligand-free form (< 1 mg/mL), the SEC-MALS results show a continuous distribution of molecular mass with an average value of 220 kDa. At higher SpeG concentration (> 1 mg/mL), the average molecular mass increases to 230 kDa. In presence of polyamines, even at low protein concentration (< 1 mg/mL), the average MW is ~ 250 kDa, which corresponds to the SpeG dodecameric state. On the other hand, the addition of AcCoA to the protein solution indicates that SpeG in complex with AcCoA has a MW distribution similar to the distribution of SpeG in a ligand-free form. The SEC-MALS data of the SpeG in ligand-free form suggests that the smaller and intermediate oligomers co-elute significantly with the larger protein dodecamers. Although the mixture of SpeG oligomers does not separate in the SEC step, the MALS data indicates that the front of the peak in enriched in dodecamer while the tail of the peak in enriched in lower molecular weight forms. One might expect that a six-fold symmetric dodecamer would dissociate to dimers and monomers and there would be very small amounts of intermediate oligomers. This is because completing the symmetric ring adds twice the interactions compared to intermediate additions. The very low concentrations of oligomers between dimer and complete dodecamer would result in at least partial separation of dodecamers from dimers and monomers, giving very asymmetric SEC peaks. In contrast, the open dodecamer of SpeG means there is no energetic advantage to the dodecamer and there will be a mixture of all oligomers with significant amounts of intermediate forms, resulting in more symmetric SEC peaks whose average MW gradually shifts with protein concentration. In this case, separation of the larger oligomers is not possible. Importantly, the addition of polyamine increases the average MW and the asymmetry of the SEC peak, indicating the increased formation of dodecamers in the solution.
Fig. 4.
SEC-MALS elution profiles and FTS melting curves. (a) The SpeG elution profiles in ligand-free form at low (<1 mg/mL) concentration, at high (>1 mg/mL) concentration, in presence of spermine and AcCoA. Molecular mass distribution of the samples is shown as bold solid lines. (b) Melting curves of SpeG vs spermine concentration. The initial binding of Sypro Orange is reduced as the polyamine concentration increases. Above 0.1 mM of spermine the significant increase in the temperature of the steep unfolding transition of the protein indicates polyamine binding.
Fluorescence thermal shift assay of the SpeG enzyme with and without polyamine
We tested spermine to see if polyamine can stabilize or destabilize the SpeG structure using a fluorescence thermal shift (FTS) assay. Melting curves of SpeG that demonstrate specific binding of the spermine are shown on Fig. 4b and S1. The addition of spermine at concentrations from 0.1 to 10 mM resulted in a significant Tm shift > 10 (°C). At low spermine concentration < 0.1 mM the melting curves indicate that although the dye (sypro orange) is able to bind to exposed hydrophobic regions of the SpeG, giving rise to the high initial relative fluorescence, the presence of low concentrations of spermine reduces that binding. Although this data can’t distinguish the mechanism of stabilization, the data do indicate that spermine interacts with SpeG and stabilizes the protein, increasing its melting temperature.
SAXS analysis of SpeG enzyme with and without polyamine
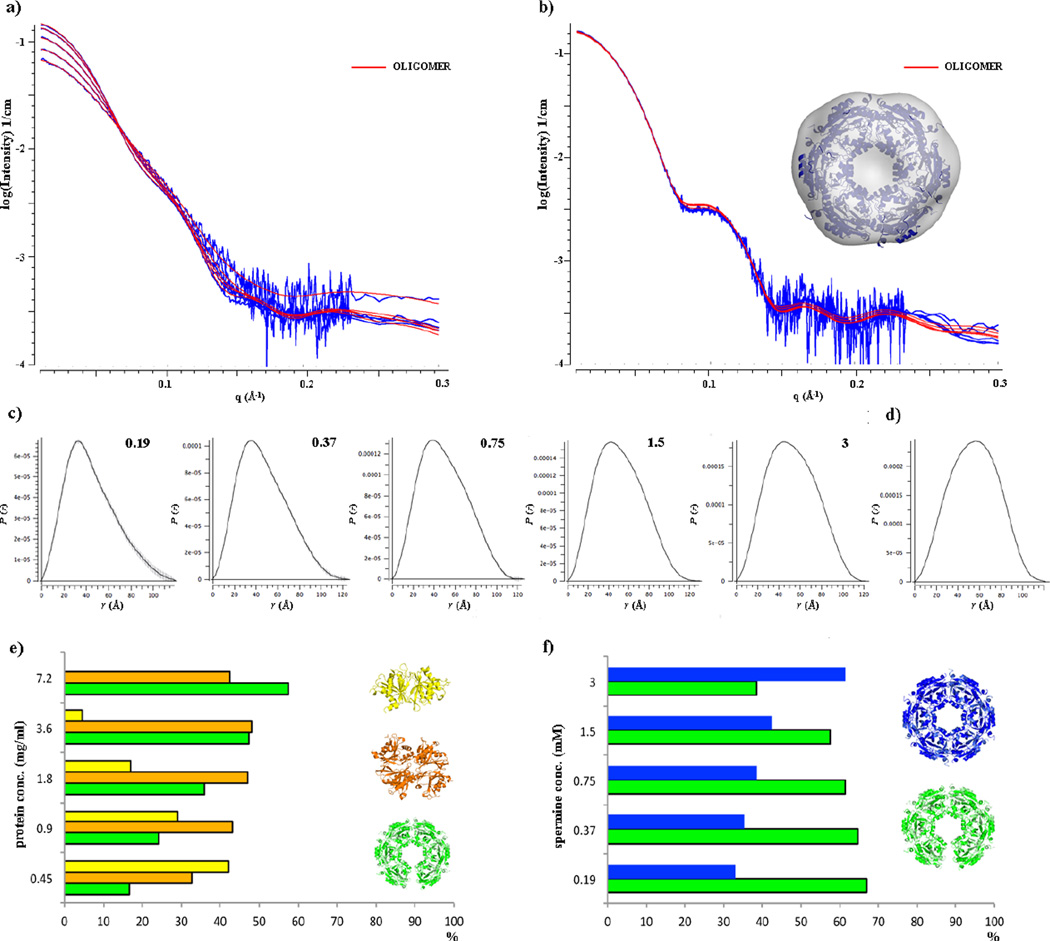
In order to verify that the dodecameric SpeG structure with its asymmetric opening could exist in solution, and was not a crystal packing artifact, we performed SAXS analysis. This is a sensitive tool for the evaluation of oligomeric state and overall size of the protein in solution. All SAXS measurements were carried out for the SpeG protein varying its concentration and in absence and presence of polyamine (spermine) (Fig. 5a, 5b).
Fig. 5.
SAXS analysis of SpeG in ligand-free form and in complex with polyamine. (a) and (b) Scattering curves for the SpeG in ligand-free form and in presence of spermine, respectively. The fit of SpeG structural models with OLIGOMER [13] to experimental data is shown in red. (c) and (d) P(r) function distributions for the SpeG in ligand-free form and in presence of the polyamine, respectively. (e) Distribution of SpeG oligomeric states in ligand-free form vs protein concentration. (f) Distribution of SpeG oligomeric states in presence of polyamine in solution vs spermine concentration.
Based on the scattering curves, we evaluated the average radius of gyration (Rg) and average MW of the protein species in solution with and without spermine addition (Table 3). We observed that for SpeG in the ligand-free form the Rg and MW values increase with the protein concentration (Fig. 5c), indicating that the protein without ligand exists in an equilibrium between different oligomeric states in solution. These results agree with the SEC-MALS analysis. To identify the volume fractions of the mixtures of SpeG oligomeric states we used the program OLIGOMER [13]. The scattering curves were fit by a linear combination of the scattering calculated from different hypothetical oligomeric forms of the SpeG structure (see Materials and methods) and are shown in Fig. 5a, 5b. The data indicate that the SpeG protein in ligand-free form exists in solution in an equilibrium between dimers, tetramers and the dodecamers in an open conformational state (Fig. 5e, Table S2, S3). At high concentration, 3.6 mg/mL and above, the equilibrium shifts to tetramers and dodecamers with an open conformation. The SAXS analysis of the species in ligand-free protein solutions has not provided evidence for SpeG dodecamers in the closed conformational state.
Table 3.
Structural parameters and molecular-mass determination of SpeG in solution.
| SpeG ligand-free form/complex with spermine | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Concentration, mg/mL | 0.45 | 0.9 | 1.8 | 3.6 | 7.2 | |||||
| Structural parameters: | free | complex | free | complex | free | complex | free | complex | free | complex |
| I(0) (cm−1) [from P(r)] | 0.043 | 0.197 | 0.072 | 0.196 | 0.099 | 0.196 | 0.123 | 0.197 | 0.144 | 0.196 |
| Rg (Å) [from P(r)] | 35 | 45.5 | 36.7 | 42.4 | 38.5 | 42.4 | 39.7 | 42.4 | 40.6 | 42.1 |
| I(0) (cm−1) (from Guinier plot) | 0.066 | 0.199 | 0.085 | 0.199 | 0.109 | 0.199 | 0.132 | 0.201 | 0.144 | 0.197 |
| Rg (Å) (from Guinier plot) | 34.2 | 43.4 | 36.4 | 43.4 | 38.6 | 43.7 | 39.6 | 43.7 | 39.9 | 43.0 |
| Dmax (Å) | 117.7 | 147.9 | 125.9 | 140.8 | 131.1 | 134.5 | 131.4 | 125.5 | 139.4 | 121.7 |
| Porod volume estimate (nm3) | 135 | 436 | 196 | 445 | 245 | 448 | 287 | 455 | 321 | 448 |
| Volume of correlation (Å3) | 537 | 1128 | 660 | 1136 | 769 | 1139 | 851 | 1145 | 909 | 1128 |
| Molecular-mass determination: | ||||||||||
| Molecular mass Mr [Vc2/(.1231×Rg)] (kD) | 68.5 | 239 | 97.5 | 241 | 124 | 241 | 149 | 244 | 168 | 240 |
| Calculated monomeric Mr from sequence (kD) | 20.7 | |||||||||
When spermine was added to the protein solution, the range of Rg and MW didn’t significantly change but rather stayed in the same range of values (Table 3). The values of Rg based on scattering data (Fig. 5d) agree with the Rg calculated from the SpeG dodecameric structure in complex with polyamines and was 42 Å. The evaluation of the different oligomeric states for SpeG in presence of spermine by the OLIGOMER program [13] shows that SpeG exists in solution only as dodecamers with an equilibrium between open and closed conformational states (Fig. 5f, Table S2, S3). The data also indicate that an increase in polyamine concentration shifts the equilibrium towards dodecamer in the closed conformation, suggesting that presence of polyamine triggers a closed dodecameric state.
Additionally, the shape of the scattering curves demonstrate that not only the oligomeric content of SpeG protein species are changing but the overall dodecameric state is modifying its shape upon binding of the polyamine. The shape of the scattering curves clearly shows differences (Fig. 5a, 5b). In the presence of the spermine, the scattering curve has a shoulder at the angle of q > 0.1 Å−1. This change could be explained by simultaneous conformational rearrangements of the loop α1–α2 localized between adjacent monomers of the SpeG hexamer [8] and the closure of the dodecameric ring that are induced by binding of the polyamine molecules into the allosteric site of the SpeG enzyme.
To reconstruct the SpeG dodecameric structure in solution in absence and presence of spermine the SASTBX [14] and/or DAMMIF [15] calculations with symmetry P6 were performed and averaged by DAMAVER [16]. The modeling of the open dodecameric structure didn’t give a reasonable envelope; this could be explained by the presence of several protein oligomeric species in solution. The ab initio model obtained for the SpeG dodecamer in presence of spermine (Fig. 5b) yielded a good fit to the experimental data with χ = 1.3. These data combine to support the dodecameric quaternary structures of SpeG observed in the crystal.
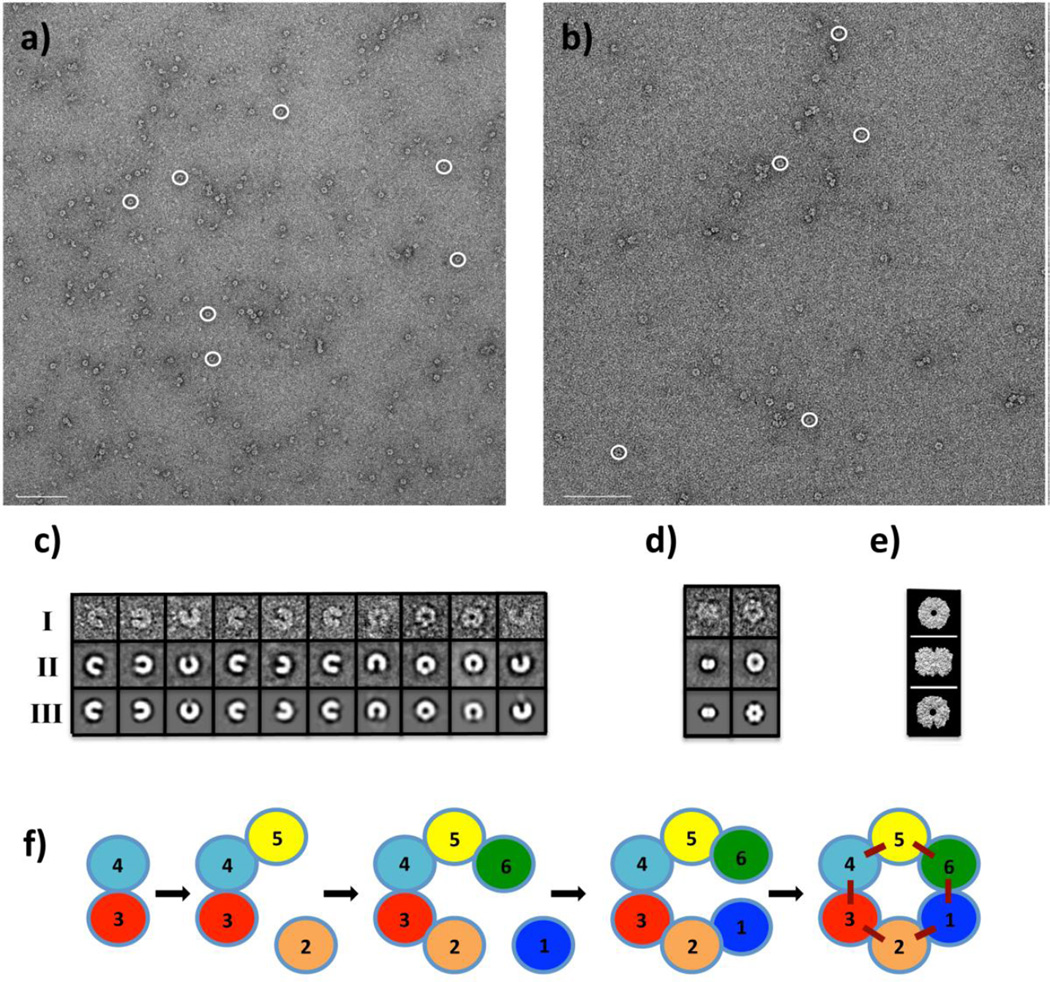
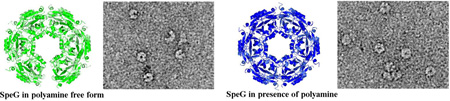
Visualization of SpeG dodecameric structure by negative stain EM
Our structural and SAXS analysis shows that the SpeG dodecamer could exist in different conformational states. To further demonstrate that the SpeG dodecamer in open form is not an artifact of our experimental procedures, we visualized SpeG dodecamers in ligand-free form (Fig. 6a) and in the presence of spermine (Fig. 6b) using negative stain EM. From a set of 3151 and 3869 SpeG particles in ligand-free form and with polyamine, respectively, that were observed on the images, the subsets of particles representing SpeG molecules in different states in the same orientation were used for single particle reconstruction. Classification of subsets of particle images yielded several class averages (Fig. 6c, 6d). Closed and open dodecamers were independently classified on images with added polyamine and without, respectively. For SpeG in ligand-free form, different homooligomers including hexamers, octamers, decamers and dodecamers were identified. Due to heterogeneity of the SpeG sample in the absence of polyamines, the identification of SpeG dimers and tetramers were difficult. From the mixed population of manually picked particles (3151) for the ligand-free protein, 679 particles represent SpeG dodecamers in the open state, while few SpeG dodecamers were observed in the closed state. Most of the SpeG particles in the absence of polyamine were found in other oligomeric states. For SpeG in the presence of spermine, only open and closed dodecamers were observed. From this set of mixed particles (3869), 3418 particles have the closed dodecameric state and only 319 SpeG particles have the open state. The projections from a 25 Å resolution-limited model obtained from the SpeG dodecameric structure in open and closed states [8] yielded three projection averages that match the side and top views of the SpeG dodecamers observed on the EM images (Fig. 6e). These data correspond very well to the SAXS analysis.
Fig. 6.
Negative stain EM analysis. (a) and (b) Negative stain EM images of the SpeG in ligand-free form and in presence of spermine, respectively. Scale bars are 200 nm. (c) SpeG oligomeric states in ligand-free form observed on the images (row I), their class averages (row II) and corresponding refined models (row III). Closed SpeG dodecamers were excluded from the classification. (d) The top and side views of SpeG dodecamer in closed state observed on the images of SpeG in presence of spermine (row I), their class averages (row II) and corresponding refined models (row III). The SpeG dodecamers in open state were excluded from the classification. (e) Projections of SpeG dodecamer in open and closed states created from a 25 Å resolution-limited model obtained from the three-dimensional structures. (f) A model for assembly of the SpeG dodecamer in the open state.
Discussion
Among GNAT family members, the SpeG dodecameric enzyme forms a distinct family of polyamine acetyltransferases that use a unique allosteric site to regulate the acetylation of polyamines [8]. Various experimental procedures including light scattering, SEC-MALS and SAXS analysis confirm that the SpeG dodecamer is likely to be the biologically relevant unit of the protein. Surprisingly, we observed that in the crystal structure, the SpeG dodecamer in a ligand-free form could exist in different conformational states: open, intermediate and closed.
The open and intermediate dodecameric forms of SpeG are unique (Fig. 3) compared to any protein dodecameric structures that have been determined so far. The conformational differences in SpeG dodecameric structures observed in the crystal could have been due to many factors including crystal packing, protein flexibility, presence of the ligand, different crystallization conditions or the protein’s biological function. The crystal contact analysis of all three dodecameric states determined in the same space group C2 shows that all have similar contacts in the crystal and most probably crystal contacts do not determine the conformational changes in the SpeG dodecamer (Table S1). Comparison of B-factor distributions of the SpeG dodecameric structures (Fig. 3a2–d2) shows that the SpeG dodecamers in ligand-free form in different conformational states have higher temperature factors compared to the closed SpeG dodecamer in presence of polyamine. Potentially this could indicate structural flexibility dictated by the dynamic nature of the enzyme. The structural data suggest that the presence of other ligands in crystallization conditions such as metal ions, sulfate ions or alcohols could alter the quaternary structure and favor the open and intermediate states of the SpeG dodecamer in the crystal environment.
The SEC-MALS and SAXS data both indicate that in the absence of polyamine the protein exists as an equilibrium mixture of oligomers, consistent with the formation of the open dodecamer. Because the open dodecamer does not have the energetic advantage of finishing a six-fold symmetric structure, the mixture of oligomers will be complex and concentration dependent. As expected, the percentage of dodecamers increases at higher protein concentrations (> 3.6 mg/mL). However, analysis of the SAXS data is not sufficient to provide evidence for the ligand-free form of SpeG dodecamers having the closed conformational state. In the presence of polyamine, the closed, symmetric dodecamer is favored and the amounts of intermediate oligomers reduced. The negative stain EM also demonstrates that the SpeG dodecamer exists in solution in an asymmetric open form (Fig. 5 and 6). Furthermore, the negative stain EM images for SpeG in ligand-free form indicate that only a small fraction of dodecamers occur in the closed state. This suggests that the distortion in the protein-protein interfaces required to close the dodecameric structure, compared to the interactions gained when the dodecamer closes, results in an equilibrium between open and closed dodecamers in the absence of polyamine that favors the open form. Although the fitting of the SAXS experimental curves looks good (Fig. 5a), it is possible that the complex mixture of states that exist in a SpeG solution of the ligand-free form could not be fully modeled by the PDB structures that were used. In addition, the identification of low MW particles corresponding to tetramers or dimers of SpeG in ligand-free form by negative stain EM analysis is difficult. Differences in concentration, sample preparation and sensitivity of the techniques could explain the variability of SpeG oligomeric states at low concentrations when SAXS and negative stain EM results are compared. Importantly, both SAXS and negative stain EM data clearly show, that in presence of polyamine (spermine) SpeG exists as dodecamer in equilibrium between the open and closed dodecameric forms. The percentage of closed dodecamers in solution is increased when the concentration of polyamines increases (Fig. 5f), indicating that polyamine binding to the intersubunit allosteric site alters the relative stability of the dodecameric subunit interactions and favors the symmetric, closed dodecamer. These data correlate very well with the SpeG crystal structures. In addition, the SEC-MALS and FTS analysis show that polyamine can affect the formation of the dodecamer and stabilize its conformation. As seen in the SpeG crystal structure in complex with spermidine or spermine [8], the binding of polyamines in the allosteric binding site induces the closed dodecameric state. In this state, the protein can quickly proceed to acetylation of polyamines in the substrate binding sites [8]. The negative electrostatic surface of the SpeG dodecameric structure (Fig. 3a3–d3) suggests that SpeG prefers to bind positively charged ligands like polyamines or metals ions at both the allosteric and substrate-binding sites.
It is very unusual for a protein to have an asymmetric structure. Usually, symmetric oligomers are observed that have point group symmetry. These symmetric arrangements are because the intersubunit interactions are maximized. One would expect the closed dodecameric state to be more stable since adding one dimeric unit to a decamer to form the dodecamer would add the contribution from two interfaces. However, in the absence of polyamine this does not happen and we see the mixture of different oligomeric states from dimers to dodecamers. In negative stain EM images we observe that a SpeG dimer can attach to the decamer creating an open SpeG dodecamer (Fig. 6c). In Figure 6f we propose a model for assembly of the SpeG dodecamer in the open state. Two dimeric units of SpeG form a tetramer and each time one dimeric unit attaches to either side of the tetramer to form hexamer etc., until the last dimeric unit will connect to the decamer to form the open dodecameric structure. At this point no more dimer units can be added. Spermidine/spermine binding to the allosteric sites at the interface between adjacent monomers of the hexamers in the dodecameric structure would propagate changes around the ring, altering the rotation angles until the rings close and the final set of intersubunit interactions are contributed. Therefore, the binding of the positively charged polyamines provide enough energy to overcome the strain and drive the closure of the asymmetric dodecameric ring to form the more stable and symmetric closed dodecameric state.
Based on our results, we predict that the open conformational state is the apo protein native state and could be functionally important. The polyamine regulation in the cell is complex and may involve additional mechanisms to regulate and coordinate polyamine concentrations. There are examples of oligomeric proteins where the breaking of symmetry plays an important role in shaping protein structures to perform required functions [17–20]. For example, the dynein motor domain reveals an asymmetric ringshaped structure [21]. The binding of nucleotide to the ATPase domain affects microtubule binding to the ATPase domain structure and induces conformational changes of the complex through long distance allosteric communication [22]. Similarly, depending on environmental conditions, the SpeG dodecamer in the open state might interact with another binding partner, which could disturb the favorable “ready for rapid catalysis” state of the enzyme and thereby coordinate the regulation of polyamine levels with other biological processes. Based on the sequence-structure alignment we hypothesize that SpeG from other pathogenic and nonpathogenic bacteria will adopt the open dodecameric state similar to the V. cholerae SpeG. We assume that the shift from dimer to dodecamer for SpeG may have physiological importance in determining polyamine concentrations in prokaryotic cells. We would also predict that the open conformational state of SpeG is important for its function and that SpeG may have additional independent functions besides spermidine acetylation.
In conclusion, we have shown that SpeG from V. cholerae exists in solution in a variety of protein homooligomeric states including dodecamers in an open state. The presence of the polyamines spermidine or spermine shifts the equilibrium to dodecamers and induces the formation of the closed, symmetric dodecamers. Our data support a model in which polyamines favor formation of the closed dodecameric state of the protein when they bind to their allosteric binding sites.
Materials and methods
Sequence-structure alignment
A search for SpeG homologs and comparison their sequences was performed with the PSI-BLAST server [23]. This produced a list of 500 sequences against the non-redundant database. A majority (90 %) of the homologs share more than 40 % sequence identity. Close homologs with high identities are primarily listed as spermidine N-acetyltransferases. Several sequences are annotated as diamine acetyltransferases, GCN5-related acetyltransferases or acetyltransferases. Proteins that did not have overlaps covering nearly the complete sequence were excluded from the sequence alignment. A set of 60 SpeG sequences were chosen and multiple sequence alignment for visualization of the sequence conservation with respect to the three-dimensional structure was generated using an in-house program “Replace_Conserved.py” developed in our laboratory (Clayton Watterson, unpublished). The SpeG monomer was colored using the program PyMOL [24] by degree of sequence conservation from red for fully conserved to blue for non-conserved residues.
Protein production and crystallization
The selenomethionine-labeled and native protein encoded by the speG gene from V. cholerae O1 biovar el tor strain N16961 (NCBI accession AAF96843, GI 9658384) was cloned, grown, expressed and purified using standard Center for Structural Genomics of Infectious Diseases protocols [25, 26]. For crystallization experiments, negative stain EM, SAXS and SEC-MALS analysis the histidine tag was removed.
Crystals of the SpeG enzyme were obtained at 19 °C using the sitting-drop vapor-diffusion method. Crystallization trials were carried out using a Phoenix robotic system (Rigaku, Art Robbins Instruments) to dispense protein and precipitant solutions (1:1 ratio) into the sitting drop that was equilibrated against a precipitant reservoir containing crystal screen solutions from ANL-2, Classics II or ComPas Crystal Screens from QIAGEN. Crystals of selenomethionine-labeled SpeG in an open dodecameric state were grown in a drop containing 0.4 µL of protein with concentration of 10 mg/mL in 100 mM Sodium Chloride, 10 mM HEPES buffer, pH 7.5 and 0.4 µL reservoir solution that contained 8 % Isopropanol, 0.1 M Tris-HCl buffer, pH 8.5. Crystals of native SpeG dodecameric structure in closed dodecameric state were grown in a drop containing 1 µL of protein with concentration of 8.5 mg/mL in 500 mM Sodium Chloride, 5 mM 2-Mercaptoethanol (BME), 10 mM Tris-HCl buffer, pH 8.3 and 1 µL reservoir solution that contained 0.05 M Ammonium Sulfate, 0.1 M tri-Sodium Citrate, 15 % PEG8000. Crystals of native SpeG in the intermediate dodecameric state were grown in crystallization conditions where drops contained 1 µL of protein with concentration of 8.5 mg/mL in 500 mM Sodium chloride, 5 mM BME, 10 mM Tris-HCl buffer, pH 8.3 and 1 µL reservoir solution that contained 0.01 M Calcium Chloride, 20 % Methanol, 0.1 M Tris-HCl buffer, pH 8.5. All crystals were harvested for data collection and flash-cooled in liquid nitrogen using 25 % Glycerol, 3.6 M Ammonium Sulfate and 25 % 2-Methyl-2,4-Pentanediol (MPD) as cryoprotectant, respectively for SpeG in open, closed and intermediate dodecameric state.
Structure solution and refinement
The X-ray crystallographic data were collected at Argonne National Laboratory (Argonne, IL) at 100° K. The single-wavelength anomalous dispersion (SAD) data set was obtained from crystals of the selenomethionine-labeled SpeG in open state at the Structural Biology Center on the beamline 19ID. The native data sets were collected from crystals of the SpeG in closed and intermediate dodecameric state at the Life Sciences Collaborative Access Team on the beamline 21ID-G and 21ID-F, respectively. All data were indexed, integrated and scaled using HKL-3000 [27]. The selenomethionine-labeled SpeG structure was solved with the HKL-3000 program suite [27]. The native SpeG structures were solved by molecular replacement with PHASER [28] from the CCP4 program suite [29]. The structure of the ligand-free form of SpeG in open state was used as the starting model. The refinement of the structures was done using REFMAC [30]. The water molecules were identified automatically with the program ARP/wARP [31]. The positions of the side chains, waters, turns and gaps in all three structures were checked and manually corrected using COOT [32]. The structures were analyzed with PROCHECK, ADIT and MolProbity [33–35]. Figures of 3D SpeG molecular structures were generated using PyMOL and CCP4MG [24, 36]. The data collection, structure determination and refinement statistics are presented in Table 2.
Coordinates and PDB accession codes
Atomic coordinates and structure factors of the validated structures have been deposited in the Brookhaven Protein Data Bank [37] with accession codes 5CNP, 4YGO (HPUB) and 4JLY for the SpeG dodecamer in the open state, in the intermediate state and in the closed state, respectively.
Size exclusion chromatography with multi-angle light scattering
SEC-MALS experiments to obtain the absolute MW of SpeG in ligand-free form and in complexes with polyamine and AcCoA were performed with a Waytt Dawn Heleos II multi-angle scattering (MALS) detector (Wyatt Technology Europe GmbH, Dernbach, Germany) coupled with Agilent Technologies 1100 LC HPLS system (Agilent Technologies, Santa Clara, CA) at Northwestern University (Evanston, IL) at the Keck Biophysics Facility. A total of 100 µL of the purified SpeG (at low 0.5 mg/mL and higher concentration 3 mg/mL), SpeG in complex with 5 mM spermine and 10 mM AcCoA in a buffer containing 500 mM Sodium Chloride, 5 mM BME in 10 mM Tris-HCl, pH 8.3 were applied on a Superdex 200 10/300 GL column (GE Healthcare, Piscataway, NJ) at a flow rate of 0.5 mL/min at 22° C. Bovine serum albumin (BSA) (Sigma-Aldrich Corp., St. Louis, MO) was used as a reference sample. The data were analyzed using ASTRA software from Wyatt Technology.
Fluorescence thermal shift assay
FTS assay was set up in the High Throughput Analysis Laboratory (HTAL) at Northwestern University (Evanston, IL) as described [38]. The assay was run using 1.9 µg/well of the SpeG (8.4 mg/mL protein stock solution) in a 10 µL assay buffer containing 100 mM HEPES, pH 7.5, 150 mM sodium chloride and 5 × sypro orange (dye). Five µL of spermine at different concentrations was dispensed to a plate wells containing 5 µL of the protein solution premixed with the dye. The thermal scan was collected from 10 to 95 °C and the fluorescence was recorded every 10 seconds. Data were analyzed with the in-house ExcelFTS software.
Small-angle X-ray scattering data collection and analysis
SAXS data for the SpeG enzyme were obtained in its ligand-free form and in complex with 3 mM spermine in buffer solution composed of 100 mM Sodium Chloride and 10 mM Tris-HCl, pH 8.3. The data were collected on the insertion device beamline of the DuPont - Northwestern – Dow Collaborative Access Team, 5ID-D, at Argonne National Laboratory (Argonne, IL). Data were collected on two CCD area detectors at 2.6 m (Mar165) and 0.29 m (Roper) from the sample, positioned above and below the direct beam path [39], which gave small and wide angle data sets overlapping slightly near q ~ 0.23 Å. A 0.25 mm by 0.25 mm beamsize with a pinhole geometry configuration was used at a wavelength of either 1.033 Å or 1.025 Å.
Each protein sample underwent five serial twofold dilutions using its buffer solution (described above) starting from roughly 7 mg/mL. The dilution buffer did not contain spermine when protein sample in complex with polyamine was measured. Data were collected on multiple 10 sec exposures from each of these dilutions and their buffers at ambient temperature while flowing at 4 µL/sec. The absence of radiation damage was confirmed by the consistency of sequential exposures. The capillary flow cell was washed with at least 1 mL of 20 % bleach, followed by at least 3 mL of water, and then at least 300 µL of the buffer solution. These washes were performed after each set of protein exposures and followed by a set of buffer exposures to ensure consistency of the background scattering. Additionally, a set of empty capillary exposures was collected before each dilution series.
The CCD images were azimuthally averaged to obtain intensity versus momentum transfer vector (4π sin θ/λ) using FIT2D [40]. The data were further corrected for transmission and incident beam variation based on the measured intensity in a beam stop mounted diode [39] and scaled to absolute units based on the small angle scattering of a water sample [41]. Bubble free exposures were averaged and the likewise averaged volume fraction corrected data from the buffer and the empty capillary exposures were subtracted from the sample solution scattering, using the concentration in mg/mL times 7.4 × 10−4 cm3/mg to estimate the protein volume fraction. This sample scattering was then divided by the concentration to produce the absolute differential scattering cross-section of the protein alone. In the case of the more concentrated samples of SpeG in complex with spermine where inter-particle interference was evident in the low-angle data, the data was extrapolated to infinite dilution by dividing out a hard-sphere structure factor equation fitted to the ratios of the concentrated samples to the most dilute [42]. Porod volume and volume of correlation [43] were calculated using the data to 0.3 Å−1. Pairwise distance distribution and real-space I(0) and Rg were calculated using GNOM from the ATSAS program suite [44]. Theoretical scattering curves were computed using CRYSOL [45], from atomic models generated from the crystal structure of the SpeG dodecamer in closed state, dodecamer in open state, dodecamer in intermediate state, decamer, octamer, two different hexameric forms, tetramer, dimer and monomer. These models, either all (Table S2) or a minimal set (Table S3), were then used in OLIGOMER [13] to estimate volume fractions of each species in solution. For samples, which showed concentration independent scattering, ab-inito models were computed with SASTBX [14] and/or DAMMIF [15]. Structure parameters are shown in Table 3. For SpeG in complex with spermine, data extrapolated to infinite dilution was used; this was not possible for the ligand-free form.
Negative stain electron microscopy
The negative stain EM data were obtained for SpeG protein without and with spermine at concentration of 5 mM using the CryoEM Facility at Northwestern University (Evanston, IL). The SpeG protein samples were diluted in a buffer solution containing 100 mM Sodium Chloride, 5 mM BME, 10 mM Tris-HCl at pH 8.3 to a final concentration of 0.002 mg/mL. To obtain images containing the complex of SpeG and polyamine, spermine was added to the protein solution and to the buffer solution to a final concentration of 5 mM. A 4 µL drop of each SpeG sample was applied to a glow-discharged carbon-coated copper grid, washed twice with buffer solution and stained in two drops of 2 % uranyl acetate. Grids were observed at room temperature using a JEOL JEM 1400 transmission electron microscope equipped with two Gatan CCD cameras: 4k by 4k CCD camera and 35 mm port CCD camera for real time screening. The instrument is operated at an acceleration voltage of 120 kV with a magnification range of 50× – 2,000,000×. Clearly visible SpeG particles were observed and selected by hand from images. The EMAN2 program was used to process and classify SpeG particles on images [46]. Chimera software was used to visualize projections created from a 25 Å resolution-limited model obtained from the SpeG dodecameric structure in open and closed states [47].
Supplementary Material
Highlights.
Spermidine N-acetyltransferase (SpeG) in ligand-free form has multiple oligomeric forms in solution.
SpeG structure revealed a unique open dodecameric state.
Polyamine alters a shift from the asymmetric open state to the closed symmetric dodecameric state.
Acknowledgments
We would like to thank Corey M. Janczak and the Keck Biophysics Facility at Northwestern University (Evanston, IL) for assistance with SEC-MALS data collection. We would like to thank Chi-Hao Luan and the HTAL at Northwestern University (Evanston, IL) for assistance with FTS assay. The negative-stain EM analysis was performed at CryoEM Facility at Northwestern University (Evanston, IL). The EM research was supported in part by the Searle Leadership Fund for the Life Sciences at Northwestern University, established by the Searle Funds at The Chicago Community Trust. The SAXS and X-ray data collection for crystal structures was done at the DND-CAT and LS-CAT, SBC-CAT beamlines, respectively, at the Advanced Photon Source Science User Facility operated for the U.S. Department of Energy (DOE), supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. This project has been funded with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contracts No. HHSN272200700058C and HHSN272201200026C (WFA), and the National Science Foundation grant MCB 1024945 (MAB).
Abbreviations
- GNAT
Gcn5-related acetyltransferase superfamily
- AcCoA
acetyl coenzyme A
- CoA
coenzyme A
- MW
molecular weight
- SEC-MALS
size exclusion chromatography/multi-angle light scattering
- FTS
fluorescence thermal shift
- SAXS
small angle X-ray scattering
- EM
electron microscopy
- PDB
protein data bank
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fukuchi J, Kashiwagi K, Takio K, Igarashi K. Properties and structure of spermidine acetyltransferase in Escherichia coli. J. Biol. Chem. 1994;269(36):22581–22585. [PubMed] [Google Scholar]
- 2.Fukuchi J, Kashiwagi K, Yamagishi M, Ishihama A, Igarashi K. Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-dificient mutant of Escherichia coli. J. Biol. Chem. 1995;270(32):18831–18835. doi: 10.1074/jbc.270.32.18831. [DOI] [PubMed] [Google Scholar]
- 3.Matsui I, Pegg AE. Induction of spermidine N1-acetyltransferase in rat kidney by treatment with folic acid. FEBS Lett. 1982;139(2):205–208. doi: 10.1016/0014-5793(82)80852-1. [DOI] [PubMed] [Google Scholar]
- 4.Limsuwun K, Jones PG. Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J. Bacteriol. 2000;182(19):5373–5380. doi: 10.1128/jb.182.19.5373-5380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carper SW, Willis DG, Manning KA, Gerner EW. Spermidine acetylation in response to a variety of stresses in Escherichia coli. J. Biol. Chem. 1991;266(19):12439–12441. [PubMed] [Google Scholar]
- 6.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vetting MW, de Carvalho LPS, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Filippova EV, Kuhn ML, Osipiuk J, Kiryukhina O, Joachimiak A, Ballicora MA, Anderson WF. A novel polyamine allosteric site of SpeG from Vibrio cholerae is revealed by its dodecameric structure. J. Mol. Biol. 2015;427(6):1316–1334. doi: 10.1016/j.jmb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn ML, Majorek KA, Minor W, Anderson WF. Broad-substrate screen as a tool to identify substrates for bacterial Gcn5-related N acetyltransferases with unknown substrate specificity. Protein Sci. 2013;22(2):222–230. doi: 10.1002/pro.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 11.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystal. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krissinel E. Crystal contacts as nature's docking solutions. J. Comput. Chem. 2010;31(1):133–143. doi: 10.1002/jcc.21303. [DOI] [PubMed] [Google Scholar]
- 13.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 14.Liu H, Hexemer A, Zwart PH. The Small Angle Scattering ToolBox (SASTBX): an open-source software for biomolecular small-angle scattering. J. Appl. Cryst. 2012;45:587–593. [Google Scholar]
- 15.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 18.Boyer PD. Molecular motors: What makes ATP synthase spin? Nature. 1999;402:247–249. doi: 10.1038/46193. [DOI] [PubMed] [Google Scholar]
- 19.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the Bacteriophage Phi 29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swapna LS, Srikeerthana K, Srinivasan N. Extent of structural asymmetry in homodimeric proteins: Prevalence and Relevance. PLoS ONE. 2012;7(5):e36688. doi: 10.1371/journal.pone.0036688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhabha G, Cheng H-C, Zhang N, Moeller A, Liao M, Speir JA, Cheng Y, Vale RD. Allosteric communication in the dynein motor domain. Cell. 2014;159(4):857–868. doi: 10.1016/j.cell.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delano WL. The Pymol Molecular Graphics System. San Carlos, CA: 2002. [Google Scholar]
- 25.Makowska-Grzyska M, Kim Y, Maltseva N, Li H, Zhou M, Joachimiak G, Babnigg G, Joachimiak A. Protein Production for Structural Genomics Using E. coli Expression. Meth. in Mol. Biol. 2014;1140:89–105. doi: 10.1007/978-1-4939-0354-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shuvalova L. Parallel Protein Purification. Meth. in Mol. Biol. 2014;1140:137–143. doi: 10.1007/978-1-4939-0354-2_10. [DOI] [PubMed] [Google Scholar]
- 27.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution – from diffraction images to an initial model in minutes. Acta Crystal. D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystal. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystal. D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystal. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystal. 1993;26:283–291. [Google Scholar]
- 34.Yang H, Guranovic V, Dutta S, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystal. D. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 35.Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 36.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystal. D. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 37.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. "The Protein Data Bank". Nucleic. Acids. Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filippova EV, Luan C-H, Dunne SF, Kiryukhina O, Minasov G, Shuvalova L, Anderson WF. Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila. J. Struct. Funct. Genomics. 2014;15(1):33–40. doi: 10.1007/s10969-014-9176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weigand S, Stillwell B, Guise WE, Quintana JPG, Keane DT. Flexibility and high throughput: supporting SAXS users at a joint industrial academic beamline. Adv. X-Ray Anal. 2009;52:58–68. [Google Scholar]
- 40.Hammersley AP, Svensson SO, Hanfland M, Fitch AN, Hausermann D. Two-dimensional detector software: From real detector to idealized image or two-theta scan. High Pressure Res. 1996;14(4–6):235–248. [Google Scholar]
- 41.Dreiss CA, Jack KS, Parker AP. On the absolute calibration of bench-top small-angle X-ray scattering instruments: a comparison of different standard methods. J. Appl. Cryst. 2006;39:32–38. [Google Scholar]
- 42.Weigand S, Filippova EV, Kiruykhina O, Anderson WF. Small angle X-ray scattering data and structure factor fitting for the study of the quaternary structure of the spermidine N-acetyltransferase SpeG. Data in Brief. 2015 doi: 10.1016/j.dib.2015.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496(7446):477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 46.Tanga G, Penga L, Baldwinb PR, Manna DS, Jiangc W, Reesa I, Ludtkea SJ. EMAN2: An extensible image processing suite for electron microscopy. J. Struc. Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids. Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids. Res. 2014;42(W1):320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.