Abstract
Although some existing epidemiological observations and molecular experiments suggested that brain disorders in the realm of psychiatry may be influenced by immune dysregulation, the degree of genetic overlap between psychiatric disorders and immune disorders has not been well established. We investigated this issue by integrative analysis of genome-wide association studies of 18 complex human traits/diseases (five psychiatric disorders, seven immune disorders, and others) and multiple genome-wide annotation resources (Central nervous system genes, immune-related expression-quantitative trait loci (eQTL) and DNase I hypertensive sites from 98 cell-lines). We detected pleiotropy in 24 of the 35 psychiatric-immune disorder pairs. The strongest pleiotropy was observed for schizophrenia-rheumatoid arthritis with MHC region included in the analysis (p = 3.9 × 10−285), and schizophrenia-Crohns disease with MHC region excluded (p = 1.1 × 10−36). Significant enrichment (>1.4 fold) of immune-related eQTL was observed in four psychiatric disorders. Genomic regions responsible for pleiotropy between psychiatric disorders and immune disorders were detected. The MHC region on chromosome 6 appears to be the most important with other regions, such as cytoband 1p13.2, also playing significant roles in pleiotropy. We also found that most alleles shared between schizophrenia and Crohns disease have the same effect direction, with similar trend found for other disorder pairs, such as bipolar-Crohn’s disease. Our results offer a novel birds-eye view of the genetic relationship and demonstrate strong evidence for pervasive pleiotropy between psychiatric disorders and immune disorders. Our findings might open new routes for prevention and treatment strategies for these disorders based on a new appreciation of the importance of immunological mechanisms in mediating risk of many psychiatric diseases.
Keywords: GWAS, Pleiotropy, Psychiatric disorder, Immune-mediated disease
1 Introduction
Psychiatric disorders are often associated with significant morbidity and mortality [47]. The estimated heritability for most psychiatric disorders is moderate to high (40%–80%), so genetic factors play a critical role in their etiology [31, 32, 55]. In the past few years, many genome-wide association studies (GWAS) have been conducted to identify genetic risk variants underlying psychiatric disorders [58]. Despite recent progress, there is much yet to be discovered regarding the genetic architecture of psychiatric disorders [15].
The relationship between psychiatric disorders and immune disorders has intrigued researchers for decades (Fig. S1). There is a moderately large body of evidence that supports a role for immune dysfunction in the development of several psychiatric disorders, including early hypothesis like the macrophage theory of depression [49], and recent findings such as the epidemiological observation of co-occurrence of rheumatoid arthritis (RA) and depression [8,35] and cross-disorder drug effects, for example some drug for psychiatric disorders have anti-inflammatory properties [36, 59]. The genetic liability underlying these observed correlations has not been well studied, with the exception that recent GWAS have repeatedly identified association between SCZ and genetic variants at the major histocompatibility locus (MHC), which also plays an important role in the immune system [22,23,41]. However, no strong evidence of shared liability was observed between Crohns disease (CD) and multiple psychiatric disorders in another study [42]. In genetics, the term pleiotropy refers to a one-to-many relationship between a gene or mutation and phenotypes [37]. In the GWAS era, pleiotropy could explain correlations among disorders, and may also boost statistical power to detect genetic associations [2, 29, 30, 40, 42, 57]. To date, pervasive pleiotropic effects have been discovered in autoimmune disorders [7] and in psychiatric disorders [15, 42] , as separate classes.
Given the public health significance of these two classes of disorders and the treatment implications of any etiological overlap, it is important to resolve the nature of genetic pleiotropy between them, to understand the underlying mechanisms of pleiotropy, and to identify specific genes and pathways driving such pleiotropy. These inquiries can only now be carried out because of the large amounts of genomic data that have become available in recent years. Large consortia have been formed to study many psychiatric disorders and immune disorders [1,5,14,16, 21,28, 40–42, 51,56]. For example, the results from a well-powered GWAS of schizophrenia [41] provided strong evidence supporting the link between schizophrenia and the immune system. Undoubtedly, the availability of high-quality omics data offers us an unprecedented opportunity to revisit the nature of the genetic connections between psychiatric disorders and immune-mediated disorders. The analysis results can deepen our understanding of the genetic architecture of complex human diseases.
Our current study takes advantage of multiple omics data resources to obtain a birds-eye view of the shared genetic components between psychiatric disorders and immune disorders. To better represent those two disorder categories while taking the data availability into account, we considered five psychiatric disorders, including schizophrenia (SCZ), bipolar affective disorder (BPD), autism spectrum disorder (ASD), attention deficit-hyperactivity (ADHD), and major depressive disorder (MDD). For immune-mediated disorders, we considered two inflammatory bowel diseases (IBDs), Crohns disease (CD) and ulcerative colitis (UC), and five other immune disorders, including multiple sclerosis (MS), psoriasis (PS), rheumatoid arthritis (RA), systemic lupus erythematosis (SLE), and insulin-dependent diabetes mellitus (T1D). For comparisons, we also included a central nervous system degenerative disease, Parkinsons disease (PD), and five traits related to education, height, and weight. We performed comprehensive genome-level analysis on psychiatric disorders and immune disorders by integrating both disorder-specific GWAS and genomic annotations, in search of common genetic liability. Our results not only confirmed previously reported genetic regions affecting disease risk for both psychiatric and immune-mediated disorders, but also implicated many novel shared genes and pathways.
2 Results
2.1 Pervasive pleiotropic effects between psychiatric disorders and immune system disorders
Previous studies have shown extensive shared genetic effects among many of the five psychiatric disorders studied by the Psychiatric Genomics Consortium (PGC) [40,42] and among multiple immune system disorders [7], separately. Consistent with those studies, we also observed pervasive pleiotropic effects among psychiatric disorders and among immune-related disorders (Table S1). Pleiotropic effects are significant (Bonferroni-adjusted p < 0.05) for all 21 pairs of immune system disorders, and for seven of the 10 pairs of psychiatric disorders (the exceptions being ASD-ADHD, MDD-ADHD, and MDD-ASD).
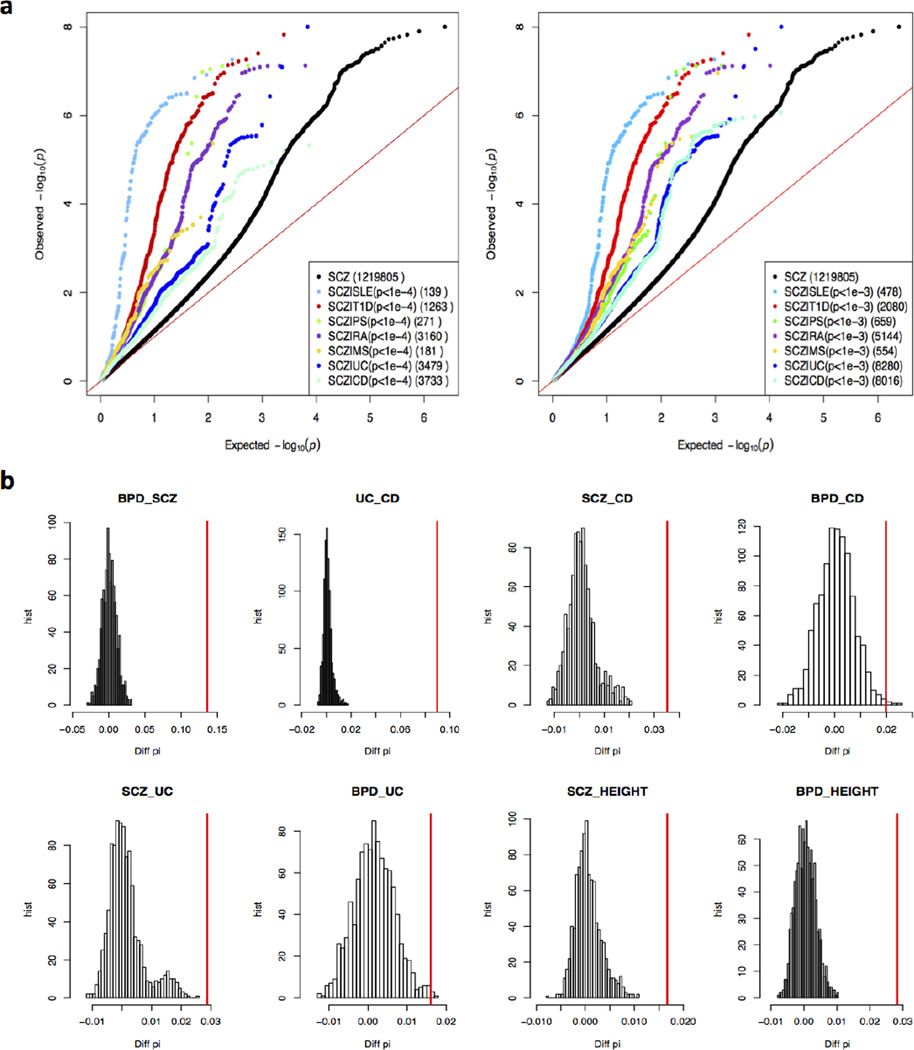
We then tested pleiotropic effects between psychiatric and immune system disorders. We first considered SCZ with seven immune-mediated disorders. The conditional Q-Q plots (Fig. 1a) suggest that all seven immune-mediated disorders share genetic liability components with SCZ.
Fig. 1. Pervasive pleiotropic effects between psychiatric and immune system disorders.
a) Conditional Q-Q plot showing pleiotropy between schizophrenia and 7 immune system disorders. Black dots represent all 1 219 805 SCZ GWAS SNPs while the other 7 colored dots represent different subsets of SNPs selected from the corresponding immune system disorder GWAS whose p < 0.0001 (left panel) and p < 0.001 (right panel), with the number of SNPs in each subset shown in brackets. (b) Chromosome-bound circular permutation to adjust LD effects for assessment of the significance of pleiotropy of eight GWAS pairs. For each of the 8 trait-pairs, 1000 times of chromosome-bound circular permutations were performed. The distribution of the test-statistic from 1,000 permutations are shown by histograms, where x-axis represents the test-statistic, Diff_PI = π̂11 − (π̂10 + π̂11)(π̂01 +π̂11) , and y-axis represents its frequency. Red vertical line denotes the observed test statistics for each trait-pair.
Conditional Q-Q plots, while simple and intuitive, suffer from arbitrary cutoffs, e.g. 1 × 10−4, and do not offer statistical assessment of pleiotropy. We then used GPA (Genetic analysis incorporating pleiotropy and annotation), a statistically rigorous approach recently developed by us [6], to quantitatively test the significance of pleiotropy between the five psychiatric disorders (SCZ, BPD, MDD, ASD, ADHD) and seven immune-mediated disorders (CD, UC, MS, PS, SLE, RA, T1D). Twenty-four of the 35 pairs were significant at Bonferroni-adjusted p-value <0.05 (Table S1), indicating pervasive pleiotropic effects between psychiatric disorders and immune mediated disorders. Consistent with previous studies [3], we observed strong pleiotropy between SCZ-MS (p = 1.3 × 10−20), but no significant pleiotropy between BPD-MS (p = 0.26), with or without the MHC region (Fig. S2).
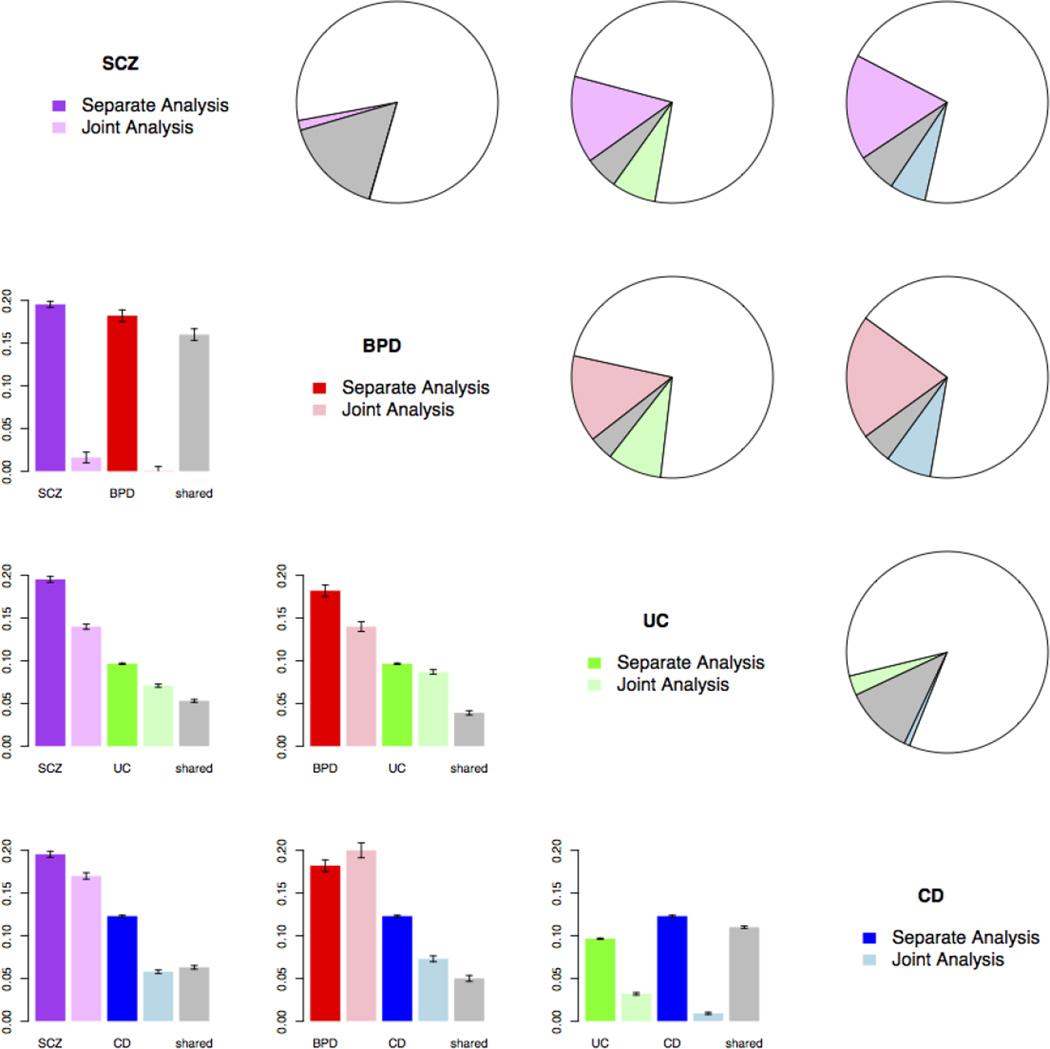
For each pair of disorders, we estimated the proportion of single nucleotide polymorphisms (SNPs) associated with both disorders vs. those associated with only one disorder (Table S1). Fig. 2 shows the results among SCZ, BPD, UC and CD. Consistent with previous studies [39, 40], most of the SCZ-associated SNPs and BPD-associated SNPs were estimated to be shared between these two disorders. Similarly, most UC-associated SNPs and CD-associated SNPs were shared between them. The proportions of SNPs shared by cross-class disorders were: SCZ-CD 0.063 (s.e.0.0021); SCZ-UC 0.053 (s.e.0.0018); BPD-CD 0.05 (s.e.0.0034); and BPD-UC 0.039 (s.e.0.0025), respectively.
Fig. 2. GPA results showing pleiotropic effects among SCZ, BPD, UC and CD.
Purple, red, green and blue represent SCZ, BPD, UC and CD; grey represents the proportion of SNPs associated with both disorders, and white represents the proportion of SNPs associated with neither disorder. Upper triangle: pie charts show proportion of SNPs associated with only one disorder, both disorders (grey), and neither disorder (white). Lower triangle: bar plots contrasting proportions of associated SNPs for each disorder when analyzed separately (first and third bar in darker color), and proportion of associated SNPs when two disorders are jointly analyzed (second and fourth bar for proportion of SNPs associated with only one disorder, and fifth grey bar for proportion of SNPs associated with both disorders). Error bars indicate one standard error.
To account for potential inflation of statistical significance in pleiotropy tests due to LD structure, pleiotropy among SCZ, BPD, CD, and UC was studied by the means of chromosome-bound circular permutations [26], as detailed in Methods. Consistent with GPA pleiotropy test results, all six disease-pairs have significant pleiotropy, with permutation-based p-values below 0.001 for SCZ-BPD, CD-UC, SCZ-CD, SCZ-UC and about 0.004 for BPD-CD and BPD-UC (Fig. 1b).
2.2 Enrichment of immune-related annotations in multiple psychiatric disorders
Observation of extensive pleiotropy naturally leads to the exploration of functional enrichment for the shared genes to better understand the underlying biology. We used central nervous system (CNS) SNPs and immune related eQTLs (see Methods) to represent the functional sites relevant to the CNS and immune system, respectively. Because 12.5% of CNS SNPs overlap immune eQTLs, we also tested enrichments excluding those overlapping SNPs.
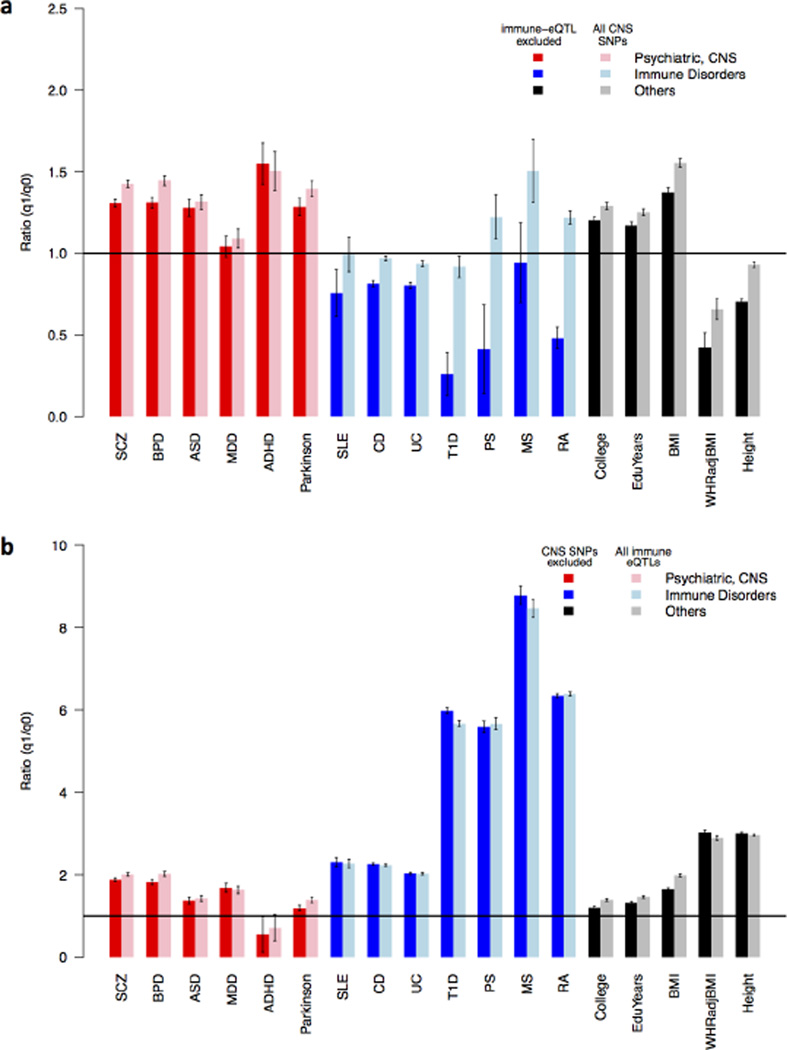
We first tested for enrichment of CNS SNPs in all 18 traits (Fig. 3a). As expected, all psychiatric disorders had modest enrichment for CNS SNPs (>1.3-fold, except for MDD, 1.09-fold). The enrichment effects could still be observed (and were even stronger for ADHD) with the MHC region and/or immune related eQTLs excluded (Fig. S3). Only three immune system disorders (MS, PS and RA) showed modest enrichment for CNS SNPs (1.5,1.2, and 1.2-fold, respectively), but not with immune eQTLs excluded (0.9, 0.4, 0.5-folds, respectively). This suggests that enrichment of CNS SNPs in immune traits was driven by overlapping immune eQTLs. We also observed enrichment of CNS SNPs for education years (1.25-fold), college completion status (1.29fold), and BMI (1.55-fold), but neither waist-to-hip-adjusted BMI nor height showed enrichment of CNS SNPs.
Fig. 3. Enrichment of CNS SNPs and immune eQTLs in 18 traits.
(a) Enrichment of CNS SNPs (comprising 18.8% of all SNPs) in 18 traits from three categories: psychiatric disorders or CNS related disorder (red), immune system related disorders (blue), and body somatic features (black). For each trait, the first bar (darker color) excludes immune eQTLs from CNS SNPs, and the second bar (light color) is for all CNS SNPs (comprising 21.4% of all SNPs). (b) Enrichment of immune eQTLs (comprising 7.5% of all SNPs) in 18 traits from three categories: psychiatric disorders or CNS related disorder (red), immune system related disorders (blue), and other body somatic features (black). For each trait, the first bar (darker color) excluded CNS SNPs from immune eQTLs, and the second bar (light color) is for all immune eQTLs (comprising 10.1% of all SNPs).
Next, we tested enrichment of immune eQTLs in the same set of 18 traits (Fig. 3b). The seven immune-mediated disorders consistently had the strongest enrichment (ranging from 2.0 to 8.5-fold). We also observed enrichment of immune eQTLs in four psychiatric disorders (SCZ, BPD, ASD, and MDD; 2.0, 2.0, 1.4, 1.6-fold, respectively), and Parkinsons disease (1.4-fold). Those enrichment effects still persisted with MHC region and/or CNS SNPs excluded, suggesting the enrichment was not solely due to eQTLs in the MHC region or overlapping with CNS SNPs (Fig. S3). We also observed immune eQTL enrichment in two education related traits, college completion (1.39-fold) and year of education (1.46-fold), and in three physical features, BMI (1.99-fold), obesity measured by waist-to-hip ratio adjusted BMI (2.90-fold), and height (2.97-fold).
To explore this hypothesis further, we tested levels of enrichment of immune related eQTLs in SNPs associated with both psychiatric disorders (SCZ, BPD, ASD, MDD, ADHD) and Crohns disease, and observed larger enrichment ratios compared with those SNPs associated with only one disease (Fig. S4). This result suggests that the shared genetic components between the five psychiatric disorders and CD are closely related to immune function. Next we tested enrichment of DNase-peak located SNPs in SCZ GWAS signals from 98 ENCODE cell lines (Table S2), and found the top cell lines were from blood elements having important roles in immune response, with the top two cell lines being CD20+ B cells and Th2 cells (CD4+ T cells) (Fig. S5). We also tested enrichment of an epigenetic marker H3K9ac (H3 lysine 9 acetylation), known to mark active enhancers and promoters, in eight tissues from the ROADMAP project [45]. We observed that both psychiatric disorders and immune-related disorders have the highest enrichment for H3K9ac markers in blood, while educational traits (years of education and colleges completion) have the highest enrichment for H3K9ac markers in brain (Fig. S6). We also observed enrichment of H3K9ac markers from fat tissue to whist-to-hip ratio adjusted BMI GWAS (Fig. S6). Those results further demonstrate the enrichment of immune specific contribution to the psychiatric disorder GWAS.
2.3 Trend of consistent effect direction between psychiatric disorders and immune system disorders
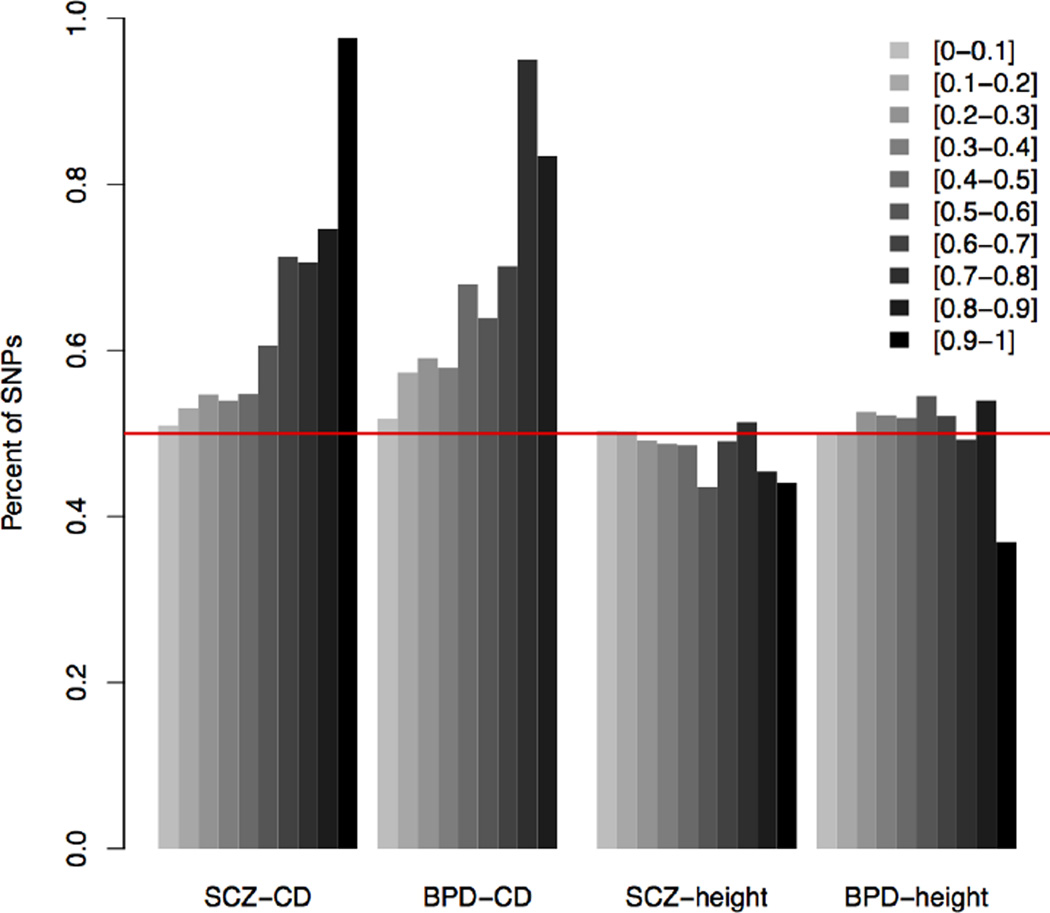
To explore the mechanism of these pleiotropic effects further, we examined effect directions. For each SNP, the same allele may increase or reduce susceptibility for the two disorders (same direction) or have opposite effects (different directions). The SCZ Q-Q plot (Fig. S7) shows interesting signals conditional on having the same effect direction with CD. We then considered four disorder pairs showing strong pleiotropy: SCZ-CD (p = 1.9 × 10−109), BPD-CD (p = 1.5 × 10−13), SCZ-Height (p = 2.0 × 10−122), and BPD-Height (p = 5.4×10−150). For each of these four pairs, there is no correlation in effect direction when all genotyped SNPs are considered (Table S3). However, trends emerged after we partitioned the SNPs into 10 groups according to their posterior probabilities of being associated with both traits. Proportions of SNPs having the same effect directions was calculated for each group. There are clear patterns for SCZ-CD and BPD-CD, but not for SCZ-height nor BPD-height as shown in Fig. 4.
Fig. 4. Trend of consistent effect directions for SCZ/BPD-CD across posterior probability groups.
Proportion of SNPs having the same effect direction for trait pairs, in each of the 10 posterior probability groups (darker colors indicate higher posterior probability), where SNPs were grouped based on posterior of being associated with both traits into 10 equal bins. Four pairs of traits: SCZ-CD, SCZ-height, BPD-CD, and BPD-height.
In general, the higher the posterior probability of a SNP being associated with SCZ (or BPD) and CD, the more likely that the SNP had the same effect direction for the pair. For SCZ-CD, among the 85 top SNPs with posterior probabilities of association with both SCZ and CD higher than 0.9 (Table S4), 97.6% of SNPs had the same effect directions (an allele either increases or reduces both SCZ and CD risks). Similarly, for BPD-CD, in the SNP group with posterior probabilities higher than 0.8, and between 0.7 and 0.8, 83% and 95% of SNPs had the same effect direction, respectively (Fig. 4). Similar patterns were also observed for SCZ-RA and BPD-RA pairs (Fig. S8). In contrast, the proportion of SCZ risk alleles that were associated with lower height was 50% for all SNP groups, regardless of their posterior probabilities of being associated with both SCZ and height. Effect direction distributions across 10 posterior groups for the BPD-height pair behaved similarly. We also investigated the influence of LD blocks on the observed effect direction trend by grouping SNPs into LD blocks based on DistiLD Database [38], as detailed in Supplementary Methods. LD blocks with high posterior probability were more likely to have a higher proportion of SNPs with same effect direction for SCZ-CD and BPD-CD, while the effect direction was less consistent for SCZ-height and BPD-height (Fig. S9).
2.4 Genome region enrichment analysis
To demonstrate the biological mechanism of the pleiotropy between psychiatric disorders and immune-mediated disorders, we tested genome-wide enrichment of potential pleiotropic SNPs in cytobands, protein-protein interaction networks, and gene ontology (GO) terms, for 28 disorder pairs between seven immune system disorders (CD, UC, MS, PS, RA, SLE, and T1D) and four psychiatric disorders (SCZ, BPD, MDD, and ASD), detailed in Methods.
In cytoband enrichment analysis, a complete list of all cytobands with enrichment odds ratio (OR)>5 and Bonferroni-adjusted p-value<0.001 in at least one disease pair were reported in Table S5. Some cytobands have significant enrichment in more than one disease pairs, such as MHC region and 1p13.2, indicating their role in affecting both psychiatric disorders and immune system disorders. Specifically, cytoband 1p13.2 was significantly enriched for the eight disorder pairs between {SCZ, BPD, MDD, and ASD} and {T1D and RA}, with Bonferroni-adjusted p-values ranging from 6.8 × 10−26 to 2.7 × 10−78, with top SNPs located in genes AP4B1, PTPN22, and PHTF1 (Fig. S10). In protein-protein interaction (PPI) network analysis, several sub-networks (Fig. S11) were highlighted in the analysis. Specifically, those protein-protein interaction clusters most responsible for shared genetic components between psychiatric disorders and immune system disorders in these data were: (1) three minor gene subunits HLA-E, HLA-F, and HLA-G, but not the three major gene subunits, interacting with TAP1, TAP2. TAP1 and TAP2 are transporters associated with antigen processing, which cooperate with MHC class I to present antigens [54]; (2) Interaction between HLA-DO, HLA-DM, and HLA-DR proteins; and (3) a set of genes with important roles in transcriptional activation, including BRD2, TUBB, ABT1, and multiple histone coding genes. In GO term enrichment analysis, the identified top terms included antigen processing and presentation, MHC protein complex, allograft rejection, and NF-kappaB binding (Table S6), which further suggests the enrichment of immune system function in shared genetic factors between psychiatric and immune system disorders.
3 Discussion
Our work demonstrates extensive pleiotropy between psychiatric disorders and immune system disorders. It is a common concern that the uneven distribution of genomic features, such as LD blocks and genes, may bias these findings. To address this issue rigorously, we performed chromosome-bound circular permutation [26] on eight trait-pairs for which we performed comprehensive analysis in this work (Fig. 1b). All eight pairs yielded highly significant permutation-based p-values, consistent with the GPA pleiotropy test results.
Beyond the evidence of pleiotropy, our results suggest how psychiatric disorders and immune system disorders are related genetically. We observed a major but not single role of MHC region in contributing to the pleiotropy between psychiatric disorders and immune system disorders. First, we observed enrichment of immune eQTLs even after the whole MHC region was removed (Fig. S3). Second, cytoband enrichment results indicate roles played by other specific genomic regions, such as 1p13.2, harboring gene PTPN22 (Protein Tyrosine Phosphatase, Non-Receptor Type 22 (Lymphoid)), which was also prioritized in our PPI analysis.
The observation of a tendency of the same effect direction for SNPs associated with either SCZ and BPD paired with CD gives some insight concerning the underlying mechanism of their shared genetic factors. Pleiotropy has been extensively reviewed [37,50,53,62], but is still not well understood in terms of its extent, mechanisms, and consequences. The Weak Hypothesis of Universal Pleiotropy (WHUP) advocated by Fisher [13] and Wright [63] is based on two assumptions that, in general, a phenotype might be influenced by many variants, and a variant might cause changes to many phenotypes. Under WHUP, extensive pleiotropy should be detected while the effect directions of shared genetic variants should be about random, which is not we observed for SCZ-CD. Our observation supports a closer genetic relationship between those two types of disorders. Various molecular mechanisms could result in pleiotropy [50]. There are biological pleiotropy, mediated pleiotropy, and spurious pleiotropy. Biological pleiotropy has separate causal paths for different phenotypes, while mediated pleiotropy has one phenotype lying on another phenotypes causal path; thus by this mechanism, one phenotype might lead to another [50]. Our results, the striking trend of shared SNPs for SCZ and CD acting in the same direction, can be best explained by mediated pleiotropy. This, together with our observation of pervasive enrichment of immune eQTLs in psychiatric disorders, and the lack of enrichment of CNS SNPs (immune eQTLs excluded) in immune-mediated disorders (except MS, which is characterized by CNS pathology), suggest that immune system disorders might mediate psychiatric disorder risk, i.e. some downstream immune dysfunctions might be a trigger to some psychiatric disorders (or subtypes).
Consistent with a recent GWAS finding that detected pathways associated with BMI mostly act in brain or peripheral nervous system [33], we observed enrichment of CNS SNPs for BMI, but a depletion of CNS SNPs for WHRadjBMI, suggesting different regulation mechanisms for body fat level and fat distribution. We also observed considerable enrichment of immune-related eQTLs in height and BMI, which are consistent with previous experiments that BMI is correlated with immune parameters [20], and that height is associated with immune response in young men [27]. Our results further confirmed the relationship between BMI and height and immune system from a genomics perspective.
Our work revealed the shared genetic factors between psychiatric and immune system disorders using novel methods and multiple omics data accumulated in recent years. We were able to show that there is pervasive pleiotropy between those two categories of disorders. Although the MHC region shows the strongest pleiotropic effects, other regions, such as cytoband 1p13.2, also contribute to the overall pleiotropy. Moreover, we found that pleiotropic SNPs for schizophrenia and Crohns disease tend to have the same effect direction for both disorders, suggesting mediated pleiotropy. Apart from cross-disorder study of GWAS summary statistics, our study included various genome annotations, including CNS SNPs, eQTLs detected in immune-related contexts, and DNase I hypertensive sites from 98 cell-lines. Study of those genome annotations provided further support for correlated genetic factors for psychiatric disorders and immune system disorders. Our work offers insights on pleiotropic mechanisms and a better understanding of pathophysiology, which may lead to improved prevention and treatment strategies for these two classes of disorders via immunological mechanisms. Although our analyses were based on results from GWAS consortia, the statistical power remains limited to identify the majority of disease associated variants for these disorders. GWAS results from larger studies and improved statistical and bioinformatics approaches will enable us to identify more shared genetic pathways between these classes of disorders, and as always despite the very high significance levels we observed for some relationships independent replication of our results is called for.
4 Materials and Methods
4.1 Genome-wide association study (GWAS) data sources
We made use of GWAS summary statistics from a set of diverse and representative traits, including major psychiatric disorders, various immune system disorders, body morphological features, and some socioeconomic measures (Table 1). The p-values were available for all traits, but only some of them have available specified alleles and their corresponding beta or odds ratios indicating effect direction.
Table 1.
Sources of GWAS summary statistics
| Traits | Trait types | Source/Reference |
|---|---|---|
| Schizophrenia (SCZ) | Psychiatric disorder | PGC [40] |
| Bipolar disorder (BPD) | Psychiatric disorder | PGC [40] |
| Autism spectrum disorder (ASD) | Psychiatric disorder | PGC [40] |
| Attention deficit-hyperactivity disorder (ADHD) | Psychiatric disorder | PGC [40] |
| Major depressive disorder (MDD) | Psychiatric disorder | PGC [40] |
| Parkinsons Disease (PD) | CNS degenerative disease | Simon-Sanchez et al. [48] |
| Crohns Disease (CD) | Immune system disorder | IIBDGC [14] |
| Ulcerative colitis (UC) | Immune system disorder | IIBDGC [1] |
| Multiple sclerosis (MS) | Immune system disorder | IMSGC [21] |
| Psoriasis (PS) | Immune system disorder | Feng BJ et al. [12] |
| Rheumatoid arthritis (RA) | Immune system disorder | Stahl et al. [52] |
| Systemic lupus erythematosis (SLE) | Immune system disorder | SLEGEN [16] |
| Insulin-dependent (Type 1) diabetes mellitus (T1D) | Immune system disorder | T1DGC [5] |
| Binary variable for college completion | Education status | Rietveld et al. [44] |
| Years of educational attainment | Education status | Rietveld et al. [44] |
| Body mass index (BMI) | Body feature | GIANT Consortium [51] |
| Waist-to-hip ratio (WHR) adjusted BMI | Body feature | GIANT Consortium [17] |
| Height | Body feature | GIANT Consortium [28] |
4.2 Genomic annotation data sources
Central nervous system (CNS) genes were identified in a previous study [43], comprising preferentially brain-expressed genes [43], neuronal-activity genes [60], learning-related genes [61], and synapse genes, defined by Gene Ontology [4]. A complete list of these genes is given in Table S7. CNS SNPs were defined as SNPs located within 50kb of CNS genes. To investigate immune system influence, we used context-specific eQTLs upon triggering immune response as detected by Fairfax et al. [11], where interferon-γ and lipopolysaccharide (LPS) were used as inflammatory proxies to stimulate innate immune effects in monocytes from volunteers of European ancestry. We used a union of cis-eQTLs detected in four distinct contexts, nave, LPS2 (monocytes exposed to 2 hours of LPS), LPS24 (monocytes exposed to 24 hours of LPS), and INF-γ (monocytes exposed to 24 hours of interferon-γ), as a set of immune-related eQTLs in our study. In total, we have 94,674 immune eQTLs and 199,202 CNS SNPs, of which 24,860 CNS SNPs are also immune eQTLs.
To investigate the impact of chromatin state, we used DNase I hypersensitivity sites extracted from ENCODE [10] DNase-seq peaks and signal of open chromatin from 125 cell lines. There were 98 cell lines after removing 27 cancer cell lines (Table S2). Although limited in cell lines from brain regions, those 98 cell lines have great coverage for various blood cells, making it suitable for studying whether there is enrichment of functional genomic regions in SCZ GWAS tissues implicated with important immune functions. DNase-Peak SNPs are SNPs located in or within 1kb from DNase-Peaks. We obtained the H3K9ac histone marker from the Roadmap project [45]. We downloaded the consolidated narrow peaks from http://egg2.wustl.edu/roadmap/data/byFileType/peaks/consolidated/narrowPeak/, and then generated our annotations based on the — log(p)-value cutoff at 6. We mainly focused on eight primary tissues, including blood (E038, E047 ,E062, E115, E116, E123, E124), brain (E067, E068, E069, E072, E073, E074, E125), breast (E027, E119), fat (E023, E025, E063), heart (E083), lung (E017, E088, E114, E128), muscle (E052, E107, E108, E120, E121), and skin (E126, E127).
4.3 Pleiotropy and annotation enrichment analysis using GPA
Pleiotropy analysis was performed via the GPA R package [6], which is a statistical approach to exploring the genetic architecture of complex traits by integrating pleiotropy and functional annotation information, including prioritizing risk genetic variants, and evaluating annotation enrichment and pleiotropy by hypothesis testing. Instead of relying on genotype-phenotype data at the individual level, it only requires the summary statistics from GWAS, which makes it useful for integrative analysis of genomic data. For each trait pair, only overlapped SNPs across the two traits were used in our analysis. For convenience, we briefly introduce the GPA model [6] and its notation here.
Consider the p-values {pi,... ,pm} obtained by performing hypothesis testing of genome-wide SNPs from one GWAS, where M is the number of SNPs. In the GPA model, these p-values are assumed to come from a mixture of null (un-associated) and non-null (associated), with probability π0 and π1= 1— π0 respectively. GPA uses the Uniform distribution on [0,1] and the Beta distribution with parameters (α, 1) to model the p-values from the null and non-null groups, respectively. Let Zj ∈ {0,1} be the latent variable indicating whether the j-th SNP is from the null or non-null group, where Zj = 0 means null and Zj = 1 means non-null. Then the GPA model for one GWAS without annotation can be written as:
| (1) |
GPA further incorporates functional annotation as follows. Let an M-dimensional vector A collect functional information from an annotation source, where Aj ∈ {0,1} indicates whether the j-th SNP is a functional unit according to the annotation source. For example, given an eQTL data, if the j-th SNP is an eQTL, then Aj = 1, otherwise Aj = 0. The relationship between Zj and Aj is described as:
| (2) |
Clearly, q0 can be interpreted as the proportion of null SNPs being annotated, q1 corresponds to the proportion of non-null SNPs being annotated, and q1 > q0 implies that there exists enrichment in this annotation.
Let θ̂={π̂0,π̂1,q̂0,q̂1,α̂}; be the collection of the estimated model parameters. Then SNPs can be prioritized based on their local false discovery rates (FDR). When there is no annotation data, the local FDR is defined as the probability that the j-th SNP belongs to the null group given its p-value, i.e., fdr̂(pj) = Pr(Zj = 0|pj;θ̂). With annotation data, the FDR can be calculated as fdr̂(pj,Aj) = Pr(Zj = 0|pj,Aj;θ̂). We can use the likelihood ratio test to assess the significance of its enrichment. Specifically, the significance of enrichment of an annotation for GWAS can be assessed by testing H0 : q0 = q1 versus H1 : q 0 ≠ q 1. Standard errors of all the parameters can also be calculated.
The extension of the above model to handle two GWAS is straightforward. Suppose the p-values from two GWAS have been collected in an M × 2 matrix p = [pjk], where pjk denotes the p-value of the j-th SNP in the k-th GWAS, k = 1,2. Let Zj ∈ {00,10,01,11} indicate the association between the j-th SNP and the two phenotypes: Zj = 00 means the j-th SNP is associated with neither of them, Zj = 10 means it is only associated with the first one, Zj = 01 means it is only associated with the second one, and Zj = 11 means it is associated with both. Then the two-groups model (1) can be extended to the following four-groups model:
| (3) |
Similarly, functional annotation information can be incorporated into the multiple GWAS model (3) in the following way:
where q00 is the probability of a null SNP being annotated, q10 is the probability of the first phenotype associated-SNP being annotated, q01 is the probability of the second phenotype associated-SNP being annotated, and q11 is the probability of jointly associated-SNP being annotated. For joint analysis of two GWAS data sets, the local FDR calculation and enrichment assessment can be done in a similar way. In addition, the pleiotropy between two phenotypes can be tested in a statistically rigourous way. When there is no pleiotropy, i.e., the signals from the two GWAS are independent of each other, testing pleiotropy can be formulated by testing the following hypothesis:
| (4) |
where π1* =π10 + π11 and π*1 = π01 + π11. likelihood ratio test statistic asymptotically follows χ2 distribution with df = 1 under the null.
4.4 Chromosome-bound circular permutation to assess the significance of pleiotropy
To fully account for the potential effects from LD structure when testing the significance of pleiotropy, chromosome-bound circular permutation [26] was adopted here. This permutation approach preserves the observed LD distribution of SNPs around the genome, and establishes a robust null distribution from which the significance of the observed pleiotropy can be calculated. To assess the significance of the pleiotropy of two traits A and B. The summary statistic set of trait A was circularly permutated within chromosomes as follows: The summary statistics were ordered according to their SNP positions. Next, a random number was generated from one to the number of SNPs in the chromosome. Then the summary statistics were shifted down by the generated random number. If their shifting status exceeded the number of SNPs in the chromosome, they resumed at the beginning position. The summary statistic set of GWAS B was unchanged. For each permutation, GPA was used to estimate the proportion of the SNPs in the four groups: SNPs associated with neither of the traits, SNPs associated with trait A, SNPs associated with trait B, and SNPs associated with both, denoted as {π̂00,π̂10,π̂01,π̂11}. Under the null hypothesis that there is no pleiotropy between A and B, the joint distribution should be the product of its marginal distribution, i.e., π̂11 = (π̂10 + π̂11)(π̂01 +π̂11). Therefore, we define our test statistic as Diff_PI = π̂11 − (π̂10 + π̂11)(π̂01 +π̂11) , and recorded the test-statistic for each permutation. The observed test statistic of the real data was compared with the null distribution obtained from chromosome-bound circular permutation, and p-values were obtained accordingly. The permutation results for eight pairs of GWAS are shown in Fig. 1b.
4.5 Effect direction analysis on the level of LD blocks
SNPs were grouped into LD blocks based on DistiLD Database [38]. For each LD block, we first calculated the number of SNPs within the block and the proportion of these SNPs having the same effect direction for both traits. For each SNP within the LD block, we evaluated its posterior probability of being associated with both traits and assigned the maximum posterior probability of these SNPs to the LD block, as the LD blocks posterior probability.
4.6 Enrichment analysis
Enrichment analysis for cytobands
Cytoband position was downloaded from the UCSC Table Browser [25], with 862 entries of cytobands in total. Enrichment tests were carried out on 28 pairs of disease pairs, between seven immune system disorders (CD, UC, MS, PS, RA, SLE, and T1D) and four psychiatric disorders (SCZ, BPD, MDD, and ASD). Fig. S12 shows the posterior probability of beging associated with both diseases for those 28 disease pairs. For each disease pair, potential shared SNPs were selected based on posterior probability Pr(Zj = 1) > 0.5. Numbers of potential shared SNPs vary from disease to disease, ranging from 0 to 4,505 (for SCZ-CD). For each cytoband, we calculated {x11,x10,x01,x00}, with x11 being the number SNPs in cytoband that are potential shared SNPs, x1010 being the number of SNPs in cytoband that are not potential shared SNPs, x01 being the number of potential shared SNPs not in cytoband, and x00 being the number of SNPs not in cytoband and are not potential shared SNPs. Under null hypothesis that there is no enrichment {x11, x10, x01, x00} follows hypergeometic distribution. The deviation from null hypothesis was tested using Fishers exact test and p-values were adjusted for multiple testing [9].
Enrichment analysis for gene ontology (GO) terms and KEGG pathways
Genome annotation enrichment was performed via DAVID [18,19] on GO [4] terms and KEGG pathways [24,34]. Enrichment tests were carried out on 28 pairs of disease pairs, between seven immune system disorders (CD, UC, MS, PS, RA, SLE, and T1D) and four psychiatric disorders (SCZ, BPD, MDD, and ASD). Gene lists were constructed with genes containing SNPs having posterior>0.8 in at least three disorder pairs.
Enrichment analysis for protein-protein interaction (PPI) network
PPI can provide independent information for prioritization of genetic findings, and thus we constructed PPI sub-networks via DAPPLE [46] in which PPI edges are overrepresented in top SNPs. Enrichment tests were carried out on 28 pairs of disease pairs, between seven immune system disorders (CD, UC, MS, PS, RA, SLE, and T1D) and four psychiatric disorders (SCZ, BPD, MDD, and ASD). For each disease pair, potential shared SNPs were selected based on posterior of being associated with both diseases Pr(Zj = 1) > 0.8 (Fig. S13).
Supplementary Material
References
- 1.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagace C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Colombel JF, Denson LA, De Vos M, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RS, Floyd JA, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot JP, Jobin G, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, Ng A, Newman W, Ophoff RA, Papi L, Palmieri O, Peyrin-Biroulet L, Panes J, Phillips A, Prescott NJ, Proctor DD, Roberts R, Russell R, Rutgeerts P, Sanderson J, Sans M, Schumm P, Seibold F, Sharma Y, Simms LA, Seielstad M, Steinhart AH, Targan SR, van den Berg LH, Vatn M, Verspaget H, Walters T, Wijmenga C, Wilson DC, Westra HJ, Xavier RJ, Zhao ZZ, Ponsioen CY, Andersen V, Torkvist L, Gazouli M, Anagnou NP, Karlsen TH, Kupcin-skas L, Sventoraityte J, Mansfield JC, Kugathasan S, Silverberg MS, Halfvar-son J, Rotter JI, Mathew CG, Griffiths AM, Gearry R, Ahmad T, Brant SR, Chamaillard M, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–252. doi: 10.1038/ng.764. URL http://www.ncbi.nlm.nih.gov/pubmed/21297633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, Rujescu D, Werge T, van de Bunt M, Morris AP, McCarthy MI, Roddey JC, McEvoy LK, Desikan RS, Dale AM, GWAS ICBP, Replication DG, Consortium PG. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. American Journal of Human Genetics. 2013;92(2):197–209. doi: 10.1016/j.ajhg.2013.01.001. URL ¡Go to ISI¿://WOS:000315179600004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, Zuber V, Bettella F, Ripke S, Kelsoe JR, Kendler KS, O’Donovan MC, Sklar P The Psychiatric Genomics Consortium Bipolar D Schizophrenia Work G., The International Multiple Sclerosis Genetics C McEvoy LK Desikan RS Lie BA Djurovic S Dale AM.,The Psychiatric Genomics Consortium PGCBD Schizophrenia Work G., The International Multiple Sclerosis Genetics Consortium I. Genetic pleiotropy between multiple sclerosis schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry. 2014 doi: 10.1038/mp.2013.195. URL http://www.ncbi.nlm.nih.gov/pubmed/24468824. [DOI] [PMC free article] [PubMed]
- 4.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Consortium GO. Gene ontology tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. URL ¡Go to ISI¿://WOS:000086884000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS Type 1 Diabetes Genetics C. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. doi: 10.1038/ng.381. URL http://www.ncbi.nlm.nih.gov/pubmed/19430480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung D, Yang C, Li C, Gelernter J, Zhao H. Gpa: A statistical approach to prioritizing gwas results by integrating pleiotropy and annotation. PLoS Genet. 2014;10(11):e1004. doi: 10.1371/journal.pgen.1004787. 787 URL http://www.ncbi.nlm.nih.gov/pubmed/25393678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, Abeca-sis GR, Barrett JC, Behrens T, Cho J, De Jager PL, Elder JT, Graham RR, Gregersen P, Klareskog L, Siminovitch KA, van Heel DA, Wi-jmenga C, Worthington J, Todd JA, Hafler DA, Rich SS, Daly MJ, Consortia FONO. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7(8):e1002. doi: 10.1371/journal.pgen.1002254. 254 URL http://www.ncbi.nlm.nih.gov/pubmed/21852963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covic T, Cumming SR, Pallant JF, Manolios N, Emery P, Conaghan PG, Tennant A. Depression and anxiety in patients with rheumatoid arthritis: prevalence rates based on a comparison of the depression, anxiety and stress scale (dass) and the hospital, anxiety and depression scale (hads) BMC Psychiatry. 2012;12(6) doi: 10.1186/1471-244X-12-6. http://www.ncbi.nlm.nih.gov/pubmed/22269280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn OJ. Multiple comparisons among means. Journal of the American Statistical Association. 1961;56(293):52–63. URL ¡Go to ISI¿://WOS:A19611734300002. [Google Scholar]
- 10.Encode Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of dna elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. URL http://www.ncbi.nlm.nih.gov/pubmed/22955616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, Mcgee C, Knight JC. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343(6175):1246. doi: 10.1126/science.1246949. >949 URL ¡Go to ISI¿://WOS:000332331500041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng BJ, Sun LD, Soltani-Arabshahi R, Bowcock AM, Nair RP, Stuart P, Elder JT, Schrodi SJ, Begovich AB, Abecasis GR, Zhang XJ, Callis-Duffin KP, Krueger GG, Goldgar DE. Multiple loci within the major histocompatibility complex confer risk of psoriasis. Plos Genetics. 2009;5(8) doi: 10.1371/journal.pgen.1000606. URL ¡Go to ISI¿://WOS:000271533500021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher RA. The genetical theory of natural selection. Oxford University Press; 1930. [Google Scholar]
- 14.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bump-stead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wij-menga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D’Inca R, Du-binsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D’Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Ver-meire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. URL http://www.ncbi.nlm.nih.gov/pubmed/21102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gratten J, Wray NR, Keller MC, Visscher PM. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci. 2014;17(6):782–790. doi: 10.1038/nn.3708. URL http://www.ncbi.nlm.nih.gov/pubmed/24866044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Ery ICSL. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in itgam, pxk, kiaa1542 and other loci. Nature Genetics. 2008;40(2):204–210. doi: 10.1038/ng.81. URL ¡Go to ISI¿://WOS:000252732900019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Magi R, Workalemahu T, White CC, Bouatia-Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpelainen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL, Moffatt MF, Molony C, Nicholson G, Schadt EE, Zondervan KT, Feitosa MF, Ferreira T, Lango Allen H, Weyant RJ, Wheeler E, Wood AR, Magic Estrada k, Goddard ME, Lettre G, Mangino M, Nyholt DR, Purcell S, Smith AV, Visscher PM, Yang J, McCarroll SA, Nemesh J, Voight BF, Absher D, Amin N, Aspelund T, Coin L, Glazer NL, Hayward C, Heard-Costa NL, Hottenga JJ, Johansson A, Johnson T, Kaakinen M, Kapur K, Ketkar S, Knowles JW, Kraft P, Kraja AT, Lamina C, Leitzmann MF, McKnight B, Morris AP, Ong KK, Perry JR, Peters MJ, Polasek O, Prokopenko I, Rayner NW, Ripatti S, Rivadeneira F, Robertson NR, Sanna S, Sovio U, Surakka I, Teumer A, van Wingerden S, Vitart V, Zhao JH, Cavalcanti-Proenca C, Chines PS, Fisher E, Kulzer JR, Lecoeur C, Narisu N, Sandholt C, Scott LJ, Silander K, Stark K, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–960. doi: 10.1038/ng.685. URL http://www.ncbi.nlm.nih.gov/pubmed/20935629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. URL ¡Go to ISI¿://WOS:000262335700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. URL ¡Go to ISI¿://WOS:000265781800006. [DOI] [PubMed] [Google Scholar]
- 20.Ilavska S, Horvathova M, Szabova M, Nemessanyi T, Jahnova E, Tulinska J, Liskova A, Wsolova L, Staruchova M, Volkovova K. Association between the human immune response and body mass index. Human Immunology. 2012;73(5):480–485. doi: 10.1016/j.humimm.2012.02.023. URL ¡Go to ISI¿://WOS:000304024500007. [DOI] [PubMed] [Google Scholar]
- 21.International Multiple Sclerosis Genetics C. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. URL http://www.ncbi.nlm.nih.gov/pubmed/17660530. [DOI] [PubMed] [Google Scholar]
- 22.International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. URL http://www.ncbi.nlm.nih.gov/pubmed/19571811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irish Schizophrenia Genomics C., the Wellcome Trust Case Control C. Genome-wide association study implicates hla-c*01:02 as a risk factor at the major histocompati-bility complex locus in schizophrenia. Biol Psychiatry. 2012;72(8):620–628. doi: 10.1016/j.biopsych.2012.05.035. URL http://www.ncbi.nlm.nih.gov/pubmed/22883433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisaa M, Goto S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The ucsc table browser data retrieval tool. Nucleic Acids Research. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. URL ¡Go to ISI¿://WOS:000188079000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kindt AS, Navarro P, Semple CA, Haley CS. The genomic signature of trait-associated variants. BMC Genomics. 2013;14(108) doi: 10.1186/1471-2164-14-108. URL http://www.ncbi.nlm.nih.gov/pubmed/23418889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krams IA, Skrinda I, Kecko S, Moore FR, Krama T, Kaasik A, Meija L, Lietuvietis V, Rantala MJ. Body height affects the strength of immune response in young men, but not young women. Sci Rep. 2014;4:6223. doi: 10.1038/srep06223. URL http://www.ncbi.nlm.nih.gov/pubmed/25164474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. URL http://www.ncbi.nlm.nih.gov/pubmed/20881960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, DeCandia TR, Ripke S, Yang J Schizophrenia Psychiatric Genome-Wide Association Study C, International Schizophrenia C Molecular Genetics of Schizophrenia C, Sullivan, PF Goddard ME, Keller MC Visscher PM Wray NR. Estimating the proportion of variation in susceptibility to schizophrenia captured by common snps. Nat Genet. 2012;44(3):247–250. doi: 10.1038/ng.1108. URL http://www.ncbi.nlm.nih.gov/pubmed/22344220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Yang C, Gelernter J, Zhao HY. Improving genetic risk prediction by leveraging pleiotropy. Human Genetics. 2014;133(5):639–650. doi: 10.1007/s00439-013-1401-5. URL ¡Go to ISI¿://WOS:000334519900014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167(11):1357–1363. doi: 10.1176/appi.ajp.2010.10020223. URL http://www.ncbi.nlm.nih.gov/pubmed/20686188. [DOI] [PubMed] [Google Scholar]
- 32.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. Common genetic determinants of schizophrenia bipolar disorder in swedish families a population-based study. Lancet. 2009;373(9659):234–239. doi: 10.1016/S0140-6736(09)60072-6. URL http://www.ncbi.nlm.nih.gov/pubmed/19150704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Magi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Hua Zhao J, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Mateo Leach I, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stancakova A, Strawbridge RJ, Ju Sung Y, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Arnlov J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Bluher M, Bohringer S, Bonnycastle LL, Bottcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Ida Chen YD, Clarke R, Daw EW, de Craen AJ, Delgado G, Dimitriou M, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. DOI 10.1038/nature14177. URL http://www.ncbi.nlm.nih.gov/pubmed/25673413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M., K., S., G., Y., S., M., K., M., F., M., T. Data, information, knowledge and principle: back to metabolism in kegg. Nucleic Acids Research. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margaretten M, Julian L, Katz P, Yelin E. Depression in patients with rheumatoid arthritis description, causes and mechanisms. Int J Clin Rheumtol. 2011;6(6):617–623. doi: 10.2217/IJR.11.6. URL http://www.ncbi.nlm.nih.gov/pubmed/22211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller N, Schwarz MJ, Dehning S, Douhe A, Cerovecki A, Goldstein-Muller B, Spellmann I, Hetzel G, Maino K, Kleindienst N, Moller HJ, Arolt V, Riedel M. The cyclooxygenase-2 inhibitor celecoxib has therapeutic effects in major depression results of a double-blind, randomized, placebo controlled, add-on pilot study to reboxetine. Mol Psychiatry. 2006;11(7):680–684. doi: 10.1038/sj.mp.4001805. URL http://www.ncbi.nlm.nih.gov/pubmed/16491133. [DOI] [PubMed] [Google Scholar]
- 37.Paaby AB, Rockman MV. The many faces of pleiotropy. Trends Genet. 2013;29(2):66–73. doi: 10.1016/j.tig.2012.10.010. URL http://www.ncbi.nlm.nih.gov/pubmed/23140989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palleja A, Horn H, Eliasson S, Jensen LJ. Distild database: diseases and traits in linkage disequilibrium blocks. Nucleic Acids Res. 2012;40(Database issue):D1036–D1040. doi: 10.1093/nar/gkr899. URL http://www.ncbi.nlm.nih.gov/pubmed/22058129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nature Reviews Genetics. 2013;14(9):661–673. doi: 10.1038/nrg3502. URL ¡Go to ISI¿://WOS:000323280100013. [DOI] [PubMed] [Google Scholar]
- 40.Cross-Disorder Group of the Psychiatric Genomics, C. Identification of risk loci with shared effects on five major psychiatric disorders a genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. URL http://www.ncbi.nlm.nih.gov/pubmed/23453885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. DOI 10.1038/nature13595. URL http://www.ncbi.nlm.nih.gov/pubmed/25056061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cross-Disorder Group of the Psychiatric Genomics C. Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, Witte JS, Absher D, Agartz I, Akil H, Amin F, Andreassen OA, Anjorin A, Anney R, Anttila V, Arking DE, Asherson P, Azevedo MH, Backlund L, Bad-ner JA, Bailey AJ, Banaschewski T, Barchas JD, Barnes MR, Barrett TB, Bass N, Battaglia A, Bauer M, Bayes M, Bellivier F, Bergen SE, Berrettini W, Betancur C, Bettecken T, Biederman J, Binder EB, Black DW, Blackwood DH, Bloss CS, Boehnke M, Boomsma DI, Breen G, Breuer R, Bruggeman R, Cormican P, Buccola NG, Buitelaar JK, Bunney WE, Buxbaum JD, Byerley WF, Byrne EM, Caesar S, Cahn W, Cantor RM, Casas M, Chakravarti A, Chambert K, Choudhury K, Cichon S, Cloninger CR, Collier DA, Cook EH, Coon H, Cormand B, Corvin A, Coryell WH, Craig DW, Craig IW, Crosbie J, Cuccaro ML, Curtis D, Czamara D, Datta S, Dawson G, Day R, De Geus EJ, Degenhardt F, Djurovic S, Donohoe GJ, Doyle AE, Duan J, Dudbridge F, Duketis E, Ebstein RP, Edenberg HJ, Elia J, Ennis S, Etain B, Fanous A, Farmer AE, Ferrier IN, Flickinger M, Fombonne E, Foroud T, Frank J, Franke B, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide snps. Nat Genet. 2013;45(9):984–994. doi: 10.1038/ng.2711. DOI 10.1038/ng.2711. URL http://www.ncbi.nlm.nih.gov/pubmed/23933821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raychaudhuri S, Korn JM, McCarroll SA, International Schizophrenia C, Alt-shuler D, Sklar P, Purcell S, Daly MJ. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6(9):e1001. doi: 10.1371/journal.pgen.1001097. URL http://www.ncbi.nlm.nih.gov/pubmed/20838587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, Westra HJ, Shakhbazov K, Abdellaoui A, Agrawal A, Albrecht E, Alizadeh BZ, Amin N, Bamard J, Baumeister SE, Benke KS, Bielak LF, Boatman JA, Boyle PA, Davies G, De Leeuw C, Eklund N, Evans DS, Ferhmann R, Fischer K, Gieger C, Gjessing HK, Hagg S, Harris JR, Hayward C, Holzapfel C, Ibrahim-Verbaas CA, Ingelsson E, Jacobsson B, Joshi PK, Jugessur A, Kaakinen M, Kanoni S, Karjalainen J, Kolcic I, Kristiansson K, Kutalik Z, Lahti J, Lee SH, Lin P, Lind PA, Liu YM, Lohman K, Loitfelder M, McMahon G, Vidal PM, Meirelles O, Milani L, Myhre R, Nuotio ML, Oldmeadow CJ, Petrovic KE, Peyrot WJ, Polasek O, Quaye L, Reinmaa E, Rice JP, Rizzi TS, Schmidt H, Schmidt R, Smith AV, Smith JA, Tanaka T, Terracciano A, van der Loos MJHM, Vitart V, Volzke H, Wellmann J, Yu L, Zhao W, Allik J, Attia JR, Bandinelli S, Bastardot F, Beauchamp J, Bennett DA, Berger K, Bierut LJ, Boomsma DI, Bultmann U, Campbell H, Chabris CF, Cherkas L, Chung MK, Cucca F, de Andrade M, De Jager PL, De Neve JE, Deary IJ, Dedoussis GV, Deloukas P, Dimitriou M, Eiriksdottir G, Elderson MF, Eriksson JG, et al. Gwas of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340(6139):1467–1471. doi: 10.1126/science.1235488. URL ¡Go to ISI¿://WOS:000320647000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518 doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ, Genetic IIBD. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. Plos Genetics. 2011;7(1):e1001. doi: 10.1371/journal.pgen.1001273. URL ¡Go to ISI¿://WOS:000286653500012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saha S, Chant D, McGrath J. A systematic review of mortality in schizophrenia: is the differential mortality gap worsening over time? Arch Gen Psychiatry. 2007;64(10):1123–1131. doi: 10.1001/archpsyc.64.10.1123. URL http://www.ncbi.nlm.nih.gov/pubmed/17909124. [DOI] [PubMed] [Google Scholar]
- 48.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao JJ, Huang XM, Wood NW, Lorenz D, Deuschl G, Chen HL, Riess O, Hardy JA, Singleton AB, Gasser T. Genome-wide association study reveals genetic risk underlying parkinson’s disease. Nature Genetics. 2009;41(12):1308. doi: 10.1038/ng.487. U68 URL ¡Go to ISI¿://WOS:000272144900012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith RS. The macrophage theory of depression. Med Hypotheses. 1991;35(4):298–306. doi: 10.1016/0306-9877(91)90272-z. URL http://www.ncbi.nlm.nih.gov/pubmed/1943879. [DOI] [PubMed] [Google Scholar]
- 50.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits challenges and strategies. Nature Reviews Genetics. 2013;14(7):483–495. doi: 10.1038/nrg3461. URL ¡Go to ISI¿://WOS:000320510000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Bieber-mann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. URL http://www.ncbi.nlm.nih.gov/pubmed/20935630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG, Consortium B, Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Lee AT, Liu X, Martin P, Morgan AW, Padyukov L, Posthumus MD, Radstake TR, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, van der Schoot CE, van Riel PL, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth BP, Consortium Y, Wijmenga C, Karlson EW, Toes RE, de Vries N, Begovich AB, Worthington J, Siminovitch KA, Gregersen PK, Klareskog L, Plenge RM. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. URL http://www.ncbi.nlm.nih.gov/pubmed/20453842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stearns FW. One hundred years of pleiotropy a retrospective. Genetics. 2010;186(3):767–773. doi: 10.1534/genetics.110.122549. URL http://www.ncbi.nlm.nih.gov/pubmed/21062962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suh WK F., C.D.M., K., F., K., W., A., P.P., B., W.D. Interaction of mhc class i molecules with the transporter associated with antigen processing. Science. 1994;264:1322–1326. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- 55.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression review and meta-analysis. Am J Psychiatry. 2000;157(10):1552–1562. doi: 10.1176/appi.ajp.157.10.1552. URL http://www.ncbi.nlm.nih.gov/pubmed/11007705. [DOI] [PubMed] [Google Scholar]
- 56.Tobacco Genetics, C. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. URL http://www.ncbi.nlm.nih.gov/pubmed/20418890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vattikuti S, Guo J, Chow CC. Heritability and genetic correlations explained by common snps for metabolic syndrome traits. Plos Genetics. 2012;8(3):e1002. doi: 10.1371/journal.pgen.1002637. URL !Go to ISI¿://WOS:000302254800093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of gwas discovery. Am J Hum Genet. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. URL http://www.ncbi.nlm.nih.gov/pubmed/22243964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker FR. A critical review of the mechanism of action for the selective serotonin reuptake inhibitors: do these drugs possess anti-inflammatory properties and how relevant is this in the treatment of depression? Neurophar-macology. 2013;67:304–317. doi: 10.1016/j.neuropharm.2012.10.002. URL http://www.ncbi.nlm.nih.gov/pubmed/23085335. [DOI] [PubMed] [Google Scholar]
- 60.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. URL http://www.ncbi.nlm.nih.gov/pubmed/18369103. [DOI] [PubMed] [Google Scholar]
- 61.Weiss LA, Shen YP, Korn JM, Arking DE, Miller DT, Fossdal R, Saemu-ndsen E, Stefansson H, Ferreira MAR, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu B, Daly MJ, Consortium A. Association between microdeletion and microduplication at 16p11.2 and autism. New England Journal of Medicine. 2008;358(7):667–675. doi: 10.1056/NEJMoa075974. URL ¡Go to ISI¿://WOS:000253127700003. [DOI] [PubMed] [Google Scholar]
- 62.Williams GC. Pleiotropy, natural-selection, and the evolution of senescence. Evolution. 1957;11(4):398–411. URL ¡Go to ISI¿://WOS:A1957XE15100002. [Google Scholar]
- 63.Wright S. Evolution and the genetics of populations. Vol. 1. University of Chicago Press; 1968. Genetics and biometric foundations. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.