Abstract
The Spindle Assembly Checkpoint (SAC) is a unique signaling mechanism that responds to the state of attachment of the kinetochore to spindle microtubules. SAC signaling is activated by unattached kinetochores, and it is silenced after these kinetochores form end-on microtubule attachments. Although the biochemical cascade of SAC signaling is well-understood, how kinetochore-microtubule attachment disrupts it remained unknown. Here we show that, in budding yeast, end-on microtubule attachment to the kinetochore physically separates the Mps1 kinase, which likely binds to the Calponin homology domain of Ndc80, from the kinetochore substrate of Mps1, Spc105 (KNL1 orthologue). This attachment-mediated separation disrupts the phosphorylation of Spc105, and enables SAC silencing. Additionally, the Dam1 complex may act as a barrier that shields Spc105 from Mps1. Together these data suggest that the protein architecture of the kinetochore encodes a mechanical switch. End-on microtubule attachment to the kinetochore turns this switch off to silence the SAC.
Introduction
The attachment of sister kinetochores to microtubules from opposite spindle poles is necessary for accurate chromosome segregation during cell division. Unattached kinetochores activate the cell cycle control known as the Spindle Assembly Checkpoint1, 2 (SAC), which arrests the cell cycle until these kinetochores form stable attachment. The SAC thus ensures accurate segregation of chromosomes into daughter cells.
The kinetochore-based biochemical cascade that generates the SAC signal is well-understood. Within unattached kinetochores, the highly conserved Mps1 kinase phosphorylates kinetochore proteins, and enables the sequential recruitment of SAC proteins3 (Fig. 1a). This cascade ultimately generates the ‘wait-anaphase’ signal and stalls the cell cycle. The formation of end-on kinetochore-microtubule attachment disrupts this cascade, presumably by interfering with one or more of its steps. Early cell biological observations of SAC silencing led to the hypothesis that a mechanical change within the kinetochore induced by end-on microtubule attachment silences the SAC4. Concurrent changes in the state of SAC signaling and the nanoscale separations between various kinetochore proteins support this hypothesis5–7. However, the causative link between specific changes in kinetochore architecture induced by microtubule attachment and the disruption of specific steps in SAC signaling is missing. This is mainly because the kinetochore is a highly complex machine containing multiple copies of more than 60 different proteins8. A change in the structure, conformation, and/or architecture of any of these proteins induced by microtubule attachment can affect SAC signaling. Consequently, the molecular basis for the mechanosensitivity of SAC signaling is unknown.
Figure 1. Cell cycle effects of anchoring Mps1 to the kinetochore using rapamycin-induced dimerization.
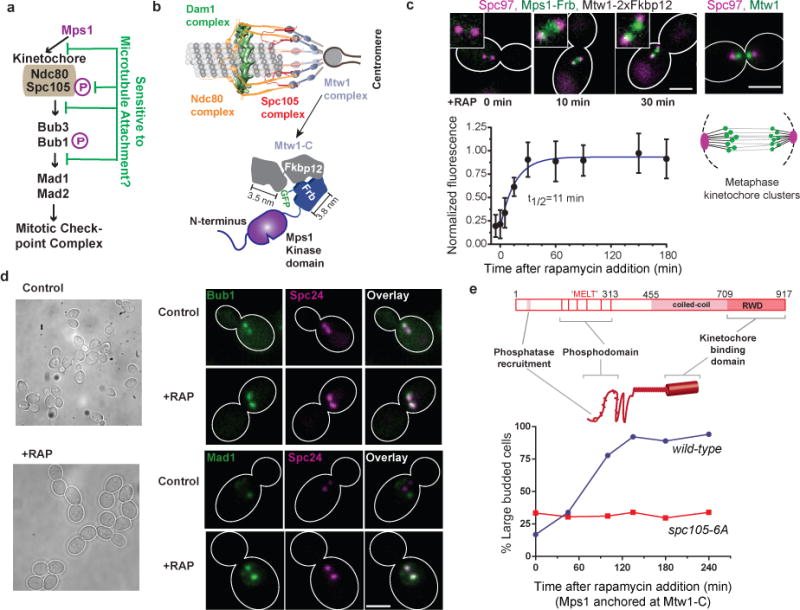
(a) The steps in the kinetochore-based signaling cascade of the SAC (magenta Ps indicate Mps1-mediated phosphorylation) that may be disrupted by microtubule attachment.
(b) Top: Protein architecture of the metaphase kinetochore-microtubule attachment23. Bottom: Schematic of the rapamycin-induced dimerization technique used to anchor Mps1 to the carboxyl terminus of Mtw1 (Mtw1-C).
(c) Top: Micrographs show the anchoring of Mps1-Frb-GFP at Mtw1-C (time after rapamycin addition indicated; scale bar ~ 3 μm). The stereotypical distribution of kinetochores in metaphase visualized with Mtw1-GFP, spindle poles visualized using Spc97-mCherry is shown in the right. Cartoon underneath depicts the metaphase spindle morphology. Bottom: kinetics of rapamycin-induced anchoring of Mps1-Frb-GFP to Mtw1-C. Error bars represent mean ± s.d. of n = 10, 11, 8, 13, 14, 18, 24, 16 and 11 kinetochore clusters analyzed from −5 to 108 min.
(d) Left: Representative transmitted-light images before and 1 hour after the addition of rapamycin to anchor Mps1 at Mtw1-C. Right: Localization of Bub1-GFP and Mad1-GFP, and kinetochores (visualized by Spc24-mCherry) in untreated cells (control) and in cells that have Mps1 anchored at Mtw1-C (+RAP). Scale bar ~ 3 μm.
(e) Top: Domain organization of Spc105. The end-to-end length of the unstructured domain of Spc105 (amino acids 1–455) is predicted to be 11.7 ± 5 nm (mean ± s.d. using the worm-like chain model60). The maximum length of it α-helical region (a. a. 455–709) is 38 nm (3.6 amino acids per turn/0.54 nm pitch). The predicted kinetochore-binding domain (RWD*) is ~ 6 nm long61. Other than these estimated dimensions, the structure and organization of Spc105 is unknown. Therefore, the depiction is not drawn to scale. The six Mps1 phosphorylation sites (consensus sequence ‘MELT’) are depicted as bars. Bottom: Cell cycle progression of asynchronous cells with the indicated genotypes observed upon anchoring Mps1 at Mtw1-C. Accumulation of large budded cells indicates mitotic arrest. Plotted points represent the average values calculated from 2 independent experiments. More than 50 cells were scored for each time point. The source data are shown in Supplementary Table 3.
Here we investigate how the architecture of the kinetochore-microtubule attachment in the budding yeast Saccharomyces cerevisiae disrupts SAC signaling. We find that the phosphorylation of Spc105 by Mps1 is both necessary and sufficient to initiate the SAC cascade. End-on kinetochore-microtubule attachment restricts Mps1 kinase activity to the outer kinetochore and maintains the phosphodomain of Spc105 in the inner kinetochore to disrupt this crucial first step in the SAC cascade to silence the SAC.
Results
Mps1, when artificially localized to the kinetochore, phosphorylates Spc105 and activates the SAC
Microtubule attachment to the kinetochore may silence the SAC by promoting the dissociation of SAC proteins from the kinetochore (Fig. 1a). If this is true, then persistent localization of key SAC proteins at the kinetochore should constitutively activate the SAC. To test this hypothesis, we used rapamycin-induced dimerization of 2xFkbp12 and Frb to artificially localize or ‘anchor’ key phosphoregulators and SAC proteins9: Mps1, Ipl1 (Aurora B), Glc7 (PP1), or Mad1 within the kinetochore (Fig. 1b). In the absence of rapamycin, each Frb-tagged protein retained its normal cellular distribution. Addition of rapamycin to the culture media rapidly anchored it to the kinetochore subunit tagged with 2xFkbp12 (Fig. 1c, right and Supplementary Fig. 1a).
Mps1 anchored at Mtw1-C in this manner led to the accumulation of large-budded cells that were arrested in metaphase (Fig. 1d–e). The kinetochores in these cells recruited both Bub1 and Mad1, indicating that the arrest was mediated by the SAC (Fig. 1d). These observations are consistent with previous reports that Mps1 fused to kinetochore proteins activates the SAC10, 11. Other SAC proteins tested: Ipl1, Mad1, and Glc7, did not delay the cell cycle when anchored to Mtw1-C (Supplementary Fig. 1b). The phosphorylation of the kinetochore protein Spc105 at one or more of its conserved ‘MELT’ motifs was necessary for the anchored Mps1 to activate the SAC12 (Fig. 1e). Importantly, these effects did not require the kinase activity of Ipl1, suggesting that the anchored Mps1 did not activate the SAC indirectly by disrupting either microtubule attachment or force generation13 (Supplementary Fig. 1c–d). This observation is consistent with data from other organisms and with the dispensability of Ipl1 for SAC signaling in budding yeast14, 15. Thus, anchoring Mps1 to the kinetochore is sufficient for constitutive SAC signaling.
SAC proteins that act downstream from Mps1 can function within attached kinetochores
The above experiments were performed in asynchronous yeast cultures. Consequently, we could not ascertain whether the anchored Mps1 activated the SAC mostly in prometaphase, before all kinetochores attach to microtubules, or if Mps1 can re-activate the SAC when anchored within stably attached kinetochores. To test this, we repressed CDC20, the gene that encodes the activating subunit of the Anaphase Promoting Complex (APC), to prevent yeast cells from entering anaphase even after all the kinetochores were attached and the SAC was satisfied16. We anchored Mps1 at Mtw1-C in such cells, released them from the arrest by inducing CDC20 expression, and then monitored cell cycle progression (Fig. 2a). We found that cells that had Mps1 anchored at Mtw1-C underwent a persistent cell-cycle arrest, whereas control cells completed anaphase within 20 minutes (Fig. 2a). Thus, Mps1 can re-activate the SAC, when it is anchored to kinetochores with stable microtubule attachments.
Figure 2. Testing the sensitivity of SAC signaling steps to the attachment status of the kinetochore.
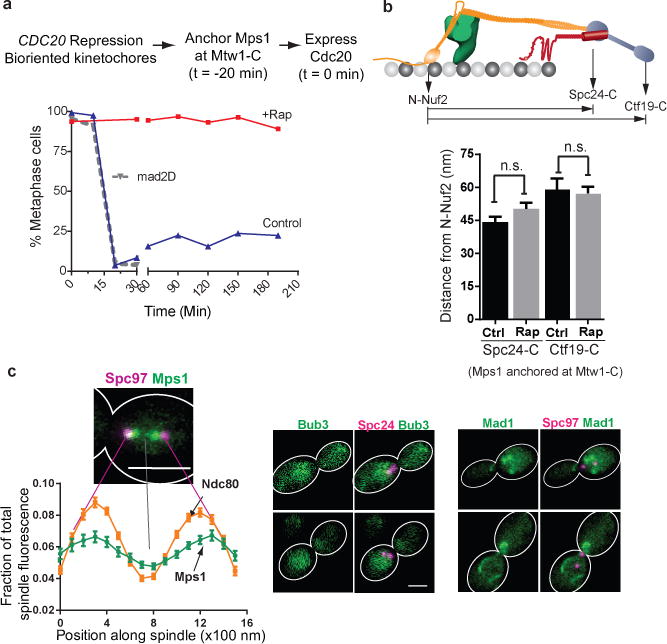
(a) Cell cycle progression of three different strains following release from metaphase arrest (methodology indicated at the top, see Methods for details). Solid lines indicate cell cycle progress of a strain expressing Mtw1-2xFkbp12 and Mps1-Frb released into media with (red) or without (blue) rapamycin. The dotted gray line indicates cell cycle progression of a mad2∆ strain similarly released from metaphase arrest. Plotted points represent the average values calculated from 2 independent experiments. The source data are shown in Supplementary Table 3.
(b) Separation between the centroids of fluorescently labeled kinetochore proteins along the spindle axis obtained by high-resolution colocalization in unperturbed metaphase cells (ctrl.) and rapamycin treated cells (rap. – rapamycin added to anchor Mps1 at Mtw1-C; mean ± s.e.m.; n = 61, 49, 19, 42 cells were analyzed (from left to right). Data were pooled from 2 independent experiments. n. s. – not significant, p-value > 0.05 using Mann-Whitney test).
(c) Left: Fractional intensity distributions of Mps1-Frb-GFP (that autonomously localizes along the spindle in the absence of rapamycin) and Ndc80-GFP along the spindle in cells arrested in metaphase using CDC20 repression (spindle pole bodies visualized using Spc97-mCherry). Error bars represent s.e.m. from n = 38 and 57 cells for Mps1 and Ndc80, respectively. The experiment was repeated twice and graph presents mean data pooled from 2 independent experiments. Right: Bub3 and Mad1 do not localize to kinetochores under the same conditions. Mad1 puncta correspond to its known localization to the nuclear envelope62. Scale bar ~ 3 μm.
These results also show that SAC proteins downstream from Mps1 can bind to and function from attached kinetochores. It is possible that the anchored Mps1 facilitates SAC protein binding by changing the overall organization of the kinetochore. However, we did not detect significant changes when we compared the nanoscale separation between key kinetochore domains in metaphase and rapamycin-treated cells using high-resolution colocalization (Fig. 2b). Even if architectural changes that facilitate SAC protein binding do occur, they can do so when the kinetochore is attached. Based on these data, we concluded that microtubule attachment to the kinetochore must hamper either Mps1 localization to the kinetochore or its kinase activity in order to silence the SAC.
Endogenous Mps1 binds to attached kinetochores
Microtubule attachment may inhibit Mps1 function by simply promoting its dissociation from the kinetochore10. Indeed, Mps1 gradually disappears from the kinetochore clusters as yeast cells progress from prometaphase to metaphase (Supplementary Fig. 1e). However, Mps1 is targeted for degradation by the APC17. This process may contribute to the disappearance of Mps1 either directly or indirectly. Consistent with this hypothesis, when we inactivated the APC using CDC20 repression, Mps1-Frb-GFP autonomously localized to attached kinetochores (Fig. 2c, left). Importantly, this autonomously-localized Mps1 did not activate the SAC, because both Bub3 and Mad1 were absent from the kinetochores (Fig. 2c, right). Furthermore, these cells entered anaphase without any detectable delay upon release from the metaphase block (Fig. 2a, dotted gray line). Finally, Mps1 was present at the kinetochore even as these cells entered anaphase (Supplementary Fig. 1f). Thus, the removal of Mps1 from the kinetochore is not necessary for either SAC silencing or anaphase onset.
It is notable that the Mps1 molecules that autonomously localize to attached kinetochores do not activate the SAC, but a similar number of Mps1 molecules anchored at Mtw1-C activate it constitutively (Supplementary Fig. 1e). The inability of the autonomously localized Mps1 to activate the SAC could be due to: (a) its inability to reach and phosphorylate Spc105 from its endogenous binding position in the kinetochore, (b) the inhibition of the kinetochore-bound Mps1 kinase, or (c) the up-regulation of Glc7 phosphatase activity in attached kinetochores18, 19. Up-regulation of Glc7 activity for SAC silencing is unlikely to be the main mechanism, because Glc7 is not necessary for anaphase onset19. Therefore, we investigated how microtubule attachment affects Mps1 kinase activity within the kinetochore.
The ability of Mps1 to activate the SAC depends on its position within the kinetochore
We first tested whether the binding position of Mps1 within the kinetochore can affect its ability to phosphorylate Spc105 and initiate SAC signaling. In metaphase, the budding yeast kinetochore spans ~ 80 nm, from the N-terminus of Ndc80 to the centromeric nucleosome20. It contains ~ 8 copies of Ndc80 complex and Spc105 molecules distributed with an average inter-molecular spacing of ~ 8 nm around the microtubule circumference21, 22, and with little inter-molecular staggering along the length of the microtubule23 (Fig. 1b). This architecture suggests that the proximity of Mps1 to Spc105 along the length of the kinetochore can affect its ability to phosphorylate Spc105.
Rapamycin-induced dimerization must stably anchor and confine Mps1 at specific kinetochore positions in order to reveal its position-specific activity. We determined this to be the case using three measurements (Fig. 3a–c). First, we found that the anchoring was stable, as indicated by negligible turn-over of Mps1-Frb-GFP anchored at Ndc80-C (Fig. 3a). Although this high stability is ideal for studying position-specific activity, it is likely to be non-physiological24. Second, Förster Resonance Energy Transfer (FRET) measurements suggested that the anchored protein is likely confined within a 10 nm region around the anchoring point (Fig. 3b). Finally, the total number of molecules anchored within the kinetochore was determined by the abundance of the anchored protein25 and also its kinetochore anchor21 (Fig. 3c). For low abundance proteins such as Mps1 and Ipl1, the entire nuclear pool was anchored at the selected kinetochore position.
Figure 3. The ability of Mps1 to activate the SAC depends on its position in the kinetochore.
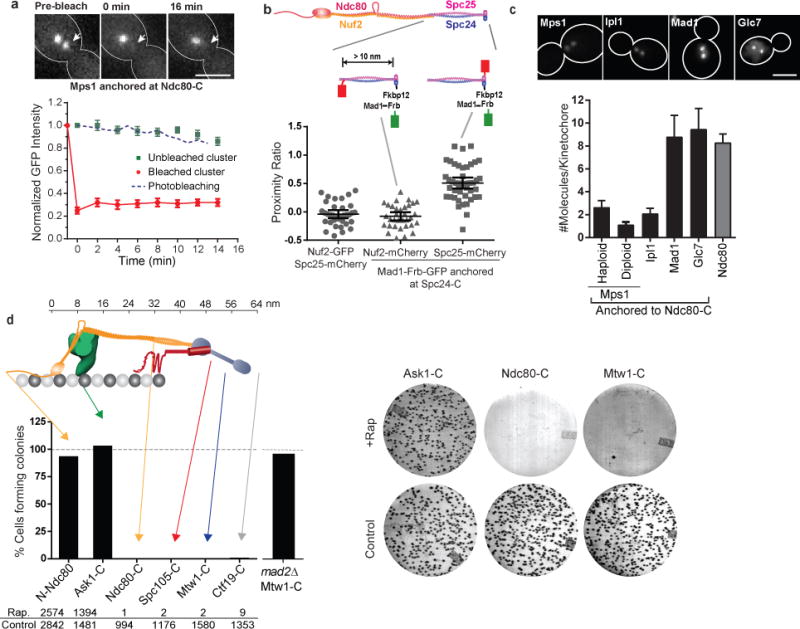
(a) Fluorescence recovery after photobleaching of Mps1-frb-GFP anchored at Ndc80-C (red circles), and loss of anchored protein from the unbleached cluster (green squares). Blue dashed line displays the expected rate of photobleaching as a result of imaging determined in cells expressing Ndc80-GFP (mean ± s.e.m. from n = 8 and 11 clusters for bleached and unbleached clusters, respectively; data pooled from 2 independent experiments). Scale bar ~ 3 μm.
(b) Top: Structure of Ndc80 complex and the positions of fluorescent tags used for FRET. Scatter plot: Proximity ratio, which is directly proportional to the FRET efficiency35, for FRET between Spc25-mCherry or Nuf2-mCherry and Mad1-Frb-GFP anchored to Spc24-C (mean ± 95% confidence interval from n = 35, 33 and 44 kinetochore clusters analyzed, from left to right. The experiment was repeated twice and graph presents mean data pooled from 2 independent experiments). Proximity ratio is defined as the acceptor fluorescence resulting from FRET normalized by the sum of direct excitation of mCherry on exciting GFP and GFP emission bleed-through into the mCherry imaging channel35. FRET between the anchored donor, Mad1-Frb-GFP, and the acceptor, Spc25-mCherry, was readily detected, but it was absent when the mCherry was fused to Nuf2-C. Spc25-C is < 3 nm away46 from Spc24-C, where the donor is anchored, whereas Nuf2-C is > 10 nm away63. We used Mad1, rather than Mps1, in this experiment to ensure that the number of donors is equal to the number of acceptor molecules (either Spc25-mCherry or Nuf2-mCherry, see (c) below) for accurate FRET quantification35.
(c) Number of protein molecules anchored at Ndc80-C, measured 30 min after rapamycin addition (mean ± s.d. from n = 25, 33, 29, 20, 41, and 30 kinetochore clusters from left to right. The experiment was performed once). Scale bar ~ 3 μm.
(d) Top: The organization of yeast kinetochore proteins along the microtubule axis20, 23. The N-terminal half of Spc105 is not drawn to scale. Bottom: Bar graph shows the number of colonies formed on rapamycin-containing plates relative to control plates. The experiment was repeated at least twice and the cumulative number of colonies scored is displayed below the graph. Right: Representative photographs of plates for three strains.
We constitutively anchored Mps1 at six distinct positions selected to sample the 80 nm length of the kinetochore-microtubule attachment (Fig. 3d, top). To assess the effects of anchoring Mps1 on the cell cycle, we plated an equal number of cells on control plates and on plates containing rapamycin, and compared the number of colonies formed in each case (Fig. 3d, right). We performed these experiments in heterozygous diploids with a wild type copy of Mps1, because Mps1 activity is also essential for other cellular functions26. Even though the wild-type, diffusible Mps1 provides these essential functions, it is not required for SAC activation (Supplementary Fig. 2a–b). Furthermore, haploids expressing only Mps1-Frb displayed identical SAC activation phenotypes (see below and Supplementary Fig. 2c).
When Mps1 was constitutively anchored at four different locations within the inner kinetochore, ranging from Ndc80-C to Ctf19-C, it completely inhibited colony growth (Fig. 3d). MAD2 deletion restored colony growth, indicating that the lack of growth was due to constitutive SAC activation (Supplementary Fig. 3a and Fig. 3d). Interestingly, Mps1 anchored at two positions located in the outer kinetochore, N-Ndc80 and Ask1-C (a Dam1 complex subunit), had no effect on colony growth (Fig. 3d). Although the number of Mps1 molecules anchored in the inner kinetochore positions was 30–50% higher than the number of Mps1 molecules anchored in the outer kinetochore, these differences did not strictly correlate with SAC activation phenotypes (Supplementary Fig. 3b). Reducing the length of the linker between Mps1 and Frb-GFP did not affect the observed phenotypes (Supplementary Fig. 3c). Finally, the observed effects were specific to Mps1: constitutive anchoring of Ipl1 or Mad1 at the same positions did not result in the same phenotypes (Supplementary Fig. 3d–g).
These data show that the position of Mps1 within the kinetochore can affect its ability activate the SAC. Since Mps1 must phosphorylate Spc105 to activate the SAC, the observed phenotypes likely reflect whether or not the anchored Mps1 can access the phosphodomain of Spc105. It is notable that Mps1 activates the SAC from different locations over a 30 nm span20 (the metaphase separation between Ndc80-C and Ctf19-C), even though its kinase activity is spatially confined to individual locations. To encounter the confined kinase activity over this wide span, the long and unstructured phosphodomain of Spc105 likely assumes variable conformations.
Mps1 anchored in the outer kinetochore does not activate the SAC
To confirm that the inability of Mps1 to activate the SAC from the outer kinetochore is only because it cannot phosphorylate Spc105, we characterized the effects of anchoring Mps1 to the C-termini of seven other subunits of the heterodecameric Dam1 complex27 (Fig. 4a). Similar to Ask1, Mps1 anchored to three other Dam1 subunits did not affect the colony growth (Fig. 4b and Supplementary Fig. 4a). Surprisingly, Mps1 anchored to four other subunits delayed colony formation, but did not appear to affect the number of colonies formed (Fig. 4b, Supplementary Fig. 4a). Slow colony growth was likely due to a transient SAC-mediated delay in the cell cycle (Fig. 4c, also Supplementary Fig. 2c). As before, reduced length of the flexible linker fusing the Mps1 kinase domain to Frb did not affect the observed cell cycle delay (Supplementary Fig. 4b).
Figure 4. The Dam1 complex defines a boundary for SAC signaling by anchored Mps1.
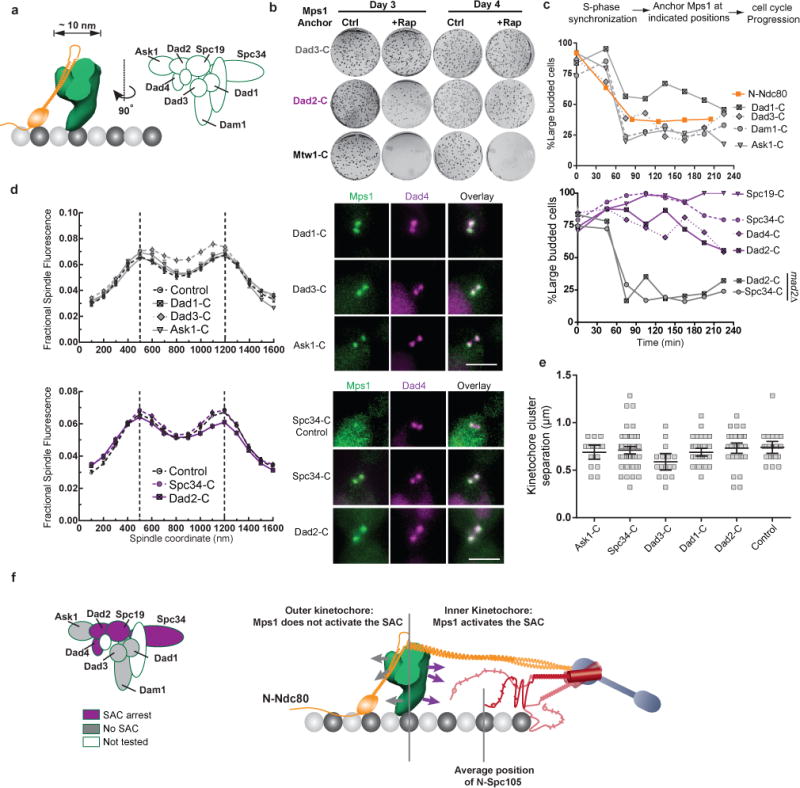
(a) Cartoon: Position of the Dam1 complex relative to Ndc80 complex23 and subunit organization within the Dam1 complex27. EMD1372 was used to infer the dimensions of the Dam1 complex.
(b) Colony growth (also see Supplementary Fig. 4a) on control (ctrl.) and rapamycin (+Rap) plates. The number of days after plating is indicated at the top; the anchoring subunit is indicated on the left.
(c) Cell cycle progression when Mps1 is anchored to a Dam1 subunit (indicated on the left) in cells released from an experimentally imposed S-phase arrest. This strategy was used to ensure that the kinetochores formed end-on attachments and loaded Dam1 complex before Mps1 was anchored 42. Plotted points represent the average values calculated from 2 independent experiments. The source data are shown in Supplementary Table 3.
(d) Normalized distribution of Dad4-mcherry on the spindle when Mps1 was anchored to the indicated positions for 1 hour (mean ± s.e.m. n = 43, 34, 28, 36, 73 and 38 cells for Dad1, Dad3, Ask1, Ctrl, Spc34 and Dad2, respectively). Control data is from untreated metaphase cells. Micrographs on the right display the localization of Dad4-mCherry relative to that of Mps1-frb-gfp anchored to the indicated subunits (scale bar ~ 3 μm).
(e) The separation between kinetochore clusters in the cells in (d), measured as the separation between maximum intensity pixels in the two Dad4-mCherry puncta in each cell; mean ± 95% confidence interval, n = 43, 16, 16, 25, 72 and 38 cells for Dad1, Dad3, Ask1, Ctrl, Spc34 and Dad2, respectively. Although there is a small decrease in spindle length when Mps1 is anchored at Dad3-C, cell cycle progression is unaffected as seen in (c).
(f) Left: Classification of Dam1 complex subunits inferred from the Mps1 anchoring experiments. Right: Activity map of the anchored Mps1 along the length of the kinetochore-microtubule attachment. Arrows from the Dam1 complex depict the proposed orientation of the C-termini of subunits used as anchors. Possible variations in the conformation of the unstructured phosphodomain of Spc105 are also depicted.
We also tested whether the anchored Mps1 in these experiments perturbed Dam1 complex localization and function, because Dam1 subunits are known Mps1 substrates27, 28. We quantified the distribution of Dad4 over the mitotic spindle after anchoring Mps1 to other Dam1 subunits (Fig. 4d). Dad4-mCherry colocalized with the anchored Mps1-Frb-GFP in every case, and its distribution was indistinguishable from Dad4 distribution in untreated cells. Thus, the association of the Dam1 complex with the kinetochore remained unaffected. The separation between kinetochore clusters in rapamycin-treated cells was also indistinguishable from the corresponding length in untreated cells (Fig. 4e). This indicates that force generation at the kinetochore, a process in which the Dam1 complex is the dominant contributor, was not affected29. Thus, the anchored Mps1 does not perturb Dam1 complex function, and the observed phenotypes reflect whether or not the anchored Mps1 can phosphorylate Spc105.
The strikingly different phenotypes induced by Mps1 anchored to Dam1 subunits are surprising. This is because dimensions of the Dam1 complex27 and its narrow distribution along the length of the kinetochore-microtubule attachment23 suggest that all of the anchoring points are confined within a ~10 nm wide zone. Although the structure of the Dam1 complex is unknown, it is conceivable that the C-termini of Dam1 subunits face towards or away from the centromere (Fig. 4f, arrows). This orientation may in turn constrain the orientation of the anchored Mps1, and determine whether or not it can phosphorylate Spc105 to activate the SAC.
Phosphorylation of Spc105 by Mps1 is sufficient to initiate SAC signaling
Our data show that the physical proximity between the Mps1 kinase and the phosphodomain of Spc105 can control the state of the SAC. Therefore, we tested whether a forced interaction between the two outside the kinetochore is sufficient to activate the SAC. We engineered a minimal, anchorable phosphodomain comprising residues 120–329 of Spc105 (referred to as Spc105120:329, Fig. 5a). It contains all 6 MELT motifs, but no known kinetochore-binding activity. When we anchored Spc105120:329 to Mps1-Fkbp12 in asynchronously dividing cells, the cells arrested in metaphase (Fig. 5b). Spc105120:329 also localized to kinetochore clusters under these conditions and recruited Mad1 (Fig. 5c, Supplementary Fig. 5a). The kinetochore-localization of Mad1 and Spc105120:329, when the latter anchored to Mps1, is likely mediated by Mps1 binding to the kinetochores. MAD2 deletion abolished the cell cycle arrest indicating that the arrest resulted from SAC activation (Fig. 5b, dashed line). When Spc105120:329:6A, the non-phosphorylatable version of Spc105120:329, was anchored to Mps1, it did not activate the SAC (Fig. 5b–c). Thus, the phosphorylation of MELT motifs in Spc105120:329 by Mps1 is necessary for the observed cell cycle arrest.
Figure 5. The phosphorylation of the Spc105 phosphodomain by Mps1 is sufficient to activate the SAC.
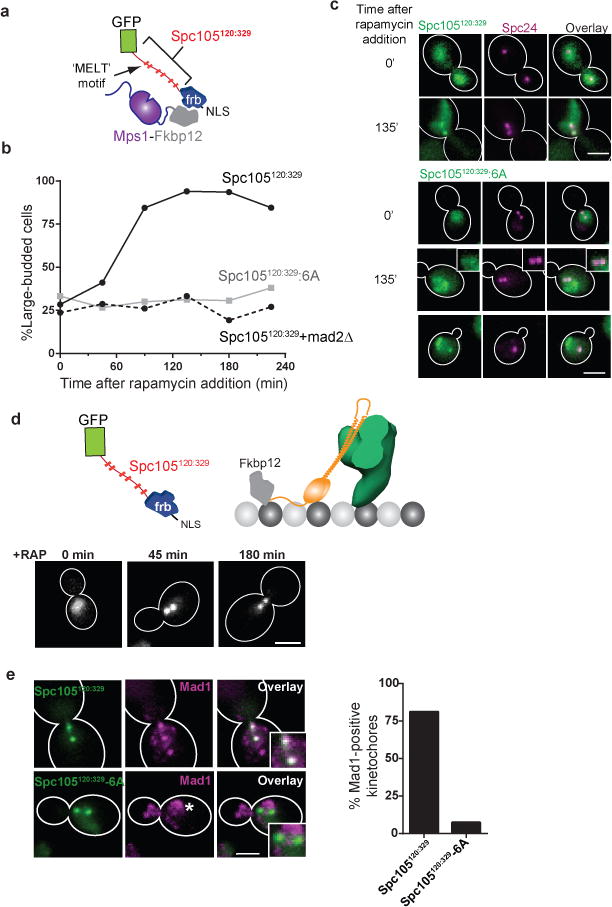
(a) Schematic of Spc105120:329: the minimal Spc105 phosphodomain. NLS – Nuclear Localization Signal used to send Spc105120:329 to the nucleus.
(b) Cell cycle kinetics following rapamycin addition to anchor the phosphorylatable (solid black line) or non-phosphorylatable Spc105120:329 (solid gray line) to Mps1-C. Dashed black line shows the cell cycle progression of the mad2Δ strain after anchoring Spc105120:329 to Mps1. Plotted points represent the average values calculated from 2 independent experiments. More than 50 cells scored for each time point in each trial. The source data are shown in Supplementary Table 3.
(c) Localization of Spc105120:329 or Spc105120:329:6A when anchored to Mps1. Scale bar ~ 3 μm.
(d) Strategy to anchor Spc105120:329 at N-Ndc80, and the localization of Spc105120:329 at indicated times after rapamycin addition. Scale bar ~ 3 μm.
(e) Recruitment of Mad1 to the kinetochore clusters when Spc105120:329 (top) or Spc105120:329:6A (bottom) is anchored at N-Ndc80. Bars represent mean data pooled from 2 independent experiments. At least 45 cells were analyzed for each sample in each trial. The source data are shown in Supplementary Table 3. Asterisk – known Mad1 localization at the nuclear envelope. Scale bar ~ 3 μm.
To test whether kinetochores contributed to the SAC signaling in the above experiment, we used cells carrying ndc10-1, a temperature sensitive allele of gene encoding the centromeric protein Ndc10 (ref. 30). At the restrictive temperature, these cells cannot assemble functional kinetochores, and are thus unable to activate the SAC. However, when Spc105120:329 was anchored to Mps1 at the restrictive temperature, ndc10-1 cells experienced a cell cycle delay similar to the delay seen in NDC10 cells under the same conditions (Supplementary Fig. 5b). Thus, the SAC signaling induced by the forced interaction between Spc105120:329 and Mps1 does not require functional kinetochores31. Together with our earlier results, these data demonstrate that the interaction between Mps1 and the phosphodomain of Spc105 is both necessary and sufficient to activate the SAC. The kinetochore may primarily serve as the scaffold that makes this interaction sensitive to microtubule attachment.
Spc105120:329 activates the SAC when anchored in the outer kinetochore, but not the inner kinetochore
Our data reveal a potential organization of Mps1 and Spc105 relative to one another that can make their interaction sensitive to the attachment state of the kinetochore. When Mps1 is anchored in the inner kinetochore, proximal to the phosphodomain of Spc105, it activates the SAC constitutively even from attached kinetochores. In contrast, if it is anchored in the outer kinetochore, distal from the phosphodomain of Spc105, it activates the SAC conditionally, only from unattached kinetochores (Supplementary Fig. 6). Therefore, to implement attachment-sensitive SAC signaling, endogenous Mps1 should bind to a site within the outer kinetochore. Consistent with this expectation, Mps1 physically interacts with the CH-domain of Ndc80, which is located in the outer kinetochore32, 33.
To test whether endogenous Mps1 binds within the outer kinetochore, we anchored Spc105120:329 at N-Ndc80, proximal to the CH-domain (Fig. 5d, top). In metaphase cells, the anchored Spc105120:329 displayed the stereotypical, metaphase kinetochore distribution: two distinct puncta separated by < 1 μm. It also recruited Mad1, and the cells remained arrested for a prolonged period (Fig. 5d–e). The cell cycle arrest was absent when Spc105120:329:6A was anchored to N-Ndc80, revealing that the phosphorylation of the MELT motifs in Spc105120:329 by kinetochore-localized Mps1 is required for SAC activation. These results demonstrate that catalytically active Mps1 binds to the outer kinetochore even after stable microtubule attachments form.
We next probed the entire kinetochore for additional Mps1 binding sites (Fig. 6a). When we anchored Spc105120:329 to Dam1 subunits expected to face towards the outer kinetochore (Ask1-C, Dam1-C, or Dad1-C, see Fig. 4f), the kinetochores recruited Mad1, and the cells arrested in mitosis (Fig. 6b top and Fig. 6c). Strikingly, Spc105120:329 was anchored to positions in the inner kinetochore, including the Dam1 subunit termini predicted to face towards the centromere (Dad4-C, Spc34-C, and Spc19-C), it had no effect on the cell cycle (Fig. 6b bottom and Fig. 6c). As expected, Spc105120:329:6A did not affect the cell cycle when anchored at any of the positions (dashed lines in Fig. 6b). These results demonstrate that catalytically active Mps1 is absent from the inner kinetochore.
Figure 6. Spc105120:329 activates the SAC only when it is anchored in the outer kinetochore.
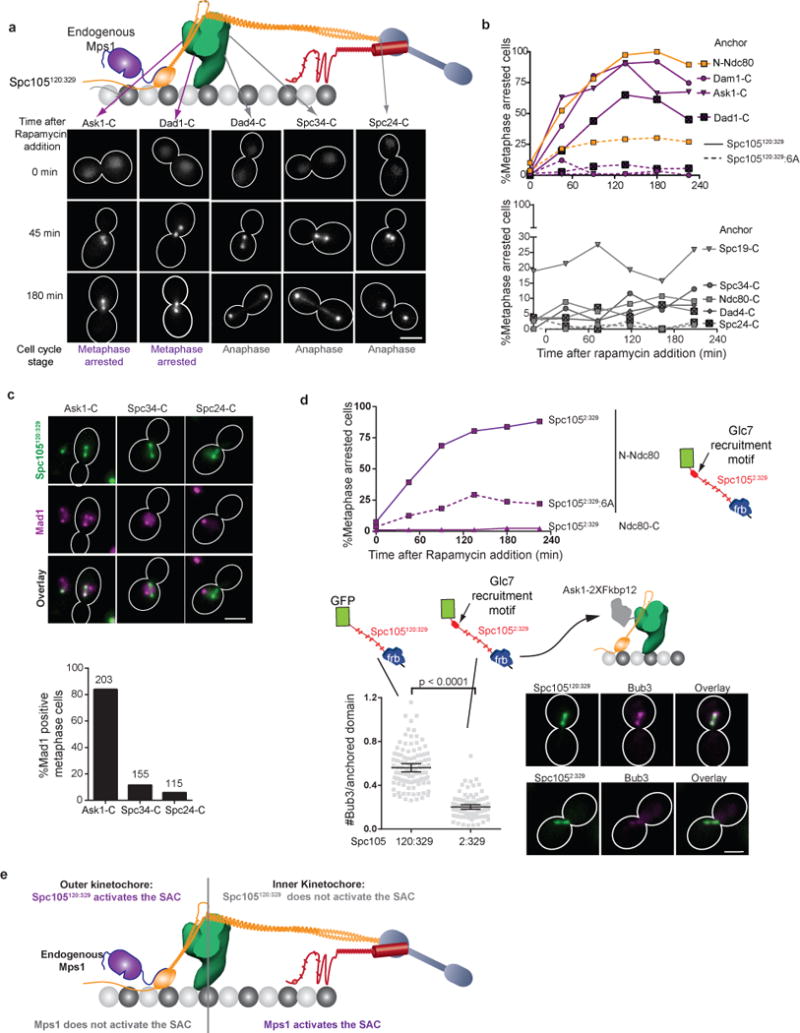
(a) Representative micrographs of asynchronously dividing cells showing the localization of Spc105120:329 and cell-cycle progression as a function of the anchoring position (indicated at the top; scale bar ~ 3 μm). Large-budded cells with < 2 μm separation between kinetochore clusters were characterized as metaphase-arrested cells.
(b) Accumulation of metaphase-arrested cells after rapamycin addition, when either Spc105120:329 (solid lines) or its non-phosphorylatable version, Spc105120:329:6A (dashed lines) was anchored at the indicated positions. The experiment was performed once, and more than 70 cells were scored for each time point (source data are shown in Supplementary Table 3).
(c) Mad1-mCherry localization after anchoring Spc105120:329 at indicated positions for 1 hour (scale bar ~ 3 μm). The bar graph shows the fraction of metaphase cells that recruit Mad1 to the kinetochores in each case. Bars present average values from 2 independent experiments. Total number of cells analyzed in each case is indicated on top of the bars.
(d) Top: Cell cycle progression as in Fig. 6a when a modified version of Spc105 phosphodomain that includes the Glc7 recruitment motif (Spc1052:329, solid lines) or its non-phosphorylatable version (Spc1052:329:6A, dashed line) was anchored at the indicated kinetochore positions. The experiment was performed once. More than 50 cells were scored for each time point and the source data are shown in Supplementary Table 3. Bottom: Micrographs (scale bar ~ 3 μm) and quantification of kinetochore-localized Bub3-mCherry 45 minutes after either Spc105120:329 or Spc1052:329 was anchored at Ask1-C in cells arrested in metaphase using CDC20 repression (mean ± 95% confidence interval from n = 102 and 100 kinetochore clusters analyzed for Spc105120:329 and Spc1052:329 anchoring, respectively). p-values computed using Mann-Whitney test.
(e) Map of the SAC activity of the anchored Spc105120:329.
The N-terminus of Spc105 localizes to the inner kinetochore and contains a Glc7 binding motif18, which is not present in Spc105120:329. Therefore, the lack of Glc7 activity in the outer kinetochore, rather than localized Mps1 activity, could also produce the observed SAC activation phenotypes. To test if this is the case, we constructed a phosphodomain that contains the Glc7 binding motif (Spc1052:329, Fig. 6d). Spc1052:329 anchored at N-Ndc80 or at Ndc80-C produced the same phenotypes as Spc105120:329 (Fig. 6d, top). We quantified Bub3-mCherry at the kinetochore, which specifically binds phosphorylated MELT motifs34, after anchoring either Spc1052:329 or Spc1052:329 to Ask1-C (Fig. 6d). Spc1052:329 recruited significantly less Bub3 confirming that it recruits Glc7 activity (Fig. 6d, bottom).
These data build an activity map for Spc105120:329 and demonstrate that catalytically active Mps1 kinase binds exclusively in the outer kinetochore even after the kinetochore establishes stable microtubule attachment. Strikingly, this map is the mirror image of the activity map for the anchored Mps1 kinase, with the Dam1 complex demarcating the boundary in both (Fig. 4f and Fig. 6e). These data strongly suggest that the Dam1 complex may contribute to SAC silencing by acting as a physical barrier that separates the phosphodomain of Spc105 from Mps1.
Separation between CH-domains of Ndc80 and N-Spc105 changes with the attachment state of the kinetochore
Our data suggest that microtubule attachment to kinetochore physically separates the CH-domains of Ndc80 and the phosphodomain of Spc105 to silence the SAC. By corollary, unattached kinetochores must bring them in close proximity to activate the SAC. To test if the separation between these two domains and the attachment state of the kinetochore are correlated, we measured FRET between N-Spc105 and either N-Nuf2 or N-Ndc80, which are proximal to the CH-domains (Fig. 7a, Supplementary Fig. 7). In both cases, FRET was undetectable in metaphase as predicted by the > 30 nm separation between N-Spc105 and the two protein termini20. In contrast, moderate FRET was detected in unattached kinetochores created by treating the cells with nocodazole indicating that mCherry and GFP fused to the respective N-termini were, on average, ~ 8 nm apart35.
Figure 7. The proximity between the CH-domains of Ndc80 and the phosphodomain of Spc105 within the kinetochore controls SAC signaling.
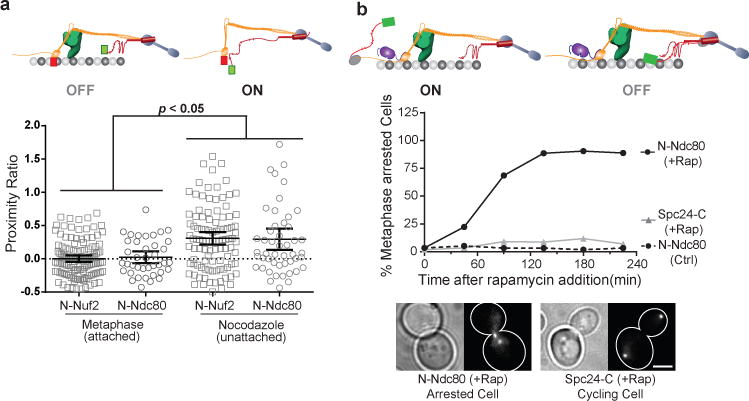
(a) Scatter plot: Proximity ratio measurements for FRET between mCherry-Nuf2 or mCherry-Ndc80 and GFP-Spc105 in attached (metaphase) and unattached (nocodazole-treated) kinetochores. It should be noted N-Ndc80 is connected to the CH-domain via a 113 amino acid long unstructured tail. Data pooled from 3 independent experiments, horizontal bars represent mean ± 95% confidence interval computed from n = 121, 37, 101 and 49 clusters (left to right). p-values were computed using Mann-Whitney test.
(b) Cell cycle kinetics after anchoring Spc105120:329 at indicated positions in strains expressing spc105-6A. Plotted points represent average values calculated from 2 independent trials. More than 70 cells scored in each trial and the source data are shown in Supplementary Table 3. Scale bar ~ 3 μm.
Proximity between the CH-domains and Spc105120:329 controls SAC signaling in attached kinetochores independently of the endogenous Spc105
Finally, we tested whether Spc105120:329 can restore the SAC in attached and unattached kinetochores in a position-dependent manner in spc105-6A strains that are SAC-deficient. The kinetochore only provides the architectural scaffold in this experiment. Consistent with the previous results, Spc105120:329 arrested the cell cycle when anchored proximal to the CH-domains (at N-Ndc80), but not when anchored distal to the CH-domains (at Spc24-C, Fig. 7b). Even within unattached kinetochores, Spc105120:329 restored the SAC when it was anchored at N-Ndc80, as expected (Supplementary Fig. 8). However, Spc105120:329 anchored at Spc24-C also activated the SAC suggesting that Mps1 can access Spc105120:329, even though its anchoring position is expected to be distal to the CH-domains. The inherent flexibility of Ndc80 and Spc105 and the presence of multiple molecules of these proteins in the kinetochore are likely responsible for this unexpected phenotype.
Discussion
Our work yields critical insights into how the protein architecture of the budding yeast kinetochore enables attachment-sensitive SAC signaling (Fig. 8a). We find that catalytically active Mps1 binds to a site located in the outer kinetochore even when the kinetochore is attached. Based on our findings and published data32, 33, 36, we propose that this site corresponds to the CH-domain of Ndc80. We also demonstrate that a persistent interaction between Spc105 and Mps1 is both necessary and sufficient to activate the SAC. These findings lead to an elegant model for the attachment-sensitive operation of the SAC (Fig. 8b–c). In unattached kinetochores, close physical proximity between the CH-domains of Ndc80 and the phosphodomain of Spc105 allows Mps1 to phosphorylate Spc105, and also enables subsequent steps in SAC signaling10, 37–41. End-on microtubule attachment to the kinetochore separates the CH-domains and the phosphodomain of Spc105 likely by pulling the CH-domains outwards and by restraining phosphodomain in the inner kinetochore. Additionally, the Dam1 complex, which is recruited after the formation of end-on attachment42, may act as a physical barrier that prevents further interaction between Mps1 and Spc105. A combination of these events leads to SAC silencing.
Figure 8. The mechanical switch model for attachment-sensitive SAC signaling.
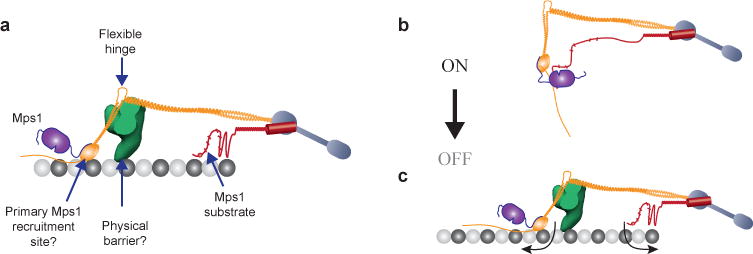
(a) Key features of the metaphase architecture of the yeast kinetochore-microtubule attachment (when SAC signaling is off), and their potential roles in executing attachment-dependent SAC signaling. Please note that the schematic displays a possible organization of Spc105 phosphodomain; the actual organization remains unknown. The microtubule-binding activity of this domain is not highlighted.
(b–c) The protein architecture of the kinetochore encodes a mechanical switch that controls SAC signaling. The CH-domain of Ndc80 and the phosphodomain of Spc105 act as the two terminals of this switch. It is in the ‘on’ state in unattached kinetochores, because the inherent flexibilities in the Ndc80 complex and Spc105 position the two terminals in close proximity. This allows Mps1 to phosphorylate Spc105 and initiate SAC signaling. Microtubule attachment toggles this switch to its ‘off’ state by physically separating the two terminals (arrows). Mps1 can no longer phosphorylate Spc105. The Dam1 complex, which is recruited to the kinetochore only after end-on microtubule attachment is established, may create a barrier that further hinders the interaction between Mps1 and Spc105. This leads to SAC silencing.
The control of SAC signaling by the physical separation of two protein domains is conceptually equivalent to the operation of a mechanical switch. As the two terminals of this microtubule-operated switch, Ndc80 complex and Spc105 must be capable of binding microtubules and changing their positions and/or conformations in response to microtubule-binding. Accordingly, the Ndc80 complex binds to microtubules via the CH-domains43. Known flexibilities in its structure should also allow it to change conformation in response to microtubule binding20, 23, 44–46. Spc105 also binds microtubules, and this may play a role in restraining its otherwise unstructured phosphodomain in the inner kinetochore47, 48. Finally, the low cellular abundance of the Mps1 kinase is crucial for the effective operation of this mechanical switch. If Mps1 is highly abundant, it can phosphorylate Spc105 through diffusive interactions, cause aberrant SAC activation, and thus effectively override the kinetochore-based switch31, 49.
While our work defines the ‘off’ state of the mechanical switch, further work is needed to define its ‘on’ state. The first key question is whether the CH-domain of Ndc80 is the only Mps1 recruitment site that is necessary for SAC signaling. Our findings and published data strongly argue for this to be the case. We find that the phosphorylation of Spc105 by Mps1 is both necessary and sufficient for SAC signaling. Therefore, the only activity that the Ndc80 complex can contribute to the SAC is the recruitment of Mps1. Accordingly, the Ndc80 complex is necessary for SAC signaling47, 50, 51, and the CH-domain binds Mps132, 33. The second key question is how the architecture of the unattached kinetochore, despite its inherent flexibility, promotes optimal interaction between Mps1 and Spc105. Answers to these questions will further validate and complete the cell biological description of the mechanical switch model for the SAC.
Whether the mechanical-switch model is applicable to the kinetochore in other eukaryotes is also an important question. Higher eukaryotes employ additional mechanisms that promote SAC silencing33, 52. Moreover, in other organisms, the forced localization of Mps1 in the outer kinetochore or Mad1 in the inner kinetochore activates the SAC11, 37, 53, 54. These differences may be because the budding yeast kinetochore stably binds exactly one microtubule in metaphase, whereas the kinetochores in most eukaryotes bind dynamically to many microtubules. A fraction of these microtubule-binding sites are unattached even in metaphase55, creating the possibility of cross-phosphorylation of SAC proteins localized in one attachment site by Mps1 localized within adjacent sites. Despite these differences, key elements of the SAC switch are highly conserved from yeast to humans. Components of the SAC switch: Mps1, the Ndc80 complex, and Spc105, and their nanoscale organization are highly conserved6. Intriguingly, even though the Dam1 complex is absent in humans, the human kinetochore recruits other microtubule-binding proteins in the same position as that of the Dam1 complex in the yeast kinetochore56–59. This striking conservation of key proteins and their architecture suggests that the kinetochore in other eukaryotes may encode a similar mechanical switch to control the SAC.
Experimental Methods
Strain and plasmid construction
Budding yeast strains and plasmids used in this study are listed in Supplementary Tables 1 and 2 respectively.
Strains used in the anchoring experiments were constructed as described in ref. 9. Briefly, we deleted FPR1 in wild-type strains to eliminate the rapamycin-binding protein product of this gene. These strains also express tor1-1, which encodes the dominant-negative, rapamycin-resistant form of the Tor1 kinase. We ensured that at least one copy of TOR1 in diploid strains was mutated to tor1-1.
Frb-GFP(S65T) (or Frb alone) was fused to the C-terminus of selected SAC proteins with a 24 or 7 amino acid linker (with the amino acid sequence ‘RIPGLINSGGGGGSGGGSGGGGAS’ or ‘SGGGGAS’ respectively). Two tandem copies of Fkbp12 (2xFkbp12) were fused to the C-terminus of kinetochore proteins with the linker coding ‘RIPGLIK’. 2xFkbp12 was fused to the N-terminus of Ndc80 via the linker sequence ‘GAAAAG’. A seven amino acid linker (sequence: ‘RIPGLIN’) was used to fuse fluorescent proteins (either GFP(S65T) or mCherry) to amine or carboxyl terminus of selected proteins.
spc105-6A strains were constructed using plasmid shuffling. Briefly, we deleted the genomic copy of SPC105 in a parent strain containing a centromeric plasmid containing SPC105 and the URA3 gene as the auxotrophic marker (pAJ274). Next, pSB1878 linearized with NsiI was integrated at the his3 locus (ref. 12). Finally, the centromeric plasmid carrying the wild-type SPC105 was kicked out by counter-selecting for URA3 on the drug 5-FOA.
Plasmids containing the minimal phosphodomain of Spc105, pAJ349 and pAJ350 were constructed by sub-cloning PCR amplification of the phosphodomain of Spc105 (amino acids: 120-329 from pSB1332 for wild-type, or from pSB1878 for the phosphonull version12) into pAFS144 carrying the Frb domain using AatII and KasI sites. These plasmids, after linearization with NsiI, were integrated at the his3 locus. For integration at LEU2 locus, the HIS3 gene in pAJ349 and pAJ350 was replaced with LEU2 to construct pAJ351 and pAJ352 respectively. The plasmids were linearized with BstEII for integration at the leu2 locus.
Cell culture
Cells were grown in yeast extract, peptone and dextrose (YPD) media at 32 °C and imaged at room temperature in synthetic media supplemented with essential amino acids and appropriate carbon source. To express N-terminally labeled kinetochore proteins from the galactose promoter (pGAL1), strains were grown in YP Raffinose media supplemented with 0.1–0.4% galactose. The galactose concentration was adjusted empirically as described previously35.
1 mg/ml stock solution of rapamycin in DMSO was diluted 1000× to achieve 1 μg/ml final concentration in all experiments involving rapamycin-induced dimerization.
To depolymerize metaphase spindle with nocodazole64, mid-log phase cells were synchronized in G1 with α-factor (2 µg/ml) for 2 hours, and then released into nocodazole-containing media (15 μg/ml) for 1.5–2 hours.
Benomyl sensitivity assay
10-fold serial dilutions log-phase cultures were frogged on YPD or plates containing (30 μg/ml) benomyl. Colonies were allowed to develop for 2–3 days at 30 °C before pictures of the plates were taken.
Metaphase arrest by CDC20 repression
Cells expressing Cdc20 from a Methionine repressible promoter (pMET3) were synchronized in G1 by treatment with α-factor (2 µg/ml) for 2 hours in synthetic media lacking Methionine. They were then released into YPD supplemented with 2 mM methionine for two hours to repress CDC20 and then treated with rapamycin for 10 minutes. Cells were washed into synthetic media lacking methionine to initiate CDC20 expression.
Inhibiting Ipl1 or Mps1 kinase activity using ATP analogs
The ATP analogs 1-NMPP1 and 1-NAPP1 (final concentration 50 μM) were used to block the activity of mps1-as1 and ipl1-as6 respectively. Cells were first synchronized in S-phase using 100 mM Hydroxyl Urea (HU) for 2.5 hours, washed with YPD, and then released into media containing the appropriate inhibitor for 15 minutes. This was followed by the addition of rapamycin to the media to anchor Mps1-Frb at Mtw1-C. We used the bud size to monitor cell cycle progress (please refer to the next section for a description of the criteria used).
To test the ability of 1-NMPP1 to block kinase activity of mps1-as1, we treated the cells with nocodazole to depolymerize the spindle and activate the SAC65. Next, we treated the cells with either 1-NMPP1 or DMSO, and monitored cell morphology. Mps1 kinase activity is necessary to maintain an active SAC and arrest the cells in mitosis. If the SAC remains active, then the cells remain arrested in mitosis as large-budded cells. However, SAC deficient cells escape the mitotic arrest and also fail in cytokinesis. They enter the next cell cycle and produce another bud thus giving rise to two-budded cells66.
To study the effect of 1-NAPP1 on ipl1-as6 activity, we measured the spindle localization of Sli15-GFP in pre-anaphase cells67. We used the bud size to find pre-anaphase cells as follows. If the bud was smaller than 50% in size as compared to the mother cell, and contained a short bar of Sli15-GFP located within the mother cell and at the bud neck, then the cell was deemed to be in pre-anaphase.
Scoring mitotically arrested cells
The cells were scored as ‘large-budded’ (e.g. Fig. 1e and Supplementary Fig. 2b), if the size of the bud was more than 2/3 the size of the mother as seen from bright-field images. Note that anaphase cells in cycling cultures will also be scored as large-budded cells by this criterion. In strains carrying fluorescent markers, we used the separation of the kinetochore clusters or spindle pole bodies to determine whether or not the cells arrested in mitosis. Large-budded cells with kinetochore-cluster separation smaller than 1 μm or spindle length smaller than 2 μm were scored as metaphase-arrested cells68.
Colony counting assays
Approximately 300 cells (estimated from the measured OD660 of liquid cultures) were plated on control and rapamycin-containing plates. After allowing the colonies to grow for 3 days at 30 °C, we determined the colony number. We ensured that the strains used in this experiment were rapamycin-resistant, by verifying that the parental haploid strains expressing either the Frb-fused SAC protein or the Fkbp12-fused kinetochore protein produced the same number of colonies on both control and rapamycin-containing plates.
Microscopy & Image acquisition
Nikon Ti-E inverted microscope with a 1.4 NA, 100×, oil immersion objective was used in imaging35. 10-plane Z-stack was acquired (200 nm separation between adjacent planes). The total fluorescence from each kinetochore cluster with GFP or mCherry tagged protein was measured using Image J, or a semi-automated MATLAB program as described earlier22. The copy numbers of kinetochore proteins and anchored proteins were calculated from the known copy number of the Ndc80 complex per kinetochore – 8 molecules per kinetochore21.
For photobleaching, an Argon-Ion laser (Photonics Instruments) beam filtered with the ET-GFP filter cube was focused on the sample by the objective. The target was manually aligned with the pre-determined location of the laser focus, and then exposed to 488 nm light for 50 ms. 5 plane Z-stacks were acquired starting immediately after bleaching for 14 minutes, at 2 minute intervals. Fluorescence was quantified from the images as above.
FRET, High resolution colocalization and Fluorescence distribution analyses were conducted as previously described20, 35, 69.
Time lapse imaging was used to follow the Mps1-Frb-GFP that autonomously bound to the kinetochore clusters in metaphase-arrested cells. Cells were released from the metaphase arrest by activating CDC20 expression, and a 6-plane Z-stack was acquired at 1 minute interval for 20 minutes. Anaphase entry was inferred from spindle elongation tracked from the spindle pole body protein (Spc97-mCherry). The change in Mps1-Frb-GFP intensity during this period was quantified, after correcting for two factors: (1) GFP photobleaching expected from imaging and. (2) Fluorescence emission from Spc97-mCherry due to cross-excitation while imaging GFP. The representative images in supplementary figure 1f have not been corrected for these factors.
Statistical Methods
Comparisons of proximity ratio measurements were conducted using the non-parametric Mann-Whitney test. The statistical tests used are described in the respective figure legends.
Supplementary Material
Acknowledgments
We thank Andrew Murray, Ben Glick, David Drubin, Mark Winey and Sue Biggins for sharing reagents, and Iain Cheeseman, Jennifer DeLuca, Mara Duncan, Richard McIntosh, and Yukiko Yamashita for comments on the manuscript. We also thank Lauren Humphrey and Simon Han for help with imaging and data analysis. APJ is supported by the Career Award at the Scientific Interface from the Burroughs-Wellcome Fund. This work was funded by R01-GM-105948. The authors have no competing financial interests.
Footnotes
Author Contributions: APJ and PA designed and performed the experiments, and wrote the manuscript; PA and AAG performed the rapamycin plating, and cell cycle kinetics experiments. APJ, PA and AAG constructed all the yeast strains.
References
- 1.Sacristan C, Kops GJ. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma. 2013;122:135–158. doi: 10.1007/s00412-013-0401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIntosh JR. Structural and mechanical control of mitotic progression. Cold Spring Harb Symp Quant Biol. 1991;56:613–619. doi: 10.1101/sqb.1991.056.01.070. [DOI] [PubMed] [Google Scholar]
- 5.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchida KS, et al. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santaguida S, Musacchio A. The life and miracles of kinetochores. Embo J. 2009 doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 10.Jelluma N, Dansen TB, Sliedrecht T, Kwiatkowski NP, Kops GJ. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J Cell Biol. 2010;191:281–290. doi: 10.1083/jcb.201003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito D, Saito Y, Matsumoto T. Centromere-tethered Mps1 pombe homolog (Mph1) kinase is a sufficient marker for recruitment of the spindle checkpoint protein Bub1, but not Mad1. Proc Natl Acad Sci USA. 2012;109:209–214. doi: 10.1073/pnas.1114647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 14.Biggins S, et al. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinrich S, Windecker H, Hustedt N, Hauf S. Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores. J Cell Sci. 2012;125:4720–4727. doi: 10.1242/jcs.110387. [DOI] [PubMed] [Google Scholar]
- 16.Yeong FM, Lim HH, Padmashree CG, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- 17.Palframan WJ, Meehl JB, Jaspersen SL, Winey M, Murray AW. Anaphase inactivation of the spindle checkpoint. Science. 2006;313:680–684. doi: 10.1126/science.1127205. [DOI] [PubMed] [Google Scholar]
- 18.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aravamudhan P, Felzer-Kim I, Joglekar AP. The budding yeast point centromere associates with two Cse4 molecules during mitosis. Curr Biol. 2013;23:770–774. doi: 10.1016/j.cub.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravamudhan P, Felzer-Kim I, Gurunathan K, Joglekar AP. Assembling the protein architecture of the budding yeast kinetochore-microtubule attachment using FRET. Curr Biol. 2014;24:1437–1446. doi: 10.1016/j.cub.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howell BJ, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 25.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Winey M. The MPS1 family of protein kinases. Annu Rev Biochem. 2012;81:561–585. doi: 10.1146/annurev-biochem-061611-090435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramey VH, et al. Subunit organization in the Dam1 kinetochore complex and its ring around microtubules. Mol Biol Cell. 2011;22:4335–4342. doi: 10.1091/mbc.E11-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimogawa MM, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraschini R, Beretta A, Lucchini G, Piatti S. Role of the kinetochore protein Ndc10 in mitotic checkpoint activation in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;266:115–125. doi: 10.1007/s004380100533. [DOI] [PubMed] [Google Scholar]
- 32.Kemmler S, et al. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nijenhuis W, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol. 2013;201:217–231. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Primorac I, et al. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife. 2013;2:e01030. doi: 10.7554/eLife.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joglekar AP, Chen R, Lawrimore JG. A sensitized emission based calibration of FRET efficiency for probing the architecture of macromolecular machines. Cell Mol Bioeng. 2013;6:369–382. doi: 10.1007/s12195-013-0290-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol. 2011;13:475–482. doi: 10.1038/ncb2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tipton AR, et al. Monopolar Spindle 1 (MPS1) Kinase Promotes Production of Closed MAD2 (C-MAD2) Conformer and Assembly of the Mitotic Checkpoint Complex. Journal of Biological Chemistry. 2013;288:35149–35158. doi: 10.1074/jbc.M113.522375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S, et al. Phosphorylation of the spindle checkpoint protein Mad2 regulates its conformational transition. Proc Natl Acad Sci USA. 2010;107:19772–19777. doi: 10.1073/pnas.1009000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 2014 doi: 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, et al. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang HW, et al. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tien JF, et al. Kinetochore Biorientation in Saccharomyces cerevisiae Requires a Tightly Folded Conformation of the Ndc80 Complex. Genetics. 2014 doi: 10.1534/genetics.114.167775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei RR, et al. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 47.Pagliuca C, Draviam VM, Marco E, Sorger PK, De Wulf P. Roles for the Conserved Spc105p/Kre28p Complex in Kinetochore-Microtubule Binding and the Spindle Assembly Checkpoint. PLoS ONE. 2009;4:e7640. doi: 10.1371/journal.pone.0007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol. 2012;196:469–482. doi: 10.1083/jcb.201111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 50.McCleland ML, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLuca JG, et al. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol. 2003;13:2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 52.Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballister ER, Riegman M, Lampson MA. Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. J Cell Biol. 2014;204:901–908. doi: 10.1083/jcb.201311113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuijt TE, Omerzu M, Saurin AT, Kops GJ. Conditional targeting of MAD1 to kinetochores is sufficient to reactivate the spindle assembly checkpoint in metaphase. Chromosoma. 2014 doi: 10.1007/s00412-014-0458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang G, et al. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125:3243–3253. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
- 57.Daum JR, et al. Ska3 Is Required for Spindle Checkpoint Silencing and the Maintenance of Chromosome Cohesion in Mitosis. Curr Biol. 2009;19:1467–1472. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varma D, et al. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol. 2012;14:593–603. doi: 10.1038/ncb2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu KS, Toda T. Ndc80 Internal Loop Interacts with Dis1/TOG to Ensure Proper Kinetochore-Spindle Attachment in Fission Yeast. Current biology. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou HX. Polymer models of protein stability, folding, and interactions. Biochemistry. 2004;43:2141–2154. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- 61.Petrovic A, et al. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol Cell. 2014;53:591–605. doi: 10.1016/j.molcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 62.Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones MH, et al. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Norden C, et al. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 67.Nakajima Y, et al. Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics. J Cell Biol. 2011;194:137–153. doi: 10.1083/jcb.201009137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marco E, et al. Scerevisiae chromosomes biorient via gradual resolution of syntely between S phase and anaphase. Cell. 2013;154:1127–1139. doi: 10.1016/j.cell.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sprague BL, et al. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys J. 2003;84:3529–3546. doi: 10.1016/S0006-3495(03)75087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


